Introduction
Echinococcosis is one of the most pathogenic helminth zoonosis worldwide caused by the larval stage of Echinococcus. Humans are infected by accidentally ingested Echinococcus eggs, which are released along with canids' feces. The parasite can reach any human organs, mainly liver and lung, causing serious life-threatening hydatid cysts (McManus et al., Reference McManus, Zhang, Li and Bartley2003; Zhenghuan et al., Reference Zhenghuan, Xiaoming and Xiaoqing2008). The infections can be asymptomatic for years and are usually found in the late stage of the disease (Piccoli et al., Reference Piccoli, Bazzocchi, Brunetti, Mihailescu, Bandi, Mastalier, Cordos, Beuran, Popa, Meroni, Genco and Cretu2013). Historically, four species have been recognized within the Echinococcus genus: E. granulosus (including 10 distinct genotypes G1–G10), E. multilocularis, E. oligarthra and E. vogeli. E. shiquicus and E. felidis were new species additionally discovered in 2005 and 2008, respectively (Boubaker et al., Reference Boubaker, Macchiaroli, Prada, Cucher and Spiliotis2013). Subsequently, in the newly taxonomic revision, previously E. granulosus species have been split into four species: E. granulosus sensu stricto (G1–G3), E. equinus (G4), E. ortleppi (G5) and E. canadensis (G6–G8, G10) (Nakao et al., Reference Nakao, Lavikainen, Yanagida and Ito2013). To date, E. granulosus s.s., E. multilocularis, E. shiquicus and E. canadensis have been identified in China (Cong-Nuan et al., Reference Cong-Nuan, Zhong-Zi, Li, Hong-Bin, David, Meng-Tong, Jin-Zhong, Yan-Lei, Jian-Qiu, Bao-Quan, Yu-Rong, Donald and Wan-Zhong2015). Both E. granulosus s.s. and E. multilocularis are widespread in western and north-western China and have long been a predominant public health and medical threat in these areas (Zhenghuan et al., Reference Zhenghuan, Xiaoming and Xiaoqing2008). Approximately 90% of human echinococcosis in China are cystic echinococcosis (CE) caused by E. granulosus s.s., whereas the remainder are from alveolar echinococcosis (AE) caused by E. multilocularis, which accounts for more than 90% of the AE burden in the world (Yang et al., Reference Yang, Liu, Zhu, Griffiths, Tanner, Bergquist, Utzinger and Zhou2014). Echinococcus canadensis is also responsible for human CE (Orkhontuul et al., Reference Orkhontuul, Batbold, Avaajigmed, Tugbayar, Tsetsegdelger, Tsogtsaikhan, Enkhsaikhan, Yanagida, Nishikawa and Ito2018), which used to be neglected in China because the limited distribution and only two human cases of E. canadensis (G6) have been reported till 2005 (Bart et al., Reference Bart, Abdukader, Zhang, Lin, Wang, Nakao, Ito, Craig, Piarroux, Vuitton and Wen2006). However, in the past 5 years human-derived G6, G7 and G10 genotypes of E. canadensis have emerged in several provinces/autonomous regions of China (Zhang et al., Reference Zhang, Yang, Zeng, Zhao, Liu, Piao, Jiang, Cai, Shen, Liu and Zhang2014; Ma et al., Reference Ma, Wang, Lin, Zhao, Li, Zhang, Ma, Zhang, Hou, Cai, Liu and Wang2015; Yang et al., Reference Yang, Zhang, Zeng, Zhao, Zhang and Liu2015; Cao et al., Reference Cao, Chen, Li, Ma, Xiao, Wang and Gao2018), which requires more research efforts.
