INTRODUCTION
Taeniids (Cestoda: Cyclophyllidea) are characteristic parasites of terrestrial mammals. Adult taeniid tapeworms occur in the small intestine of typically carnivorous definitive hosts, and their cystic larvae (metacestodes) develop in tissues or body cavities of herbivorous or omnivorous intermediate hosts (Abuladze, Reference Abuladze and Skrjabin1964). A predator–prey relationship between the definitive and intermediate hosts maintains the transmission of taeniids.
The family Taeniidae Ludwig, 1886 is undergoing a taxonomic revision due to the development of molecular diagnostic methods and increasingly available DNA sequence data (Lavikainen et al. Reference Lavikainen, Haukisalmi, Lehtinen, Henttonen, Oksanen and Meri2008, Reference Lavikainen, Laaksonen, Beckmen, Oksanen, Isomursu and Meri2011). Currently, the largest taeniid genus, Taenia Linnaeus, 1758, consists of approximately 45 valid species (see Hoberg, Reference Hoberg2006; Lavikainen et al. Reference Lavikainen, Haukisalmi, Lehtinen, Henttonen, Oksanen and Meri2008; Rossin et al. Reference Rossin, Timi and Hoberg2010; Haukisalmi et al. Reference Haukisalmi, Lavikainen, Laaksonen and Meri2011), and in addition, mitochondrial DNA (mtDNA) evidence has revealed at least 3 previously unrecognized species (Lavikainen et al. Reference Lavikainen, Haukisalmi, Lehtinen, Henttonen, Oksanen and Meri2008; Galimberti et al. Reference Galimberti, Romano, Genchi, Paoloni, Vercillo, Bizzarri, Sassera, Bandi, Genchi, Ragni and Casiraghi2012). Felids serve as the definitive hosts for a minimum of 14 species of Taenia (Loos-Frank, Reference Loos-Frank2000). In addition, several other Taenia spp. can occasionally parasitize felid hosts (Abuladze, Reference Abuladze and Skrjabin1964; Jones and Pybus, Reference Jones, Pybus, Samuel, Pybus and Kocan2001).
Lynx spp. are Holarctic felids, occurring especially in boreal and temperate forests. As medium-sized cats, lynx prey on a wide range of mammals including rodents, lagomorphs and even cervids. Therefore, lynx are involved in the life cycles of several species of Taenia (Abuladze, Reference Abuladze and Skrjabin1964; Jones and Pybus, Reference Jones, Pybus, Samuel, Pybus and Kocan2001). Recently, endoparasites of the Eurasian lynx (Lynx lynx) were examined in Finland by analysing a considerable number of fecal and intestinal samples (Deksne et al. Reference Deksne, Laakkonen, Näreaho, Jokelainen, Holmala, Kojola and Sukura2012). Tapeworms recovered from the survey were not in adequate condition for morphological examination, and identifications were limited to the generic level. Based on rostellar hooks and the size of the strobilae, however, at least 2 species of Taenia were observed: Taenia laticollis Rudophi, 1819 and a much larger Taenia sp., in which specific identity has remained unclear. The focus of the present study is the molecular identification of Taenia tapeworms in lynx from Finland. In the current study we explore the diversity and phylogenetic placement of these Taenia spp., emphasizing the status of a putative unknown species.
MATERIALS AND METHODS
Intestinal samples and helminth collection
The Taenia tapeworms were collected from intestines of 296 lynx (L. lynx), which were shot during the hunting season 2010–2011 (from the beginning of December to the end of February) in Finland. This represented 80% of the legally hunter-harvested lynx, and included lynx from all of the 15 game management districts of Finland.
Skinned lynx carcasses were stored frozen until dissection and necropsy. Intestines were removed from the thawed carcasses and intestinal contents were squeezed out and washed. The helminths were preserved in 70% ethanol. The tapeworms were macroscopically identified at the generic level.
