Introduction
Crustaceans are a diverse group of organisms, including commercially important species such as shrimp, lobsters and crabs, in addition to a variety of parasitic species associated with both vertebrate and invertebrate hosts. Parasitic crustaceans include the isopods of the superfamilies Bopyroidea and Cryptoniscoidea, which are specialized for the parasitism of other crustaceans (Williams and Boyko, Reference Williams and Boyko2012). The family Bopyridae is a highly diversified group of isopods, with more than 600 species in nine subfamilies (Boyko et al., Reference Boyko, Moss, Williams and Shields2013). One bopyrid genus, Probopyrus, includes isopods that are typically ectoparasites in the branchial chamber of paleomonid prawns (Masunari et al., Reference Masunari, Da Silva Castagini and Oliveira2000). The species of this genus include P. bithynis Richardson, 1904, P. buitendijki (Horst, 1910), P. floridensis Richardson, 1904, P. markhami Román-Contreras, 1996, P. pacificensis Román-Contreras, 1993 and P. pandalicola (Packard, 1879), which are known to parasitize prawns of the genera Macrobrachium, Palaemon and Palaemonetes (Lemos de Castro, Reference Lemos de Castro1974; Masunari et al., Reference Masunari, Da Silva Castagini and Oliveira2000; Román-Contreras, Reference Román-Contreras2004; Brinton and Curran, Reference Brinton and Curran2015a; Gopalakrishnan et al., Reference Gopalakrishnan, Raja, Trilles, Rajkumar, Rahman and Saravanakumar2017; Ribeiro et al., Reference Ribeiro, Horch and Williams2019; de Barros et al., Reference de Barros, da Silva Neto and Calado2021).
Macrobrachium amazonicum Heller, 1862 is a freshwater prawn with a wide distribution in South America, and is the native prawn species with the most widespread occurrence in the inland waters of Amazonia (Odinetz-Collart and Moreira, Reference Odinetz-Collart and Moreira1993). Despite being endemic to the Amazon region (Odinetz-Collart, Reference Odinetz-Collart1991), M. amazonicum is also found in the basins of the Paraná and São Francisco rivers (Bialetzki et al., Reference Bialetzki, Nakatani, Baumgartner and Bond-Buckup1997; Sampaio et al., Reference Sampaio, Silva, Santos and Sales2007), as well as many other hydrographic basins in South (Kensley and Walker, Reference Kensley and Walker1982; Melo, Reference Melo2003; Valencia and Campos, Reference Valencia and Campos2007) and Central America (Vergamini et al., Reference Vergamini, Pileggi and Mantelatto2011).
Macrobrachium amazonicum is the definitive host of P. bithynis, and a number of studies have focused on the relationship between these two species. Odinetz-Collart (Reference Odinetz-Collart1990), for example, found evidence of a stable interaction between the two species, supported by data on the infestation rates and life cycle of the host, based on specimens collected on the lower Tocantins River, in the Brazilian state of Pará, given that the body length of the female isopods correlated positively with that of the prawn host. More recently, Corrêa et al. (Reference Corrêa, Sousa, Silva, Adriano, Oliveira and Tavares-Dias2018) described histopathological alterations in the gills of M. amazonicum specimens collected from the lower Amazon River, in Pará state, caused by P. bithynis infestation. These authors concluded that the alterations were consistent with the ingestion of the branchial tissue by P. bithynis, which would have a negative impact on the respiratory capacity of the host. Infestation by Probopyrus females may also induce the castration of the host, the feminization of the males (Beck, Reference Beck1980), reduction of the development of the nutritional conditions of the host (de Barros et al., Reference de Barros, da Silva Neto and Calado2021), and even predator−prey interactions, through the reduction in the capacity of the host to camouflage itself, leaving it more susceptible to predation (Brinton and Curran, Reference Brinton and Curran2015b).