Since different Echinococcus species have diverse diseases treatment and prognosis (Brunetti et al., Reference Brunetti, Kern and Vuitton2010; McManus et al., Reference McManus, Gray, Zhang and Yang2012), species genotyping is essential, especially in China where co-endemic has occurred. A number of molecular approaches based on DNA detection have been designed to accurately identify different Echinococcus species. PCR-sequencing (Nakao et al., Reference Nakao, Sako, Yokoyama, Fukunaga and Ito2000; Xiao et al., Reference Xiao, Qiu, Nakao, Li, Yang, Chen, Schantz, Craig and Ito2006a) or PCR-RFLP (Xiao et al., Reference Xiao, Nakao, Qiu, Budke and Giraudoux2006b; Chaabane-Banaoues et al., Reference Chaabane-Banaoues, Oudni-M'rad, M'rad, Amani, Mezhoud and Babba2016) method can identify the genotype of Echinococcus species PCR amplicons by sequencing or restriction fragment length polymorphism (RFLP) in a relatively slow and costly way. Uniplex PCR (uPCR) can identify its target Echinococcus species based on only PCR which relatively simplify the genotyping (Boufana et al., Reference Boufana, Umhang, Qiu, Chen, Lahmar, Boué, Jenkins and Craig2013). By contrast, mPCR which can simultaneously detect more than one pathogens in a single PCR mixture is more rapid and have been widely applied in both clinical and laboratory research (Henegariu and Al, Reference Henegariu and Al1997). Some mPCR approaches have been developed for detecting certain Echinococcus species (Dinkel et al., Reference Dinkel, Kern, Brinker, Oehme, Amélie, Giraudoux, Mackenstedt and Romig2011; Boubaker et al., Reference Boubaker, Macchiaroli, Prada, Cucher and Spiliotis2013; Cong-Nuan et al., Reference Cong-Nuan, Zhong-Zi, Li, Hong-Bin, David, Meng-Tong, Jin-Zhong, Yan-Lei, Jian-Qiu, Bao-Quan, Yu-Rong, Donald and Wan-Zhong2015), but to our knowledge, no studies aiming at simultaneous identification of the three species (E. granulosus s.s., E. multilocularis and E. canadensis) that can cause human echinococcosis in China have been performed. Moreover, those methods were mostly applied on feces or animal tissues for epidemiological investigation purpose, and the effect on detection of human-derived parasite samples was seldom assessed. Therefore, in this study we aimed to set up a single-tube mPCR approach that can be applied for the accurate identification of E. granulosus s.s., E. multilocularis and E. canadensis (G6–G8, G10) to assist the diagnosis of human echinococcosis.
Materials and methods
Sample collection and extraction
In total, 72 parasite tissues were collected post-operatively from echinococcosis patients received surgical treatment from the West China Hospital and Sichuan Provincial People's Hospital in Sichuan Province from 2015 to 2017. Adults of Taenia solium, Taenia saginata and Taenia asiatica were kindly provided by Tiaoying Li (Professor of Cysticercosis Control and Research Department, Sichuan Center for Disease Control and Prevention, Sichuan, China). Other parasite individuals isolated from pikas, voles or dogs were collected by our group during routine parasite disease surveillance in Sichuan Province. In addition, blood samples were also collected from 38 healthy people ruled out parasitic infections during routine physical examinations. All the parasite samples above were listed in Table 1 and confirmed by morphological identification and PCR-based sequencing. One E. granulosus s.s., one E. multilocularis and one E. canadensis (G6) isolates were selected from the 72 patient samples, respectively, as standard strains for the establishment of mPCR and the rest (54 E. granulosus s.s., 14 E. multilocularis and one E. canadensis) were employed for subsequent reliability test of the mPCR.
Table 1. Information on parasite samples used in the study

Note: Parasites marked with * were used to check the specificity of the mPCR assay.
Total genomic DNA of samples was extracted by using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, GER) according to the manufacturer's instructions. The DNA concentration was measured by using a NanoDrop One (Thermo Fisher Scientific, Wisconsin, USA), to confirm the quality of DNA extractions. Then the extracted DNA samples were stored at −20 °C until use.
Primer design
Complete mitochondria genes of all the Echinococcus genus, including E. granulosus s.s. (accession no. NC008075), E. multilocularis (accession no. NC000928) and E. canadensis (G6) (accession no. NC011121) as well as the remaining six Echinococcus species namely E. shiquicus (accession no. NC009460), E. oligarthrus (accession no. NC009461), E. equinus (accession no. NC020374), E. ortleppi (accession no. NC011122), E. vogeli (accession no. NC009462) and E. felidis (accession no. NC021144) were retrieved from GenBank (https://www.ncbi.nlm.nih.gov/gene/) and aligned by MEGA7.0 (https://www.megasoftware.net/). A number of primer pairs specific for E. granulosus s.s., E. multilocularis and E. canadensis were designed manually on conserved regions of E. canadensis (Boufana et al., Reference Boufana, Umhang, Qiu, Chen, Lahmar, Boué, Jenkins and Craig2013) and then the primer pair with maximum specificity was pre-selected by aligned with sequences in the GenBank (https://www.ncbi.nlm.nih.gov/gene/). Primer sequences were subsequently synthesized by Shanghai Invitrogen Biotechnology Co Ltd.