Selection of Taenia specimens for analyses
The majority of the Taenia tapeworms resembled T. laticollis by their general appearance and size. Length and shape of a few observed rostellar hooks supported this diagnosis. Since the worms were so numerous, it was not feasible to identify all specimens based on molecular characters. To create a general view of the species diversity among the T. laticollis -like specimens, we decided to identify strobilate cestodes or their fragments from one third of the infected lynx from each game management district. To cover the geographical area thoroughly, the lynx were selected in relation to maximum distances between collection sites separating host specimens, trying to avoid individuals from the same or neighbouring municipalities. Furthermore, both mildly (<10 worms per animal, n = 37 lynx) and heavily (11–112 worms per animal, n = 30 lynx) infected animals were selected assuming that different infection intensities might be related to different species of Taenia. One or 2 tapeworms per selected lynx (2 worms if more than 1 were present) were subjected to molecular identification.
Molecular identification was also performed for all specimens of Taenia, which differed macroscopically from typical T. laticollis and could be suspected to represent another species. This included 7 specimens from 5 lynx. Six of the specimens (from 4 lynx) represented the unknown large Taenia tapeworms, having obviously wider and longer strobilae than T. laticollis. Furthermore, a single tapeworm fragment having a strange dark colour was subjected to molecular identification.
Two short anterior fragments with scoleces were found in one of the lynx infected with the large unknown Taenia sp. The rostellar hooks indicated that these belonged to T. laticollis. These specimens were further identified by sequencing to demonstrate a possible mixed infection.
Comparative materials and morphological examination
Since mtDNA sequence data of T. laticollis have not been published, we used well-preserved morphologically verified strobilate adults as reference specimens. The samples (8 isolates) were collected in routine necropsies at the Finnish Food Safety Authority Evira from 6 Finnish lynxes found dead during the winter of 2008–2009. In addition, 2 specimens of Taenia taeniaeformis (Batsch, 1786) in a Finnish lynx, collected during a necropsy at Evira in 2009, were used for morphological and molecular comparison.
We also analysed a reference specimen of Taenia omissa Lühe, 1910, a Nearctic species of typically large dimensions which infects felids, mainly the cougar (Puma concolor). The sample was collected in a necropsy at the Canadian Cooperative Wildlife Health Centre from an adult male cougar, which was accidentally trapped in a coyote snare and died in the town of Pincher Creek, southern Alberta, in the spring of 2011.
Reference specimens (T. laticollis, T. omissa and T. taeniaeformis) were preserved in 70% ethanol following collection. These specimens and others representing the unknown species of Taenia were stained with Mayer's haemalum or Semichon's acetic carmine, cleared in eugenol and mounted in Canada balsam. The rostella were cleared in Berlese's medium. Comparative morphological studies, focusing on structural characteristics of the proglottids and rostellar hooks, were completed based on this series of specimens. The hooks were drawn with the aid of a camera lucida, and measurements were taken from these drawings using a calibrated ruler. Specimens were identified morphologically according to Verster (Reference Verster1969), Rausch (Reference Rausch1981) and Loos-Frank (Reference Loos-Frank2000). Vouchers for entire specimens, including those that substantiate the definitive identification for sequences used in the study have been deposited in the Finnish Museum of Natural History, University of Helsinki, under code MZH 123001-123141.
Molecular identification and phylogenetic analysis
The molecular identification was based on 2 alternative mtDNA regions, namely partial sequences of the cytochrome c oxidase subunit 1 (cox1) and the NADH dehydrogenase subunit 1 (nad1) genes (396 bp and 491 bp, respectively). In addition, for the phylogenetic analysis, the complete nad1 (894 bp) was sequenced for T. omissa and the unknown Taenia sp.
