Introduction
Tapeworms (Cestoda) of the order Diphyllobothriidea are a unique group that occurs in a wide host spectrum, including all groups of tetrapodes (amphibians, reptiles, birds and mammals), with an almost cosmopolitan distribution in fresh and sea water, but also in the terrestrial environment (Kuchta et al., Reference Kuchta, Scholz, Brabec and Bray2008). Systematics and phylogenetic relationships of the Diphyllobothriidea are not definitely resolved, because many taxa have not yet been molecularly analyzed (Waeschenbach et al., Reference Waeschenbach, Brabec, Scholz, Littlewood and Kuchta2017). A total of 70 species of 18 genera and 3 families are currently recognized as valid in this order (Kuchta and Scholz, Reference Kuchta, Scholz, Caira and Jensen2017). Diphyllobothriideans also include several important human parasites, such as some species of the genera Dibothriocephalus, Diplogonoporus and Spirometra (Kuchta et al., Reference Kuchta, Scholz, Brabec and Narduzi-Wicht2015; Waeschenbach et al., Reference Waeschenbach, Brabec, Scholz, Littlewood and Kuchta2017). Human disease caused by Dibothriocephalus (formerly Diphyllobothrium) tapeworms, commonly referred to as ‘broad tapeworms,’ is the most important fish-borne zoonosis (Scholz et al., Reference Scholz, Garcia, Kuchta and Wicht2009), with up to 20 million people infected worldwide (Chai et al., Reference Chai, Darwin Murrell and Lymbery2005).
Dibothriocephalus latus is considered the principal and most frequent human fish-borne cestode in Europe (Scholz and Kuchta, Reference Scholz and Kuchta2016). Recent records and surveys in Europe indicate that this broad tapeworm is now less common in the former historical endemic areas, i.e. Scandinavian countries, but its frequency persists or even has an increasing tendency in the area of Alpine lakes (Switzerland, eastern France and northern Italy) (Kuchta et al., Reference Kuchta, Scholz, Brabec and Narduzi-Wicht2015; Gustinelli et al., Reference Gustinelli, Menconi, Prearo, Caffara, Righetti, Scanzio, Raglio and Fioravanti2016; Radačovská et al., Reference Radačovská, Bazsalovicsová, Blasco Costa, Orosová, Gustinelli and Králová-Hromadová2019). Moreover, plerocercoids of D. latus have recently been reported from rainbow trout (Oncorhynchus mykiss) from Argentina (Kuchta et al., Reference Kuchta, Radačovská, Bazsalovicsová, Viozzi, Semenas, Arbetman and Scholz2019).
The life cycle of D. latus is complex and requires 3 hosts to complete, including planktonic crustaceans serving as the first intermediate host. Freshwater fish: mainly perch (Perca fluviatilis), pike (Esox lucius) and burbot (Lota lota) serve as second intermediate hosts, and fish-eating mammals (including humans) represent its definitive host (Kuchta et al., Reference Kuchta, Scholz, Brabec and Narduzi-Wicht2015).
Although the zoonotic cestode D. latus has been attracting scientists for a long time and biological information, including the complete genome sequence (International Helminth Genomes Consortium, 2019), has accumulated in the literature, the karyotype and chromosome characteristics remain unknown. A few published karyological studies of members of the family Diphyllobothriidae have shown that the haploid (i.e. basal) chromosome number (n) is identical in all studied species, n = 9 (Wolcott, Reference Wolcott1959; Wikgren and Gustafsson, Reference Wikgren and Gustafsson1965; Sasada, Reference Sasada1978; Petkevičiūtė, Reference Petkevičiūtė1992; Petkevičiūtė, Reference Petkevičiūtė1996; Okino et al., Reference Okino, Ushirogawa, Matoba, Nishimatsu and Saito2017). Most of them have 2 sets of chromosomes with diploid number 2n = 18, but 2 species, Spirometra erinaceieuropaei and S. mansonoides, have 3 sets of chromosomes and are described as triploids, with 3n = 27 (Sasada, Reference Sasada1978; Liu and He, Reference Liu and He1988; Okino et al., Reference Okino, Ushirogawa, Matoba, Nishimatsu and Saito2017). Karyotypes of all studied species are highly symmetrical in relation to chromosome morphology, consisting only of bi-armed small- to medium-sized chromosomes (Špakulová et al., Reference Špakulová, Orosová and Mackiewicz2011). An exception to this rule is Schistocephalus solidus that has nearly half of the chromosome pairs mono-armed (Petkevičiūtė, Reference Petkevičiūtė1996). All published karyological analyses were restricted to classical staining techniques without the use of more advanced cytogenetic techniques, such as fluorescence in situ hybridization (FISH). In addition, no information is yet available on spermatogenesis in these fish-borne zoonotic cestode species.