Primer selection experiments
The designed primers were combined into various mixtures, each including three primer pairs specific for E. granulosus s.s., E. multilocularis and E. canadensis, respectively, and can generate expected amplicons of distinct size for each species. These primer sets were selected by testing with single and pooled standard DNA, as well as other related species. The primer set that allows accurate discrimination of E. granulosus s.s., E. multilocularis and E. canadensis in both single and pooled target templates without generating any nonspecific products was retained for the mPCR approach (presented in Table 2).
Table 2. Characteristics of primers used in mPCR

Multiplex PCR conditions
The mPCR was conducted using a SmpliAmp thermal cycler (Applied Biosystems, California, USA) in a 25 µL reaction system containing 20 ng standard DNA of each Echinococcus species, 6 µL of double distilled water, 12.5 µL of GoTaq Hot Start Polymerase mixture (Promega, Wisconsin, USA) and 7.5 µ m (each) of Gd3-4F, Gd3-4R, Mtb2F, Mtb2R, Cox1F and Cox1R primers. Annealing temperatures were optimized before the assay and 56 °C was confirmed to be with the best specificity and sensitivity of the method. So that the optimized thermal cycling conditions were as follows: an initial denaturation at 94 °C for 2 min, followed by 35 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 40 s, and a final extension at 72 °C for 5 min.
For all experiments, both positive and negative controls (double distilled water) were included. To avoid extraneous DNA contamination, the reaction mix was prepared with dedicated equipment, and the operation area was exposed to UV light for at least half an hour prior to the assay.
Identification of PCR products
Five microlitres of PCR products were separated by electrophoresis in a 2% agarose gel stained with Super Red (Biosharp, Hefei, CN) and visualized by using Universal Hood II UV transilluminator (Bio-Rad, California, USA) with Quantity One analyst software (Bio-Rad, California, USA).
Amplicons generated from standard DNA of E. granulosus s.s., E. multilocularis and E. canadensis in the mPCR were sequenced by Shanghai Invitrogen Biotechnology Co Ltd., and then compared with existing sequences in GenBank (http://www.ncbi.nlm.nih.gov/BLAST/) for further confirmation of the genotype.
Specificity evaluation of mPCR
To test the possible unspecific cross binding with other parasite species, 30 ng template DNA derived from 18 other parasite isolates including five samples of one Echinococcus species (E. shiquicus), 12 samples of seven closely related Taenia species and one non-related Toxascaris leonina were applied in individually performed standard mPCRs. The parasite samples for the assessment are presented in Table 1 marked with an asterisk.
Sensitivity evaluation of mPCR
Five-fold serial dilutions of three target DNA with the concentrations ranging from 1000 to 0.0128 pg were individually used to evaluate sensitivity of the mPCR approach. The lowest detection limit of each target species was determined according to the lowest amount of DNA that can yield a clearly visible band.
Reliability evaluation of mPCR
To confirm the reliability of the newly developed mPCR, a total of 69 parasite samples removed from patient livers were tested. These samples have been genotyped to be 54 E. granulosus s.s., 14 E. multilocularis and one E. canadensis before according to PCR-based sequencing described by Nakao et al. (Reference Nakao, Sako, Yokoyama, Fukunaga and Ito2000). Reliability was assessed by comparing the consistency of mPCR approaches and sequencing results. Besides, in order to exclude nonspecific reaction which may be caused by the host (human) DNA, 38 healthy human DNA extracted from blood samples was also investigated. This blood was alternative for liver tissue because healthy human liver tissue was not available in this study.
Results
Identification of PCR products
PCR products amplified with single and pooled standard DNA of E. granulosus s.s., E. multilocularis and E. canadensis are shown in Fig. 1. Expected amplicons of 167, 237 and 441 bp were observed for E. granulosus s.s., E. multilocularis and E. canadensis, respectively. The respective diagnostic products were also detected when checked with different DNA mixtures of E. granulosus s.s., E. multilocularis and E. canadensis (Fig. 1, lanes 4–7). Fragment generated by each target species was highly similarity (98–99%) with respective reference sequences in GenBank: E. granulosus s.s. (GenBank accession no. MG672293), E. multilocularis (GenBank accession no. KY205670) and E. canadensis (GenBank accession no. MH274989), which showed 1–4 base pair differences between them.