DNA extractions, enzymatic amplifications and sequencing were performed as reported previously (Lavikainen et al. Reference Lavikainen, Lehtinen, Meri, Hirvelä-Koski and Meri2003, Reference Lavikainen, Laaksonen, Beckmen, Oksanen, Isomursu and Meri2011). The mtDNA regions were amplified using previously published primers (partial cox1: 5′-TTTTTTGGGCATCCTGAGGTTTAT-3′ and 5′-TAAAGAAAGAACATAATGAAAATG-3′ by Bowles et al. (Reference Bowles, Blair and McManus1992); partial nad1: 5′-AGATTCGTAAGGGGCCTAATA-3′ and 5′-ACCACTAACTAATTCACTTTC-3′ by Bowles and McManus (Reference Bowles and McManus1993); complete nad1: 5′-TATTAAAAATATTGAGTTTGCGTC-3′ and 5′-TCTTGAAGTTAACAGCATCACGAT-3′ by Hüttner et al. (Reference Hüttner, Nakao, Wassermann, Siefert, Boomker, Dinkel, Sako, Mackenstedt, Romig and Ito2008)). For the partial cox1 and nad1, the sequencing primers were the same as used for the primary PCR. For the complete nad1, the PCR primers and also a reverse sequencing primer (5′-CCATTAAACAAGCCTCAAACCT-3′ by Lavikainen et al. (Reference Lavikainen, Haukisalmi, Lehtinen, Henttonen, Oksanen and Meri2008)) were used.
Sequences were compared with previously published mtDNA data assembled for various species of Taenia. For the phylogenetic analyses, the partial cox1 and complete nad1 sequences were aligned separately using ClustalW2 (Chenna et al. Reference Chenna, Sugawara, Koike, Lopez, Gibson, Higgins and Thompson2003). In addition to the new sequences, 25 previously published cox1 sequences representing 20 species of Taenia (Bowles and McManus, Reference Bowles and McManus1994; Okamoto et al. Reference Okamoto, Bessho, Kamiya, Kurosawa and Horii1995; Le et al. Reference Le, Blair, Agatsuma, Humair, Campbell, Iwagami, Littlewood, Peacock, Johnston, Bartley, Rollinson, Herniou, Zarlenga and McManus2000; Nakao et al. Reference Nakao, Sako and Ito2003, Reference Nakao, McManus, Schantz, Craig and Ito2007; Jeon et al. Reference Jeon, Lee, Kim, Hwang and Eom2005, Reference Jeon, Kim and Eom2007; Zhang et al. Reference Zhang, Hu, Jones, Alsopp, Beveridge, Schindler and Gasser2007; Lavikainen et al. Reference Lavikainen, Haukisalmi, Lehtinen, Henttonen, Oksanen and Meri2008, Reference Lavikainen, Haukisalmi, Lehtinen, Laaksonen, Holmström, Isomursu, Oksanen and Meri2010, Reference Lavikainen, Laaksonen, Beckmen, Oksanen, Isomursu and Meri2011; Jia et al. Reference Jia, Yan, Guo, Zhu, Wang, Shi, Chen, Zhan, Zhang, Fu, Littlewood and Cai2010; Liu et al. Reference Liu, Lin, Li, Liu, Liu, Yuan, Song, Zhao, Zhang and Zhu2011; Galimberti et al. Reference Galimberti, Romano, Genchi, Paoloni, Vercillo, Bizzarri, Sassera, Bandi, Genchi, Ragni and Casiraghi2012) and 13 nad1 sequences representing 9 species of Taenia (Le et al. Reference Le, Blair, Agatsuma, Humair, Campbell, Iwagami, Littlewood, Peacock, Johnston, Bartley, Rollinson, Herniou, Zarlenga and McManus2000; Nakao et al. Reference Nakao, Sako and Ito2003, Reference Nakao, McManus, Schantz, Craig and Ito2007; Jeon et al. Reference Jeon, Lee, Kim, Hwang and Eom2005, Reference Jeon, Kim and Eom2007; Jia et al. Reference Jia, Yan, Guo, Zhu, Wang, Shi, Chen, Zhan, Zhang, Fu, Littlewood and Cai2010, Reference Jia, Yan, Lou, Ni, Dyachenko, Li and Littlewood2012; Hüttner et al. Reference Hüttner, Siefert, Mackenstedt and Romig2009; Liu et al. Reference Liu, Lin, Li, Liu, Liu, Yuan, Song, Zhao, Zhang and Zhu2011) were included. The GenBank Accession numbers or references for the previously published sequences are presented in Fig. 2. The alignments were manually adjusted. Gaps and codons with ambiguous sites were deleted. The final alignments contained 366 and 891 nucleotides for the partial cox1 and complete nad1, respectively. Echinococcus oligarthrus (Diesing, 1863) and Taenia mustelae Gmelin, 1790 were used as outgroups for cox1, and E. oligarthrus for nad1 data sets. The phylogenetic trees were constructed by the neighbour-joining (NJ) method in PAUP* v4.0b10 (Swofford, Reference Swofford2002) using Kimura 2-parameter distances (Kimura, Reference Kimura1980), and assessed with 10 000 bootstrap replicates. Furthermore, to resolve the phylogenetic relationships of the unknown Taenia sp. and the closest taxa, a nad1 sequence (894 bp) data set of 5 species (see Fig. 2C, Taenia multiceps Leske, 1780 as an outgroup) was addressed with the maximum likelihood (ML) method. The substitution model (HKY85 + I) and its parameters were determined for the ML analysis by Akaike Information Criterion implemented in Modeltest 3.7 (Posada and Crandall, Reference Posada and Crandall1998). Heuristic search was used to estimate the ML tree, which was tested by bootstrapping with 10 000 replicates.