While these records of bopyrid parasitism on M. amazonicum all refer to specimens collected in the coastal region of the Amazon (Maciel and Valenti, Reference Maciel and Valenti2009), no evidence has been found of this phenomenon, up to now, in the inland waters of the Amazon basin. The present study not only provides records of the occurrence of Probopyrus sp. parasitism in specimens of M. amazonicum collected in areas that are approximately 650 km from the mouth of the Amazon River, but also reports on molecular analyses that indicate distinct host–parasite relationships between the inland and coastal regions of the Amazon basin. The implications of these findings for the taxonomy of some Probopyrus species are also discussed.
Materials and methods
Study area
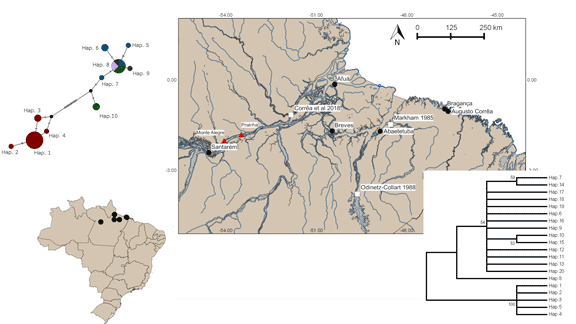
The study area includes both coastal and inland regions of Brazilian Amazonia. Inland, specimens were collected on the left margin of the Amazon River in the municipality of Santarém, Pará (Brazil), in an area of várzea swamp known as Pixuna do Tapará (02°24.98′ S, 54°33.9′ W). Local prawn fishermen indicated the presence of large numbers of parasitized M. amazonicum in this area. In the coastal region, specimens of M. amazonicum, both with and without parasites, were collected from the municipalities of Abaetetuba, Afuá, Augusto Corrêa, Bragança and Breves (Fig. 1), all located in the Brazilian state of Pará.

Fig. 1. Location of the sampling points in the inland and coastal regions of Brazilian Amazonia (Black circles), previous records of Probopyrus bithynis (white squares) and geographic boundaries of the continental and coastal genetic lineages of the host Macrobrachium amazonicum (red triangles). More details are presented in the discussion.
Sampling
In the Santarém region, prawns were collected in a type of trap known locally as the covo, which is used by the local shrimpers. These traps are made of semi-fixed frames of wood or iron (2 m × 1.5 m) covered with a wire or nylon mesh, and set in the direction of the current. There is a lateral rectangular opening at each extremity, large enough to allow individual prawn to enter the structure, where they are trapped (Castro e Silva and Cavalcante, Reference Castro e Silva and Cavalcante1994). In the coastal region, the prawns were collected using baited traps known locally as the matapí (see Maciel and Valenti, Reference Maciel and Valenti2009). Each parasitized prawn was placed in an individual plastic bag to avoid losing or mixing the parasites. In the laboratory, the parasites were removed from each host and preserved in 1.5-ml microtubes containing 100% ethanol.
Molecular analyses
The total genomic DNA of the host was obtained from abdominal tissue using the ammonium acetate protocol of Bruford et al. (Reference Bruford, Hanotte, Brookfield, Burke and Hoelzel1998). The DNA of the parasites was obtained from the males using the QIAamp DNA Investigator kit (QIAGEN), following the maker's instructions. The sample included 28 parasites and 29 M. amazonicum specimens (Table 1). Part of the sample material was deposited in the scientific collection of the Museum of Biological Diversity – Zoology (Museu de Diversidade Biológica – área Zoologia, MDBio – Zoologia) of the Campinas State University (Universidade Estadual de Campinas – UNICAMP) with the ZUEC CRU 4381, 4382 and 4383 vouchers. Information on new vouchers also can be obtained from the authors or from the museum's curatorship.
Table 1. Number of specimens of the host (Macrobrachium amazonicum) and parasite (Probopyrus sp.) analysed in the present study at each sampling point.