To fill this gap in cytogenetic knowledge, we present a detailed analysis of the D. latus karyotype. A particular interest was paid to variable chromosome numbers observed in our preliminary research, suggesting possible polyploidization of the karyotype. First, the numerical variation was critically reexamined in the chromosome set of D. latus, including a detailed study of mitotic chromosome morphology. Then FISH was applied with the 18S rDNA probe to determine the number and location of major ribosomal DNA clusters, which are suitable and readily available molecular markers of mitotic as well as meiotic chromosome sets. FISH was also performed with a (TTAGGG)n probe to find out if this ancestral telomeric motif of Metazoa is conserved at the end of D. latus chromosomes. The eventual presence of interstitial telomeric sequences (ITSs) was examined by FISH with tyramide signal amplification (TSA-FISH) with high detection sensitivity (Carabajal Paladino et al., Reference Carabajal Paladino, Nguyen, Šíchová and Marec2014).
Material and methods
Origin of plerocercoid larvae
Plerocercoids of D. latus were collected from 132 European perch (P. fluviatilis) originating from Lake Iseo (Italy) in May 2018. Lake Iseo is the fourth largest of the glacial lakes in the subalpine region in the north of the country (45°43′N, 10°05′E). The fish were collected by professional fishermen using a gill net. The perch were immediately placed on ice and transported to the laboratory within 16 h, where their musculature was examined for parasites by parasitological necropsy (detailed filleting of the whole musculature). All obtained plerocercoids of D. latus were examined under a stereomicroscope for morphological characteristics and placed in 0.9% NaCl solution before hamster infection.
According to their morphology, all plerocercoids were identified as D. latus larvae. Ten plerocercoids were fixed in 96% ethanol for subsequent molecular genotyping. Genomic DNA was extracted using the QIAamp® DNA mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer's instruction and then used as a template for polymerase chain reaction (PCR). PCR amplification of the D. latus cytochrome c oxidase subunit 1 (cox1, 437 bp) was performed according to Wicht et al. (Reference Wicht, Yanagida, Scholz, Ito, Jiménez and Brabec2010). Multiplex PCR of the COI gene confirmed that the plerocercoid larvae belong to D. latus.
Infection of experimental animals
The hamster, Mesocricetus auratus, which was used as an experimental host in this study, has not been confirmed to serve as the definitive host for D. latus under natural conditions; however, the animals were successfully infected in several previous studies (Andersen, Reference Andersen1972, Reference Andersen1978; Andersen and Halvorsen, Reference Andersen and Halvorsen1978; Dick and Poole, Reference Dick and Poole1985). All 24 experimental hamsters were maintained in the animal holding facilities of the Faculty of Agrobiology, Food and Natural Resources, Czech University of Life Science Prague (CZU), and kept under the following standard conditions: humidity of 55 ± 60%, temperature of 22 ± 2°C, photoperiod of 12 h light : 12 h dark. The hamsters were given standard rodent food and water ad libitum. Infection was performed in full consciousness of the animals by a single oral administration of 2 plerocercoids (per host); care was taken to ensure that each hamster swallowed both plerocercoids.
Dibothriocephalus latus specimens
Host animals were anaesthetized with isoflurane (i.e. halogenated ether) and sacrificed by injecting the commercial preparation T-61 at designated time (20–22 days postinfection). The small intestine was removed from freshly killed hosts. Only 2 out of 24 infected hamsters were positive. Two alive tapeworms of D. latus (430- and 452-mm long) were isolated, rinsed several times in saline solution, and processed immediately for cytogenetic analysis.
Chromosome preparations
For cytogenetic analysis, whole live specimens were treated with 0.025% colchicine solution in saline for 1 h to arrest mitosis in dividing cells at the metaphase stage, and placed in hypotonic solution (0.075 M KCl) for 4 h for swelling at room temperature. During this treatment, the worms were torn along their entire length (each proglottid) using an insulin syringe. The torn worms were placed in freshly prepared cold fixative solution (methanol: acetic acid = 3:1) for 30 min (with 2 replacements of the fixative) and then stored at −20°C until further use. Chromosome preparations were obtained by the spreading technique (Orosová and Špakulová, Reference Orosová and Špakulová2018). Briefly, glass slides were incubated in acid ethanol for 1 h. A small piece of fixed parenchyma with testes was placed on a dry-cleaned slide in a drop of 60% acetic acid and macerated using tungsten needles, or alternatively, a larger piece (1 proglottid) was transferred to an Eppendorf tube with 60% acetic acid and homogenized. Then, a few drops of macerated or homogenized suspension were applied to the slide and spread using a heating plate at 45°C. Finally, the preparations were passed through an ethanol series (70, 80, and 100%, 1 min each) and stored at −20°C until further use. Before each staining, slides were removed from the freezer, passed through an ethanol series, and air-dried.