The new DNA sequences of this study have been deposited in the DDBJ/EMBL/GenBank databases under the Accession numbers JX860621-JX860633.
RESULTS
Molecular identification
Taenia tapeworms were found in two-thirds (201/296) of the lynx, ranging from 1 to 112 worms per animal. The total number of individual worms recovered from all hosts exceeded 2700. Altogether 135 tapeworms from 72 lynx were subjected to PCR-based molecular identification. This is 5% of the Taenia tapeworms recovered, and 36% of the lynx infected with Taenia. Most of the analysed samples were from central, southern and eastern Finland (Fig. 1) coincidental with the distribution of the harvested lynx.

Fig. 1. Geographical origins of the genetically analysed cestode specimens from lynx collected during the hunting season of 2010–2011 in Finland. Lynx individuals infected with identified cestodes are indicated with the following symbols: dot, Taenia laticollis; open circle, Taenia sp.; open circle with a dot inside, mixed infection with T. laticollis and Taenia sp.; triangle, Diphyllobothriidae. The origin of one lynx infected with Taenia sp. was not documented.
Two species of Taenia were differentiated using mtDNA sequence data. The most abundant species was T. laticollis represented by 127 specimens from 67 lynx. In contrast, a second putative species occurred at low prevalence and intensity, being represented by 5 specimens from 4 lynx. These latter cestodes were obviously distinguished from T. laticollis by the larger dimensions of the strobila, and did not correspond in mtDNA sequences with any of the reference specimens of this study nor those previously published. In a single case, a mixed infection with both species was confirmed. The lynx harbouring the unknown Taenia sp. were shot in western and southern Finland, whereas T. laticollis was found in all game management districts (Fig. 1). One specimen, classified as the unknown species macroscopically, remained unidentified due to the ambiguous sequencing result.
Among the reference specimens evaluated in the present study, T. omissa and T. laticollis showed characteristic sequences differing from previously published mtDNA data for other species of Taenia. The sequences of the T. taeniaeformis specimens were identical with those of an isolate in a domestic cat from Finland (Lavikainen et al. Reference Lavikainen, Haukisalmi, Lehtinen, Henttonen, Oksanen and Meri2008).
Two specimens were misidentified macroscopically as Taenia tapeworms before the molecular identification. One of them aroused suspicion because of its dark colour while another was erroneously classified within the group of T. laticollis-like specimens. Both of the tapeworm specimens were short fragments of strobila. The specimens shared identical cox1 sequences (396 bp), which were very different from those of Taenia spp. According to the results of a BLAST nucleotide database search (basic local alignment search tool, http://blast.ncbi.nlm.nih.gov), the cox1 sequence showed highest similarity (89%) with those of Sparganum proliferum (Ijima, 1905) and Spirometra erinaceieuropaei (Rudolphi, 1819) (GenBank Accession numbers AB015753 and JQ267473, respectively) suggesting that these specimens may represent a taxon within the family Diphyllobothriidae Lühe, 1910.