The extracted DNA was processed by polymerase chain reaction (PCR) to isolate and amplify different regions of the cytochrome oxidase C subunit I (COI) gene. The A and F primers (Palumbi and Benzie, Reference Palumbi and Benzie1991) were used to amplify the COI of M. amazonicum, while the HCO2198 and LCO1490 primers (Folmer et al., Reference Folmer, Black, Hoeh, Lutz and Vrijenhoek1994) were used for the parasites.
The molecular identification of the parasites was based on sequences of the 18S rDNA gene, obtained using the ‘ai’ and ‘bi’ primers of Whiting et al. (Reference Whiting, Carpenter, Wheeler and Wheeler1997). Despite being a highly conserved region, this is the gene with the largest number of bopyrid sequences deposited in GenBank. Given this, samples of parasites were selected randomly from each sampling locality. Similarly, the COI database of the parasites included two sequences of P. pandalicola (GenBank id: MH087672 and MK308333).
The quality of the extracted DNA and the PCR products was evaluated by electrophoresis in 1% agarose gel to which GelRed (Biotium) was added. The sequences were obtained using an ABI 3500 (Applied Biosystems) automatic sequencer with the Big Dye 3.1 kit (Applied Biosystems), following the maker's instructions. The sequencing reactions were run in both directions using the PCR primers.
Data analysis
The sequences obtained were aligned in CodonCode Aligner v7.1.2 (CodonCode Corporation) for the visualization and editing of reading errors. Three databases were compiled, one for the hosts (COI) and two for the parasites (COI and 18S rDNA).
The number of haplotypes (unique sequences) was obtained from these three databases using DNAsp v5.10.1 (Librado and Rozas, Reference Librado and Rozas2009). For the hosts, a haplotype network was constructed in PopART (Leigh and Bryant, Reference Leigh and Bryant2015), based on the median joining networks method (Bandelt et al., Reference Bandelt, Forster and Röhl1999). In the case of the parasites, a neighbour-joining tree was constructed in MEGA v7.0 (Kumar et al., Reference Kumar, Stecher and Tamura2016) using the p distance model. This program was also used to obtain the mean genetic (p) distances between the populations. The 18S rDNA sequences were used for a basic local alignment search tool (BLAST) search (MegaBLAST), with a similarity of at least 98% being considered valid, with e-values near or equal to zero.
Results
18s rDNA
An 18S rDNA sequence of 975 base pairs (bps) was obtained from ten bopyrid individuals (four from Santarém, two from Abaetetuba and Augusto Corrêa, and one each from Afuá and Breves). All the sequences were 100% identical and the BLAST search demonstrated that all the parasites collected from both study areas belong to the genus Probopyrus, with sequences closest to the species Probopyrus pacificiensis, P. pandalicola and P. buitendijki (Table 2).
Table 2. Results of the BLAST search of the 18S rRNA sequences of the Probopyrus specimens obtained in the present study

COI gene
The COI database for the parasites included 28 sequences of 603 bps, which included 20 haplotypes, considering all the populations analysed. Haplotypes 1–5 were recorded in individuals from Santarém (inland population), while haplotypes 6–20 were recorded exclusively in the coastal populations (Fig. 2).

Fig. 2. Neighbour-joining tree obtained from the COI sequences showing the relationships among haplotypes 1–20 retrieved from the Probopyrus parasites collected in Brazilian Amazonia. The values above the nodes represent the bootstrap significance of the clades. Bootstrap values of less than 50 are not shown here.
These findings were further corroborated by the mean genetic distances. The parasite sequences from Santarém are approximately 16% different from those of the other localities (Table 3), whereas the differences between the coastal populations ranged from only 0.41% (between Breves and Afuá) to 0.81% between Abaetetuba and Augusto Corrêa. In comparison with the sequences of P. pandalicola, the genetic distance was 16.68% from the Santarém population, and up to 18.37%, in the case of the population of Abaetetuba.