Fig. 1. PCR products of single and pooled DNA of E. granulosus s.s., E. multilocularis and E. canadensis in mPCR assay. Lanes 1–3, amplicons of E. granulosus s.s., E. multilocularis and E. canadensis, respectively; lane 4, amplicons of mixed DNA of E. granulosus s.s. and E. multilocularis; lane 5, amplicons of mixed DNA of E. granulosus s.s. and E. canadensis; lane 6, amplicons of mixed DNA of E. canadensis and E. multilocularis; lane 7, amplicons of mixed DNA of E. granulosus s.s., E. multilocularis and E. canadensis, N, negative control; M, DNA marker.
Specificity of the mPCR
The mPCR was proved to be 100% specific when tested with other closely related Taenia species and one non-related T. leonina (Fig. 2, lanes 9–21), as no products were observed or their products were significantly larger than the expected bands. The only exception was E. shiquicus (Fig. 2, lanes 4–8), which cross react with multiple primers and generate 3–4 slightly visible bands among target bands region.

Fig. 2. Specificity of the mPCR. Lanes 1–3, positive controls (amplicons of E. granulosus s.s., E. multilocularis and E. canadensis, respectively); lanes 4–20, amplicons of other parasite DNA samples: E. shiquicus (lanes 4–8), T. asiatica (lane 9), T. saginata (lanes 10 and 11), T. solium (lanes 12 and 13), T. hydatigena (lane 14), T. taeniaeformis (lanes 15–18), T. serialis (lane 19), T. multiceps (lane 20) and T. leonina (lane 21); M, DNA marker (100–600 bp); N, negative control.
Sensitivity of the mPCR
The results showed the detection limits of mPCR assay varied among species. The lowest limit for the DNA detection of E. granulosus s.s. and E. canadensis can both reach as less as 0.32 pg, and the lowest detection limit for E. multilocularis was 1.6 pg (Fig. 3), which revealed the high sensitivity of the method.
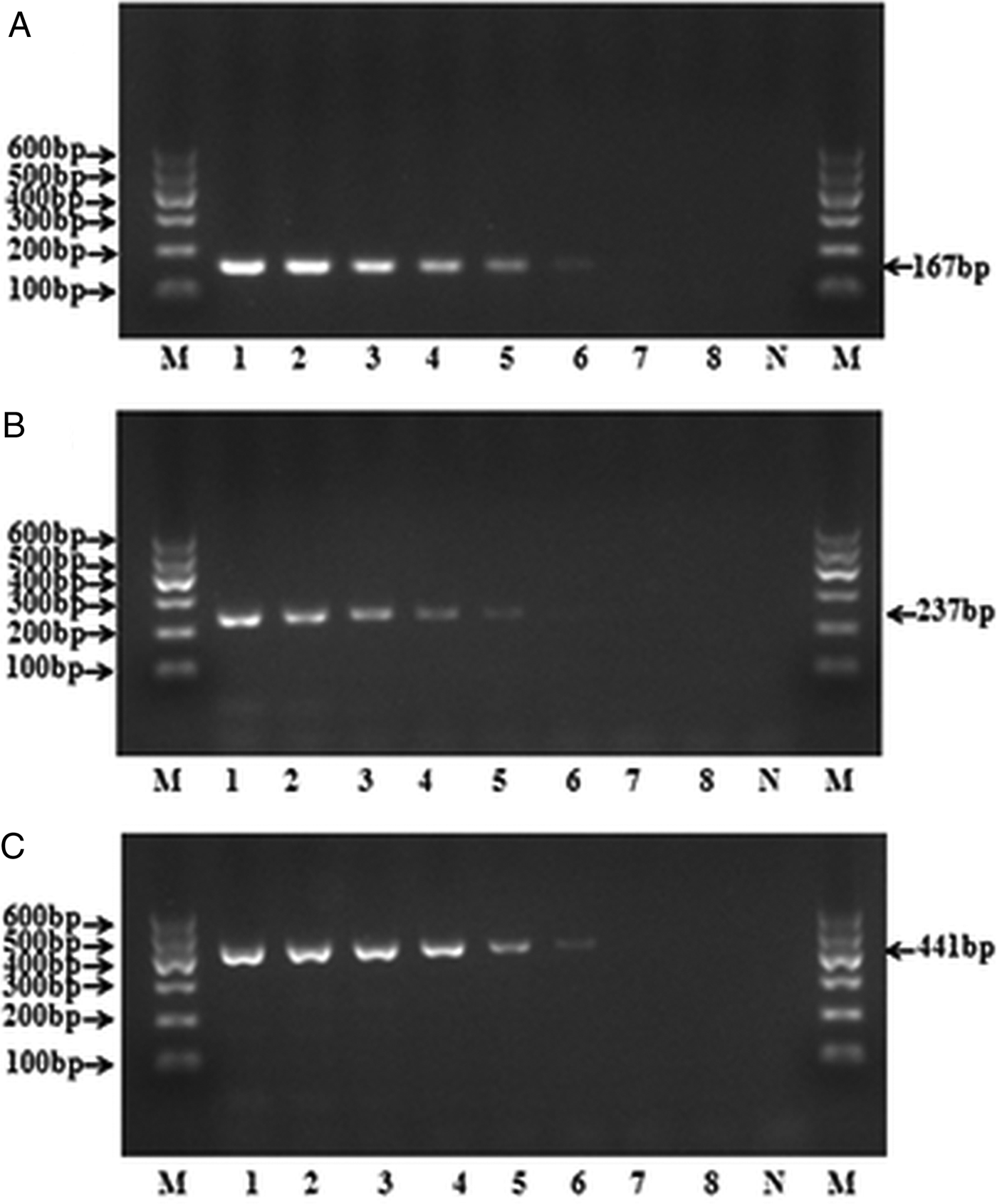
Fig. 3. Sensitivity of the mPCR for three targeted species, respectively. The DNA detection limit using a serial dilution of standard DNA of (A) E. granulosus s.s., (B) E. multilocularis and (C) E. canadensis, respectively. Lanes 1–8, 5-fold serially diluted DNA templates (1000, 200, 40, 8, 1.6, 0.32, 0.064, 0.0128 pg); M, DNA marker; N, negative control.
Validation of the mPCR