Sequence variation and phylogenetic relationships
Taenia laticollis
The identifications of 70 T. laticollis specimens were based on the partial sequence of the cox1 gene. Since several samples remained PCR negative, an alternative region (partial nad1) was applied for 57 specimens. Among these, 4 cox1 haplotypes (designated as A, B, C and D; see Fig. 2) and 3 nad1 haplotypes (NA, NB and NC) were detected. In addition, a reference specimen of T. laticollis had a nad1 haplotype of its own (ND). The sequence variations were 0·3–0·5% in cox1 (corresponding to 1–2 nucleotide differences per 396 bp) and 0·2–0·8% in nad1 (1-4/491 bp).
In the phylogenetic analysis of the cox1 dataset, T. laticollis formed a clade with Taenia macrocystis (Diesing, 1850) (Fig. 2A). The branching order among the haplotypes of T. laticollis, as well as phylogenetic relationships of T. laticollis with Taenia parva Baer, 1926 and T. taeniaeformis, were not well resolved. The haplotypes of T. laticollis differed in cox1 sequence (366 bp) from T. macrocystis by 9·8–10·4%, and from the other analysed Taenia spp. by 10·9–15·8%.

Fig. 2. (A-C) Dendrograms for Taenia spp. based on 2 mtDNA genes. (A) Neighbour-joining tree inferred with Kimura 2-parameter distances from the partial cox1 gene, Echinococcus oligarthrus and T. mustelae as outgroups. (B) Neighbour-joining tree inferred with Kimura 2-parameter distances from the complete nad1 gene, E. oligarthrus as an outgroup (not shown). (C) Maximum-likelihood tree inferred from the nad1 sequences of selected taxa, T. multiceps as an outgroup (not shown). Accession numbers or references of the previously published sequences are shown in parentheses. The sequences obtained in the present study are marked with an asterisk. Dots indicate species using primarily felids as definitive hosts. Bootstrap values >50% are shown. The scale bars are proportional to the number of substitutions per site.
Unknown Taenia sp
No intraspecific variation was detected in the partial cox1 sequence (396 bp) of the unknown Taenia sp. The complete nad1 (894 bp) was sequenced only for a single specimen, and thus it is not possible to analyse its intraspecific variation. In the phylogenetic analyses of the partial cox1 and complete nad1 sequence datasets, the unknown species was located as a close sister either to Taenia hydatigena Pallas, 1766 or to a clade formed by T. hydatigena and Taenia regis Baer, 1923 (Fig. 2). A well-supported monophyletic group including Taenia sp., T. hydatigena and T. regis formed a clade with T. omissa. This clade appears as a polytomy with the other main clades of Taenia when nodes with bootstrap support below 50% are ignored.
The nucleotide sequences of Taenia sp. differed from those of T. hydatigena and T. regis by 6·8–7·7% in cox1 and 8·3–10·4% in nad1 (Table 1). The level of divergence was similar between T. hydatigena and T. regis. All 3 species differed from T. omissa by more than 10% in the cox1 sequence and about 20% in nad1.
Table 1. Pairwise comparison of percentage nucleotide sequence differences in nad1 (891 bp, above the diagonal) and cox1 (366 bp, below the diagonal) among the unknown Taenia sp. and closely related species
1 Selected previously published sequences, for Accession numbers, see Fig. 2A and B.
Morphological characters of Taenia sp. and differential diagnosis
Specimens of the unknown Taenia sp. were not in adequate condition for morphological examination. Strobilae could not be measured exactly due to the fragmentation of the specimens. The estimated length of the strobila in individual cestodes was at least 100 cm and the maximum width 7–8 mm. The samples were decomposed, and the only internal structures detected were the cirrus sac, terminal vagina and a vaginal sphincter. The cirrus sac is short, not overlapping the ventral longitudinal osmoregulatory canal. The vagina is dilated before the sphincter. Only 3 scoleces with a few rostellar hooks were found. Altogether 5 large and 6 small well-aligned hooks were measured. The large hooks are 202–217 μm and the small 124–132 μm in length. The large hooks are characteristic in shape having a short strongly curved blade and a long guard (Fig. 3). The hooks can easily be distinguished from those of the reference samples (T. laticollis, T. omissa and T. taeniaeformis) by shape and size (Fig. 3).