Table 3. Mean genetic distances (p distance in %) estimated from the COI sequences of Probopyrus (dark grey, lower) and Macrobrachium amazonicum (light grey, upper) between the sampling localities in eastern Brazilian Amazonia

a The values within parentheses represent the intra-population variation of the host and parasite, respectively.
The 29 COI sequences of M. amazonicum included 10 haplotypes, of which haplotypes 1–4 were recorded exclusively in Santarém, and haplotypes 5–10 were found only in the coastal populations (Fig. 3). The sequences from Santarém were also approximately 3% different from those of the other localities, with extremely low variation (0.1–0.3%) being found among the coastal populations (Table 3).

Fig. 3. Haplotype network based on COI sequences of the host Macrobrachium amazonicum collected in Brazilian Amazonia.
Discussion
The present study provides the first evidence of the infestation of an inland population of M. amazonicum by a parasite of the genus Probopyrus. The molecular analyses also revealed that the coastal and inland populations of M. amazonicum are parasitized by different Probopyrus species.
Parasites of the genus Probopyrus have a well-established relationship with a number of different representatives of the family Palaemonidae, in particular, the prawns of the genus Macrobrachium (Markham, Reference Markham1985; Saito et al., Reference Saito, Shokita and Naruse2010). In the specific case of M. amazonicum, records of infestation by P. bithynis are restricted to the coastal region of the Amazon basin, in areas near the Tucuruí hydroelectric dam on the Tocantins River, which is approximately 300 km from the Atlantic Ocean (Odinetz-Collart, Reference Odinetz-Collart1988, Reference Odinetz-Collart1990). More recently, records of this parasitism were obtained from the lower Amazon, near the community of Maruim, in the municipality of Gurupá, around 400 km from the Atlantic Ocean, in the Brazilian state of Pará (Corrêa et al., Reference Corrêa, Sousa, Silva, Adriano, Oliveira and Tavares-Dias2018). The inland population analysed here is from Santarém, approximately 650 km from the ocean (see Fig. 1).
In the case of the earliest record, Odinetz-Collart (Reference Odinetz-Collart1988) observed parasitized prawns only downstream from the Tucuruí dam, and none from the reservoir itself, and a similar lack of parasitism in prawn specimens collected from the region of Manaus (Central Amazonia) and the Ucayali River in Peru (Odinetz-Collart, Reference Odinetz-Collart1990). Given the evidence, this author concluded that bopyrid parasites are limited by the distribution of the brackish water copepods that act as intermediate hosts. In the laboratory, Dale and Anderson (Reference Dale and Anderson1982) found that Acartia tonsa Dana, 1849 was the intermediate host of P. bithynis, even in the presence of other copepods present in the mixed zooplankton cultures experiment made by the authors. Acartia tonsa is a common calanoid copepod found in estuarine environments (Figueroa et al., Reference Figueroa, Figueroa and Hicks2020). Given this, it is very likely that A. tonsa acts as an intermediate host in the coastal populations of Probopyrus analysed in the present study. This does not apply, however, to the parasites collected in the inland area (Santarém), which must rely on a different calanoid copepod intermediate host, still unidentified.
In addition to the fact that this is a freshwater region, and thus outside the geographic range of A. tonsa, the analysis of the COI sequences indicated a clear separation (with no gene flow) of the parasites collected in Santarém from those obtained in the coastal municipalities (Afuá, Abaetetuba, Augusto Corrêa and Breves), with a mean genetic distance (16.2%) that is consistent with the presence of two distinct Probopyrus species parasitizing M. amazonicum. The lack of gene flow between the populations of the definitive host found in Santarém and the coastal areas (Abaetetuba, Augusto Corrêa, Afuá and Breves) further reinforces this conclusion. While still preliminary, the few available genetic data clearly indicate a lack of gene flow between the coastal and inland populations of M. amazonicum (Vergamini et al., Reference Vergamini, Pileggi and Mantelatto2011; Iketani et al., Reference Iketani, Pimentel, Torres, Rêgo and Sampaio2021). Iketani et al. (Reference Iketani, Pimentel, Torres, Rêgo and Sampaio2021) obtained COI sequences from a number of M. amazonicum populations distributed along the length of the Amazon River, and found that the coastal group was restricted to the lower Amazon, below Prainha (approximately 550 km from the Atlantic Ocean), while the inland populations were located upriver from the municipality of Monte Alegre (around 620 km from the Atlantic) (see Fig. 1).