In addition, 57 D. latus plerocercoids obtained from 109 European perch were also cytogenetically processed. A total of 92 chromosome slides were prepared and screened. However, all these preparations showed no dividing cells.
Karyological analysis
Slides were stained with 5% Giemsa (Merck, New Jersey, USA) solution in phosphate buffer (pH 6.8) for 25 min and rinsed with distilled water. Morphometric measurements of metaphase chromosomes were carried out on captured images. The chromosomes were arranged according to the centromere position and a karyotype was assembled. Chromosome numbers were counted in 272 mitotic cells from 2 individuals. Absolute length (AL, chromosome long q (L) + short p (S) arm length in μm), relative length (=AL × 100%/half-length of all chromosomes in metaphase spread) and the centromeric index [=S/(L + S)] were measured and calculated in 15 best mitotic spreads with clearly distinguishable chromosomes. The mean and standard deviation were calculated using a Microsoft Excel spreadsheet. Chromosomes were classified according to Levan's nomenclature method (Levan et al., Reference Levan, Fredga and Sandberg1964).
Silver nitrate staining
Silver nitrate (AgNO3) staining was performed following the standard protocol commonly used for cestodes (Ráb and Roth, Reference Ráb, Roth, Balíček, Forejt and Rubeš1988). Briefly, 6 drops of 50% AgNO3 solution and 3 drops of 2% gelatine in 1% formic acid were mixed on the surface of the chromosome spread preparation and covered with a coverslip. Then the slide was incubated on a heating plate at 45°C until the preparation turned light brown. Slides were rinsed with flowing distilled water and air-dried.
DAPI staining
Chromosome preparations were stained with DAPI (4',6-diamidino-2-phenylindole; Sigma-Aldrich, St. Louis, MO, USA) as described in Traut et al. (Reference Traut, Sahara, Otto and Marec1999). Briefly, slides were rinsed in phosphate-buffered saline (1 × PBS) containing 1% Triton X-100 for 5 min and stained with DAPI (0.5 μg mL−1 in PBS containing 1% Triton X-100) for 15 min in the dark, washed in 1% Kodak-PhotoFlo in 1 × PBS for 3 min, and then briefly washed in 1% Kodak-PhotoFlo in miliQ water, all at room temperature. Finally, the slides were mounted in an antifade based on 1,4-diazabicyclo[2.2.2]octane (DABCO; Sigma-Aldrich).
Probe preparation
The 18S rDNA probe, about 1500 bp long, was generated by PCR from D. latus genomic DNA extracted using the QIAamp® DNA mini Kit. PCR was performed using 2 primers, 18S-WormA forward (5′- GCGAATGGCTCATTAAATCAG-3′) and 18S-WormB reverse (5′- CTTGTTACGACTTTTACTTCC-3′), as described by Littlewood and Olson (Reference Littlewood, Olson, Littlewood and Bray2001). Telomeric probe (TTAGGG)n was generated by non-template PCR as described previously by Sahara et al. (Reference Sahara, Marec and Traut1999). Probes were labeled with biotin-16-dUTP (Roche Diagnostics, Mannheim, Germany) using the improved nick translation procedure described by Kato et al. (Reference Kato, Albert, Vega and Birchler2006) with some modifications. The 20 μL reaction contained 1 μg of unlabeled 18S rDNA probe, 50 μ m of dATP, dCTP and dGTP, 10 μ m of dTTP, 20 μ m of labelled nucleotides, 1× nick translation buffer (50 mm of Tris-HCl, pH 7.5; 5 mm of MgCl2; 0.005% of bovine serum albumin, BSA), 10 mm of ß-mercaptoethanol, 0.005 of U DNase I and 20 U of DNA polymerase I (both ThermoFisher, Waltham, MA, USA). The reaction time was carried out for 50 min at 15°C for 18S rDNA probe and 1 h at 15°C for telomeric probe.