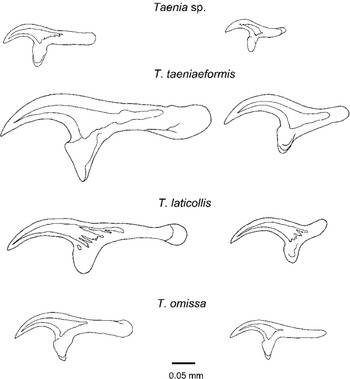
Fig. 3. Large and small hooks of Taenia sp., T. taeniaeformis and T. laticollis from lynx, and T. omissa from the cougar.
Taenia sp. can be distinguished from those species that occur in felids across the Holarctic region (Table 2). This includes 9 valid species (Verster, Reference Verster1969; Loos-Frank, Reference Loos-Frank2000) and 1 species of undetermined status, Taenia wyolagus Shults, Reference Shults1982 (published as a master's thesis), which had passed into oblivion and was omitted from the latest revision of the genus (Loos-Frank, Reference Loos-Frank2000). Taenia sp. is distinct from those species by its relatively small rostellar hooks. Dimensions of the small hooks overlap with those of Taenia pisiformis (Bloch, 1780) and Taenia bubesei Ortlepp, 1938 (considered as a synonym of T. regis by Verster, Reference Verster1969), the latter of which has been recorded once in a Caspian tiger, Panthera tigris virgata (Petrov and Potekhina, Reference Petrov and Potekhina1953); the large hooks are clearly different in structure relative to these species. Additionally, 4 species, T. bubesei, T. omissa, T. rileyi Loewen, 1929 and T. taeniaeformis have a vaginal sphincter.
Table 2. Taenia spp. of felids recorded in the Holarctic region

1 Considered as a synonym of Taenia regis Baer, 1923, by Verster (Reference Verster1969).
Taenia kotlani Murai, Gubányi & Sugár, 1993, which has been described based only on the metacestode, differs from Taenia sp. by the hook shape, although the hooks are similar in length; the blade of the large hooks is clearly shorter in Taenia sp. Although T. hydatigena is genetically closely related and has similar-sized hooks, it differs from Taenia sp. by having longer and less curved blades in the large hooks. In addition, T. hydatigena lacks a vaginal sphincter. This structure is present in another genetically closely related species, T. regis. The measurements of the small hooks of Taenia sp. overlap those of T. regis if T. bubesei is considered conspecific. The large hooks of T. regis sensu lato, however, are highly variable in shape (Verster, Reference Verster1969) and are longer than those of Taenia sp.
DISCUSSION
In the current survey, the presence of 2 species of Taenia in lynx collected during the hunting season 2010–2011 in Finland was demonstrated by mtDNA sequencing and comparative morphology. Taenia laticollis, a lynx tapeworm with a Holarctic distribution (Loos-Frank, Reference Loos-Frank2000), was the numerically dominant species in this material. A second, and apparently undescribed species, was rare, and its unique mtDNA sequences clearly differed from all available sequences of Taenia (22 species; including T. laticollis and T. omissa, sequences of which were obtained in the present study). In addition, this unknown Taenia sp. is clearly distinct from the other species of Taenia recorded in felids (including Lynx) from the northern hemisphere, based on the morphology of the rostellar hooks. In a recent study, this putative new species was not found in wolves (Canis lupus) from Finland or Sweden, nor brown bears (Ursus arctos) in Finland (Lavikainen et al. Reference Lavikainen, Laaksonen, Beckmen, Oksanen, Isomursu and Meri2011). In the 4 infected lynx of the present study, the strobilae appeared to be fully developed. Evidently, lynx serve as a primary definitive host for this species. According to the present results, Taenia sp. demonstrated here has not been reported previously or described. Well-preserved adult specimens and metacestodes, however, would be needed to further compare this species with the previously described members of the genus.