Sequences of COI are widely used for the molecular identification of species using the DNA barcode approach proposed by Hebert et al. (Reference Hebert, Cywinska, Ball and deWaard2003), with many studies applying this method successfully in crustaceans, in particular, those of the order Decapoda (Lefébure et al., Reference Lefébure, Douady, Gouy and Gibert2006; Costa et al., Reference Costa, deWaard, Boutillier, Ratnasingham, Dooh, Hajibabaei and Hebert2007; da Silva et al., Reference da Silva, Creer, Dos Santos, Costa, Cunha, Costa and Carvalho2011; Raupach and Radulovici, Reference Raupach and Radulovici2015). Raupach and Radulovici (Reference Raupach and Radulovici2015) reviewed the literature from the period between 2003 and 2014 and found 164 papers on the DNA barcode of crustaceans, although only six of these studies focused on the order Isopoda. Given that the largest interspecific genetic distances found in decapod crustaceans range from 20.92% (da Silva et al., Reference da Silva, Creer, Dos Santos, Costa, Cunha, Costa and Carvalho2011) to 22.66% (Costa et al., Reference Costa, deWaard, Boutillier, Ratnasingham, Dooh, Hajibabaei and Hebert2007), and those in isopods, from 12.01% to 27.17% (see S1 table of Raupach et al., Reference Raupach, Barco, Steinke, Beermann, Laakmann, Mohrbeck, Neumann, Kihara, Pointner, Radulovici, Segelken-Voigt, Wesse and Knebelsberger2015), the divergence of up to 16.31% observed in the present study between the inland (Santarém) and coastal populations (Afuá, Abaetetuba, Breves and Augusto Corrêa) of Probopyrus parasites infesting M. amazonicum would appear to be consistent with the presence of two distinct parasite species in the two regions (Fig. 2 and Table 3). By contrast, the level of genetic differentiation (0.41–0.81%) between the coastal Probopyrus populations is only slightly higher than that observed in M. amazonicum (0.1–0.3%), which indicates the occurrence of gene flow between the populations of both parasites and hosts in this coastal sector.
The morphology of Probopyrus has been the subject of considerable controversy in recent decades. In an analysis of the morphology of the cryptoniscus larvae of Probopyrus bithynis, P. pandalicola and P. floridensis, Dale and Anderson (Reference Dale and Anderson1982) concluded that the morphological characteristics of these larvae were sufficient to confirm the validity of the three species, which, despite their morphological similarities as adults, can be distinguished based on larval morphometric parameters. However, the Probopyrus species from the Western Atlantic was synonymized with P. pandalicola by Markham (Reference Markham1985) based solely on adult morphology. At the present time, P. bithynis, P. pandalicola and P. floridensis are all considered to be valid by the World Register of Marine Species (WoRMS, 2021).