For all 69 patient-derived parasite DNA samples, a clearly genotype-specific binding pattern was observed. Among them, 54 samples were identified as E. granulosus s.s., 14 samples were E. multilocularis and one sample was E. canadensis according to the sizes of PCR products (part electrophoresis results are presented in Fig. 4A). Therefore, the results obtained by mPCR were completely in accordance with that of the sequencing. Moreover, no target band was observed from 38 normal human DNA samples (part electrophoresis results are shown in Fig. 4B), which indicates that host tissue did not affect PCR outcomes.

Fig. 4. Validation results of mPCR. Reliability was assessed by tested with human-derived parasite DNA that has been sequenced (A) and healthy human DNA (B). (A) Lanes 1–26, parasite DNA from humans, including one E. canadensis (lane 10), eight E. multilocularis (lanes 4, 8, 11, 14, 17, 22, 23 and 26) and the rest were E. granulosus s.s. (B) Lanes 27–51, healthy human DNA; M, DNA marker; P, positive control (DNA mixture of E. granulosus s.s., E. multilocularis and E. canadensis); N, negative control.
Discussion
Multiplex polymerase chain reaction (mPCR) is a method that can detect more than one target species by using multiple primer pairs in a single reaction tube. Since its first description in 1988, this method has been successfully applied in many areas of DNA identification, such as bacteria, viruses, fungi or parasites (Henegariu and Al, Reference Henegariu and Al1997; Elnifro et al., Reference Elnifro, Cooper, Ashshi and Klapper2000). Primer design is undoubtedly a key factor for successfully establishing a PCR approach. In general, it is not very difficult to design primers for conventional (uniplex) PCR, but the design of mPCR primers is much more complicated because it is not a simple combination of several uPCRs. When more than one pair of primers are mixed in the same reaction tube, the primers can be randomly paired in the mixture to form a new reaction system that is far more complex than the original separated ones. At this point, the design of mPCR primers needs to overcome many difficulties, including poor sensitivity and specificity, different annealing temperatures of each primer, preferential amplification of one sequence over others (Polz and Cavanaugh,Reference Polz and Cavanaugh1998), easily formation of primer dimer and so on (Elnifro et al., Reference Elnifro, Cooper, Ashshi and Klapper2000). There are no other means to predict the performance characteristics of multiple primer pairs system except empirical testing and a trial-and-error approach.