Consistent with a morphological phylogeny by Hoberg et al. (Reference Hoberg, Alkire, de Queiroz and Jones2001), T. macrocystis is the closest relative of T. laticollis in the present phylogenetic analysis. A close relationship of the new Taenia sp. with a tapeworm of canids, T. hydatigena, and a tapeworm of Panthera spp., T. regis, is shown in the present analysis. Although closely related, the level of sequence difference, as well as the morphological characters, supports their recognition as distinct species. A monophyletic group containing these 3 species is distantly related to a tapeworm of the cougar, T. omissa. The phylogenetic position of T. hydatigena among those species of Taenia characteristic of felids suggests that host-switching to canids has occurred during the evolutionary history of T. hydatigena. Such is consistent with the developing view of the importance of guild structure and foraging habits and host colonization in the context of regional faunas as determinants of diversification among taeniid tapeworms (e.g. Hoberg, Reference Hoberg2006).
Both T. laticollis and T. macrocystis are adapted to a predator–prey relationship between felids and lagomorphs (Rausch, Reference Rausch1981; Zyll de Jong, Reference Zyll de Jong and van1966). The intermediate host of the new Taenia sp. remains unknown. The lynx harbouring Taenia sp. were from southern and western Finland, where the preferred prey include hares (Lepus spp.), European roe deer (Capreolus capreolus) and an introduced cervid species, the white-tailed deer (Odocoileus virginianus) (Pulliainen, Reference Pulliainen1981; Pulliainen et al. Reference Pulliainen, Lindgren and Tunkkari1995). In contrast, in eastern Finland the mountain hare (Lepus timidus) forms the most important part of the diet of the lynx. Judging by the phylogenetic position of Taenia sp., the intermediate host might be a ruminant. Close phylogenetic relationships with species parasitizing ruminants, however, do not automatically indicate that similar intermediate hosts should be involved in transmission. For example, Taenia serialis (Gervais, 1847), which occurs in lagomorphs as a metacestode, is closely related to species parasitizing ungulates based on phylogenetic trees inferred from both mt and nuclear DNA data (e.g. Zhang et al. Reference Zhang, Hu, Jones, Alsopp, Beveridge, Schindler and Gasser2007; Lavikainen et al. Reference Lavikainen, Haukisalmi, Lehtinen, Henttonen, Oksanen and Meri2008; Knapp et al. Reference Knapp, Nakao, Yanagida, Okamoto, Saarma, Lavikainen and Ito2011).
The prevalence of the putative new species is 1·4% in the present material. The exact prevalence of T. laticollis cannot be calculated since only a subsample of the specimens was subjected to molecular identification. The prevalence would be about 66%, if the remaining and morphologically similar specimens all represented T. laticollis. In previous studies, T. laticollis has been frequently found in Lynx spp. from Europe and North America (e.g. Smith et al. Reference Smith, Addison, Joachim, Smith and Quinn1986; Valdmann et al. Reference Valdmann, Moks and Talvik2004), even occurring as the most common species (Zyll de Jong, Reference Zyll de Jong and van1966). In addition to T. laticollis and the new species, T. taeniaeformis evidently occurs sporadically in lynx from Finland, as 2 of our reference samples represented this species. Specimens of T. taeniaeformis, however, were not found in the material collected in 2010–2011.