FISH with biotin-labeled probes
Fluorescence in situ hybridization was performed according to the standard protocol used for insects (Sahara et al., Reference Sahara, Marec and Traut1999) with slight modifications for cestodes (Orosová and Špakulová, Reference Orosová and Špakulová2018). Preparations were treated with proteinase K (20 mg mL−1) in 1 × PBS for 5 min at 37°C, washed twice in 1 × PBS for 5 min each, and then digested by 100 μg mL−1 RNase A in saline sodium citrate (2 × SSC) for 1 h at 37°C and washed twice in 2 × SSC for 5 min each. Then the slides were incubated in 5 × Denhard's solution for 30 min at 37°C. Chromosomal DNA was denatured in 70% formamide in 2 × SSC for 3 min and 30 s at 68°C. The probe cocktail for each slide (10 μL; 50% deionized formamide, 10% dextran sulfate, 2 × SSC) contained ~50 ng of biotinylated 18S rDNA probe and 25 μg of sonicated salmon sperm DNA (Sigma-Aldrich). The probe was denatured at 90°C for 5 min. Hybridization at 37°C for 20 h was followed by stringent washes, which included 50% formamide in 2 × SSC (3 × 5 min, 46°C) (Fluka, Buchs, Switzerland), 2 × SSC (5 × 2 min, 46°C), 0.1 × SSC (3 × 5 min, 62°C) and 4 × SSC containing 0.1% Tween 20 (3 × 3 min, 37°C). Hybridization signals were amplified and visualized by 3-step detection, Cy3-conjugated streptavidin (Jackson ImmunoRes. Labs. Inc., West Grove, PA, USA), biotinylated anti-streptavidin (Vector Labs. Inc., Burlingame, CA, USA) and Cy3-conjugated streptavidin. The preparations were counterstained with 0.5 μg mL−1 DAPI in a DABCO-based antifade.
FISH with tyramide signal amplification
The telomeric probe was prepared by nick translation as described above and purified using Sephadex (Illustra Sephadex G-50 fine DNA grade, GE healthcare, Chicago, IL, USA). For TSA-FISH we used the procedure described in Zrzavá et al. (Reference Zrzavá, Hladová, Dalíková, Šíchová, Ounap, Kubíčková and Marec2018). In particular, chromosome slides were treated with 10 mm HCl for 10 min at 37°C and incubated in 1% hydrogen peroxide for 30 min at RT to quench endogenous peroxidase activity. Then the preparations were digested with 100 μg mL−1 RNase A for 1 h at 37°C and blocked with 5 × Denhardt's solution for 30 min at 37°C. Chromosomes were denatured in a probe mix containing 10–30 ng of the labeled telomeric probe, 50% deionized formamide, and 10% dextran sulfate in 2 × SSC for 5 min at 70°C and hybridized overnight. Hybridization signals were enhanced by Antifluorescein–horseradish peroxidase conjugate (PerkinElmer, Waltham, MA, USA) diluted by 1:1000 concentration and incubated with tyramide solution (TSA Plus Fluorescein system, PerkinElmer) for 5–7 min. The preparations were counterstained and mounted in DABCO-based antifade containing 0.5 μg mL−1 of DAPI.
Microscopy and image processing
The stained preparations were observed using 2 fluorescence microscopes, Leica DM 4000 B equipped with a digital camera DFC 450 C and Olympus B51 equipped with a DP70 CCD camera, and photographed separately for each fluorescent dye. Black-and-white fluorescent images were pseudocolored (light blue for DAPI, red for Cy3) and merged using Adobe Photoshop, version 7.0.
Results
Basic mitotic karyotype
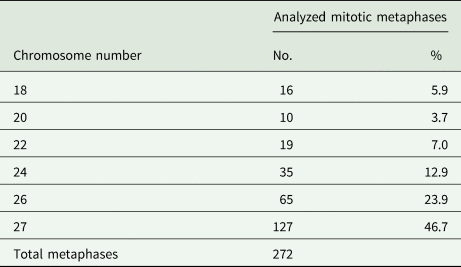
Analysis of dividing mitotic cells from 2 adult worms showed high variation in the number of chromosomes. Table 1 shows the range of chromosome counts of the 272 mitotic metaphases evaluated (135 from the first and 137 from the second tapeworm specimen) with clearly distinguishable chromosomes. Both specimens showed 6 cell types that differed in chromosome number, ranging from 18 to 27 chromosomes. The most frequently detected chromosome count in mitotic cells from testes, ovary and vitellaria of adult worms was 27 (46.7%, Fig. 1), which is the number corresponding to the triploid set, 3n = 27. The remaining mitotic metaphases evaluated were therefore aneuploid without 1–9 chromosomes (Table 1). No significant differences in the length of the 3 chromosomes of each triplet were determined by post hoc test. Similarly, no significant differences in the karyotype structure were found between the 2 evaluated adult worms. The evident predominance of the 3n = 27 cytotype in both D. latus specimens clearly indicated the triploid nature of this Alpine lake population (Fig. 2).