The present study successfully developed an mPCR method to detect and differentiate human-derived DNA of E. granulosus s.s., E. multilocularis and E. canadensis (G6–G8, G10). This method can work well with single or mixed target templates and generate specific amplicons of expected length that are highly similarity with their intended target Echinococcus species. It achieved a high degree of species specificity because no predicted product was detected from any other tested helminths, except for the amplification of E. shiquicus. Although E. shiquicus has been found on foxes and plateau pika in Qinghai-Tibet plateau region of China (Xiao et al., Reference Xiao, Qiu, Nakao, Li, Yang, Chen, Schantz, Craig and Ito2006a; Ma et al., Reference Ma, Wang, Lin, Zhao, Li, Zhang, Ma, Zhang, Hou, Cai, Liu and Wang2015), to date human infection of E. shiquicus has not been reported (McManus et al., Reference McManus, Gray, Zhang and Yang2012; Nakao et al., Reference Nakao, Lavikainen, Yanagida and Ito2013). Thus, cross-reactivity with E. shiquicus is unlikely to occur in human echinococcosis detection. In addition, E. shiquicus can generate 3–4 weakly visible bands in the mPCR, so that it would not interfere with the detection of single or double infected samples. Confusion can only appear when encountering 3-plex infection, but this is very unlikely to happen and can be easily resolved by further sequencing the PCR products. Therefore, the effect of E. shiquicus on this method can be neglected. In the most recent taxonomic revision, the genus Echinococcus was divided into nine species, including E. felidis, E. equinus, E. oligarthra, E. vogeli, E. ortleppi, E. granulosus s.s., E. multilocularis, E. canadensis and E. shiquicus (Nakao et al., Reference Nakao, Lavikainen, Yanagida and Ito2013). DNA samples of the former five Echinococcus species are unavailable as they have not been found in any part of China. Despite this, in order to minimize the possibility of unspecific interactions with these five closely related species, comparison of the primer target sequences for the five species with those of the three target species has also been conducted, which showed 6–11 base pair differences between them. According to Liu (Cong-Nuan et al., Reference Cong-Nuan, Zhong-Zi, Li, Hong-Bin, David, Meng-Tong, Jin-Zhong, Yan-Lei, Jian-Qiu, Bao-Quan, Yu-Rong, Donald and Wan-Zhong2015), it is highly unlikely to produce any amplicon from the five species during the mPCR assay due to such large differences, thus further illustrating the strict species specificity of the method.
Previous studies show that mPCR assay has lower sensitivity compared to uPCR assay (Dong et al., Reference Dong, Cheng and Jian2000; Bharathi et al., Reference Bharathi, Rameshkumar, Ramakrishnan, Reddy, Shivkumar and Ramesh2013; Boufana et al., Reference Boufana, Umhang, Qiu, Chen, Lahmar, Boué, Jenkins and Craig2013). Nevertheless, the method designed in this study is proved to be highly sensitive, with a detection threshold of as less as 0.32 pg for E. granulosus s.s. and E. canadensis, and 1.6 pg for E. multilocularis. The detection limit is slightly lower than that was described in uPCR assays developed by Boufana et al. (Reference Boufana, Umhang, Qiu, Chen, Lahmar, Boué, Jenkins and Craig2013) (2–10 pg), and is significantly lower than the detection limits of mPCR that were previously reported by Boubaker et al. (Reference Boubaker, Macchiaroli, Prada, Cucher and Spiliotis2013), and Cong-Nuan et al. (Reference Cong-Nuan, Zhong-Zi, Li, Hong-Bin, David, Meng-Tong, Jin-Zhong, Yan-Lei, Jian-Qiu, Bao-Quan, Yu-Rong, Donald and Wan-Zhong2015) (0.1–5 ng and 10–20 pg, respectively). With the implementation of control programmes and large-scale population echinococcosis screening in China, the majority of newly discovered patients are in the early infection stage with small lesions. Therefore, only minute amounts of tissue samples can be collected in most cases, which puts higher requirements on the sensitivity of DNA detecting methods. The mPCR method developed herein can meet its need due to its high sensitivity.