The present results contrast with those in the Baltic countries, where T. pisiformis, primarily a parasite of canids (Loos-Frank, Reference Loos-Frank2000; Jones and Pybus, Reference Jones, Pybus, Samuel, Pybus and Kocan2001), is common in lynx (Kazlauskas and Matuzevicius, Reference Kazlauskas and Matuzevicius1981; Bagrade et al. Reference Bagrade, Vismanis, Kirjušina and Ozolinš2003; Valdmann et al. Reference Valdmann, Moks and Talvik2004). In Latvia, T. pisiformis was detected in all examined lynx (42 individuals), and no other species of Taenia were found (Bagrade et al. Reference Bagrade, Vismanis, Kirjušina and Ozolinš2003). In Estonia, T. pisiformis was also found in all examined lynx (37), but T. laticollis was common as well with a prevalence of 41%, and single cases of T. taeniaeformis and T. hydatigena were detected (Valdmann et al. Reference Valdmann, Moks and Talvik2004). In 39 lynx from Lithuania, T. taeniaformis and T. pisiformis were the most common species, and Taenia crassiceps (Zeder, 1800), T. krabbei and T. laticollis were rare (Kazlauskas and Matuzevicius, Reference Kazlauskas and Matuzevicius1981). In all these studies, the specimens were identified based on morphology. Different diets cannot explain the difference between the lynx tapeworms in Finland and the Baltic countries nearby, since T. pisiformis uses the same intermediate hosts as T. laticollis, i.e. lagomorphs. Instead, these observations suggest that T. pisiformis is absent from Finland and may demonstrate the distinct nature of the helminth fauna in this region of northern Europe. Similar patterns of species diversity for taeniids in wolves have also been observed (Lavikainen et al. Reference Lavikainen, Laaksonen, Beckmen, Oksanen, Isomursu and Meri2011), perhaps reflecting some general historical and ecological mechanisms that have influenced diversity.
Tapeworms examined during the present study were damaged due to freezing, thawing and processing, and morphological characters were mostly unidentifiable. Such challenges are not uncommon when conducting surveys of helminths among wildlife species. Consequently, we applied a molecular-based method, mtDNA sequencing, for accurate identification of Taenia spp. Although mtDNA sequencing has been successfully applied to identification of taeniids (e.g. Zhang et al. Reference Zhang, Hu, Jones, Alsopp, Beveridge, Schindler and Gasser2007; Lavikainen et al. Reference Lavikainen, Laaksonen, Beckmen, Oksanen, Isomursu and Meri2011), it is relatively costly and time consuming, which limits its use when a large number of specimens are under evaluation. In large-scale epidemiological studies, another method, for example multiplex PCR (e.g. Al-Sabi and Kapel, Reference Al-Sabi and Kapel2011) or restriction fragment length polymorphism PCR (e.g. Hüttner et al. Reference Hüttner, Siefert, Mackenstedt and Romig2009) would be more practical, if the sole need is to provide an accurate diagnosis. Sequencing is essential, however, for characterization of an unknown species, phylogenetic analyses, or validation of other molecular methods. For example, recently, an unknown species of Taenia was discovered by mtDNA sequencing in brown bears and elk (Alces alces) from Finland (Lavikainen et al. Reference Lavikainen, Haukisalmi, Lehtinen, Laaksonen, Holmström, Isomursu, Oksanen and Meri2010, Reference Lavikainen, Laaksonen, Beckmen, Oksanen, Isomursu and Meri2011). Subsequently it was described as a new species, Taenia arctos Haukisalmi, Lavikainen, Laaksonen & Meri, Reference Lavikainen, Laaksonen, Beckmen, Oksanen, Isomursu and Meri2011, and was shown to have a considerably broad distribution across the Holarctic. The present discovery in lynx suggests that the true diversity of the genus Taenia, although being the focus of extensive studies since the 1700s, is still insufficiently known. Further, our study clearly highlights the need for integrated analyses using molecular and morphological characters, and the assembly of archival collections that form the foundations for assessment of biotic structure and faunal history (e.g. Hoberg et al. Reference Hoberg, Galbreath, Cook, Kutz and Polley2012).
ACKOWLEDGEMENTS
We thank Jorma Korhonen, Anita Kenttälä and Sanna Kokko (Finnish Game and Fisheries Research Institute, Taivalkoski field station) for their contribution in sample collection and Ilpo Kojola (Finnish Game and Fisheries Research Institute) for sample collaboration. Alberta Sustainable Resources Development is acknowledged for submitting a cougar carcass for necropsy.
FINANCIAL SUPPORT
V.H. has been supported by an NSF PBI award Nos 0818696 and 0818823.