Based on the available data (Odinetz-Collart, Reference Odinetz-Collart1988, Reference Odinetz-Collart1990; Corrêa et al., Reference Corrêa, Sousa, Silva, Adriano, Oliveira and Tavares-Dias2018), it would be reasonable to assume that the species found in the coastal populations is P. bithynis, given that the inland population appears to represent a new species of Probopyrus. Probopyrus bithynis presents differences in the larval morphology that can be used to distinguish this species from other Probopyrus taxa (see Dale and Anderson, Reference Dale and Anderson1982), in addition to some of the traits of the adult female (see Ribeiro et al., Reference Ribeiro, Horch and Williams2019). As larvae were not collected in the present study and the research team has limited practical experience with the morphology of Probopyrus species, it was not possible to provide a more conclusive diagnosis of the morphology of the specimens collected here. Given this, it is necessary to describe the morphological characteristics of the material collected in the present study. It is also necessary to expand the number of sampling points, especially in the inland region and to conduct analyses to determine the prevalence and other parameters of the population dynamics of the parasites and their hosts.
Ribeiro et al. (Reference Ribeiro, Horch and Williams2019) recently recorded the occurrence of P. cf. pandalicola in the Brazilian state of Bahia and provided insights into the morphology of the females and distinctions from the description of Markham (Reference Markham1985), while also emphasizing the need for molecular data and the analysis of the larval morphology for more reliable identification of the species. Ribeiro et al. (Reference Ribeiro, Horch and Williams2019) also reinforce the need for molecular data to delimit the different species of the group and determine whether P. pandalicola is a single, widely dispersed species or a complex of cryptic species. The use of COI sequences was suggested by the authors and the present work has done that by analysing COI sequences obtained from different populations of Probopyrus species.
Few molecular data are available on the parasites of the family Bopyridae, and up to now, most studies have sequenced the 18S rDNA gene, which has a much lower mutation rate than COI. Dreyer and Wägele (Reference Dreyer and Wägele2001) used the sequences of this gene to evaluate the phylogenetic relationships of the family Bopyridae, while Boyko et al. (Reference Boyko, Moss, Williams and Shields2013) expanded the dataset to the superfamilies Bopyroidea and Cryptoniscoidea. In the specific case of the genus Probopyrus, 18S rDNA sequences are available for P. pacifiensis (Dreyer and Wägele, Reference Dreyer and Wägele2001), P. buitendijki (Boyko et al., Reference Boyko, Moss, Williams and Shields2013) and P. pandalicola (Cho, Reference Cho2012), in addition to those described in the present study. Wu et al. (Reference Wu, Xiong and Yu2015) evaluated the taxonomic resolution of the 18S gene in copepods and determined that the limit between the intra- and inter-specific similarity was close to 100%. In the authors’ words this value ‘is unrealistic when attempting to achieve a high rate of successful identification, owing to potential PCR or sequencing errors’. From this perspective, the levels of similarity observed in the BLAST search run in the present study (Table 3) are of limited interpretative value, indicating only that the sequences analysed all belong to representatives of the genus Probopyrus.
While the 18S rDNA gene is not useful for the molecular identification of species, the COI sequences presented here indicate that this marker can be extremely valuable, not only for the molecular identification of species, but also for the study of the population genetics of Probopyrus species, which should contribute to a better understanding of the taxonomy and phylogenetic relationships within the genus. These data will, in turn, provide important insights for the understanding of the parasite–host relationships involving these isopods.
Data
Nucleotide sequences obtained in this paper are available in the GenBank database under accession numbers MZ686260 (Probopyrus, 18S rRNA) MZ687054 – MZ687073 (Probopyrus, COI) and MZ674502 – MZ674511 (M. amazonicum, COI).
Acknowledgements
We would like to thank the local shrimp fishermen for helping with the fieldwork. We also thank Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for the master's scholarship to RISP and the Graduate Program in Natural Resources of the Amazon (PGRNA) for financing part of the costs of field trips. The authors also thank the anonymous referee who provided useful and detailed comments on an earlier version of the manuscript.
Author contributions
GI conceived and designed the study. CRM and RISP conducted the field sampling. RISP and GI gathered data and wrote the article.
Financial support
This study was supported by Fundação Amazonia de Amparo a Estudos e Pesquisas – FAPESPA (ICAAF N° 002/2018).
Conflict of interest
None.
Ethical standards
Not applicable.