In the application test of our mPCR method, a large number of patients and healthy human samples were investigated. For all previously gene-sequencing determined patient samples, the genotypes could be successfully re-confirmed by mPCR, thus demonstrating the high accuracy and reliability of the method. Meanwhile, no amplicon was observed from any healthy human DNA samples, which indicates that the method was not interfered by contaminating DNA from the host (human). On the other side, only two human-derived E. canadensis (G6) samples were available for this study, because of its very low prevalence in China. Although the sample size and genotypes of E. canadensis were insufficient in the application test, our BLAST analyses of E. canadensis revealed that most E. canadensis (G6–G8, G10) isolates listed in GenBank can full-match with E. canadensis primers, which confirmed our method could successfully amplify G6–G8 and G10 genotypes. Compared to other identification methods for human echinococcosis in China, such as PCR-based sequencing, PCR-RFLP or histopathology (Bart et al., Reference Bart, Abdukader, Zhang, Lin, Wang, Nakao, Ito, Craig, Piarroux, Vuitton and Wen2006; Li et al., Reference Li, Ito, Nakaya, Qiu, Nakao, Zhen, Xiao, Chen, Giraudoux and Craig2008), the mPCR method presented herein is more advantageous: it is more rapid and less costly. Thereby, it clearly simplifies the identification of human echinococcosis in China and is suitable for routine tests.
The current clinical diagnosis of echinococcosis primarily relies on imaging techniques, and serological test plays a complementary role (McManus et al., Reference McManus, Gray, Zhang and Yang2012; Sarink et al., Reference Sarink, Koelewijn, Slingerland, Tielens, Van Genderen and Van Hellemond2018). Imaging diagnosis can distinguish the two medically important CE and AE lesions (Brunetti et al., Reference Brunetti, Kern and Vuitton2010), but it fails to discriminate the CE lesions caused by E. granulosus s.s. and E. canadensis, respectively. In addition, misdiagnosis or uncertain cases can easily occur in imaging examination when encountering atypical pathological features, early stage or similar lesions, or inexperienced inspectors (Zhang and McManus, Reference Zhang and McManus2006; HołodyZaręba et al., Reference HołodyZaręba, Zaręba and Kędra2013; Mesut et al., Reference Mesut, Gulbiz, Sabri, Erdem, Ravza, Dilek, Murat and Oguz2016). Although serology is a helpful tool for early diagnosis of echinococcosis, questions remain with regards to its specificity and effectiveness for clinical detection. Till now, there are no available standardized echinococcosis diagnostic kits that are generally accepted by clinical physicians (Zhang and McManus, Reference Zhang and McManus2006; McManus et al., Reference McManus, Gray, Zhang and Yang2012). These all illustrate the widely used non-invasive tests could not reach accurate species identification. Invasive molecular biology technique base on the detection of parasite-specific DNA provides a more accurate approach for differential diagnosis of echinococcosis and is now considered to be a gold standard (Schweiger et al., Reference Schweiger, Grimm, Tanner, Müllhaupt, Bertogg, Müller and Deplazes2012). Possible due to the need for invasive examination, its development and application on clinical diagnosis is still limited. Nevertheless, treatment and prognosis of echinococcosis are diverse among species (Brunetti et al., Reference Brunetti, Kern and Vuitton2010; McManus et al., Reference McManus, Gray, Zhang and Yang2012), precise differential diagnosis is fundamental for determining the optimal treatment plan with the least amount of morbidity and mortality. Therefore, an efficient molecular diagnostic method is essential and should to be employed more widely or even taken as a routine clinical diagnostic tool. The mPCR established in this study provides such a method, which can simultaneously differentiate and accurately identify E. granulosus s.s., E. multilocularis and E. canadensis. It has a high application potential and may greatly contribute to the confirmatory diagnosis of human echinococcosis in China. Moreover, due to the high accuracy and reliability, this method can be a useful tool for evaluating the results of imaging and serological tests, which in turn can further develop the detection accuracy of non-invasive diagnosis.
Acknowledgements
We would like to acknowledge the West China Hospital and Sichuan Provincial People's Hospital for the support in the patient samples collection. Thanks to Tiao-Ying Li for kindly providing parasite material.
Financial support
This work was supported by the Sichuan Medical Association (Fan Chen et al., grant numbers Q16062); Science and Technology Department of Sichuan Province (Qian Wang et al., grant number 2018SZ0116) and the Health Planning Committee of Sichuan (Fan Chen et al., grant number 18PJ584).
Conflict of interest
None.
Ethical standards
The study was approved by the Ethics Committee of Sichuan Center for Disease Control and Prevention. All the samples were collected according to the procedures and guidelines of the ethics committee.