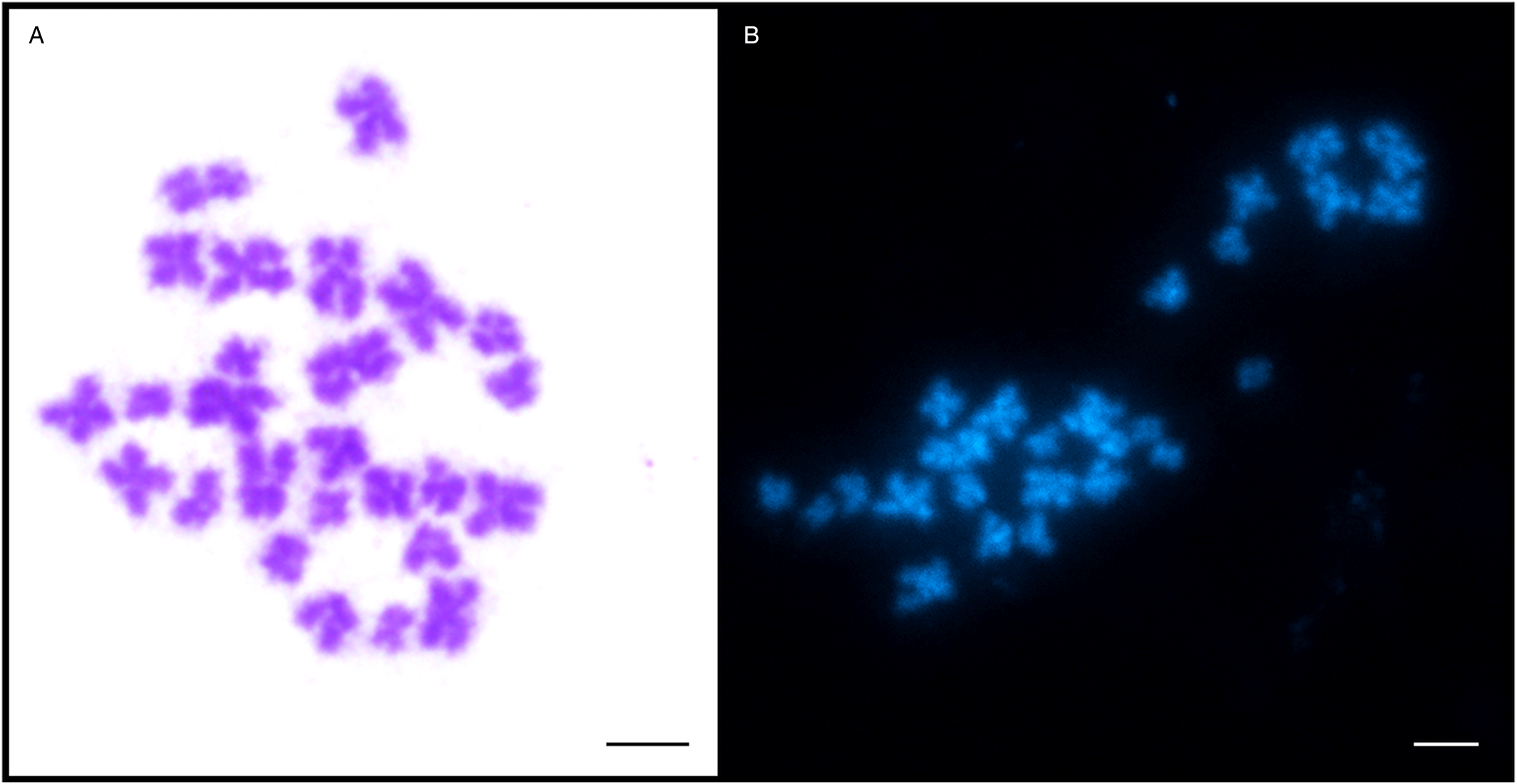
Fig. 1. Metaphase chromosomes of Dibothriocephalus latus. (A) Giemsa and (B) DAPI staining (3n = 27; scale bar = 10 μm).
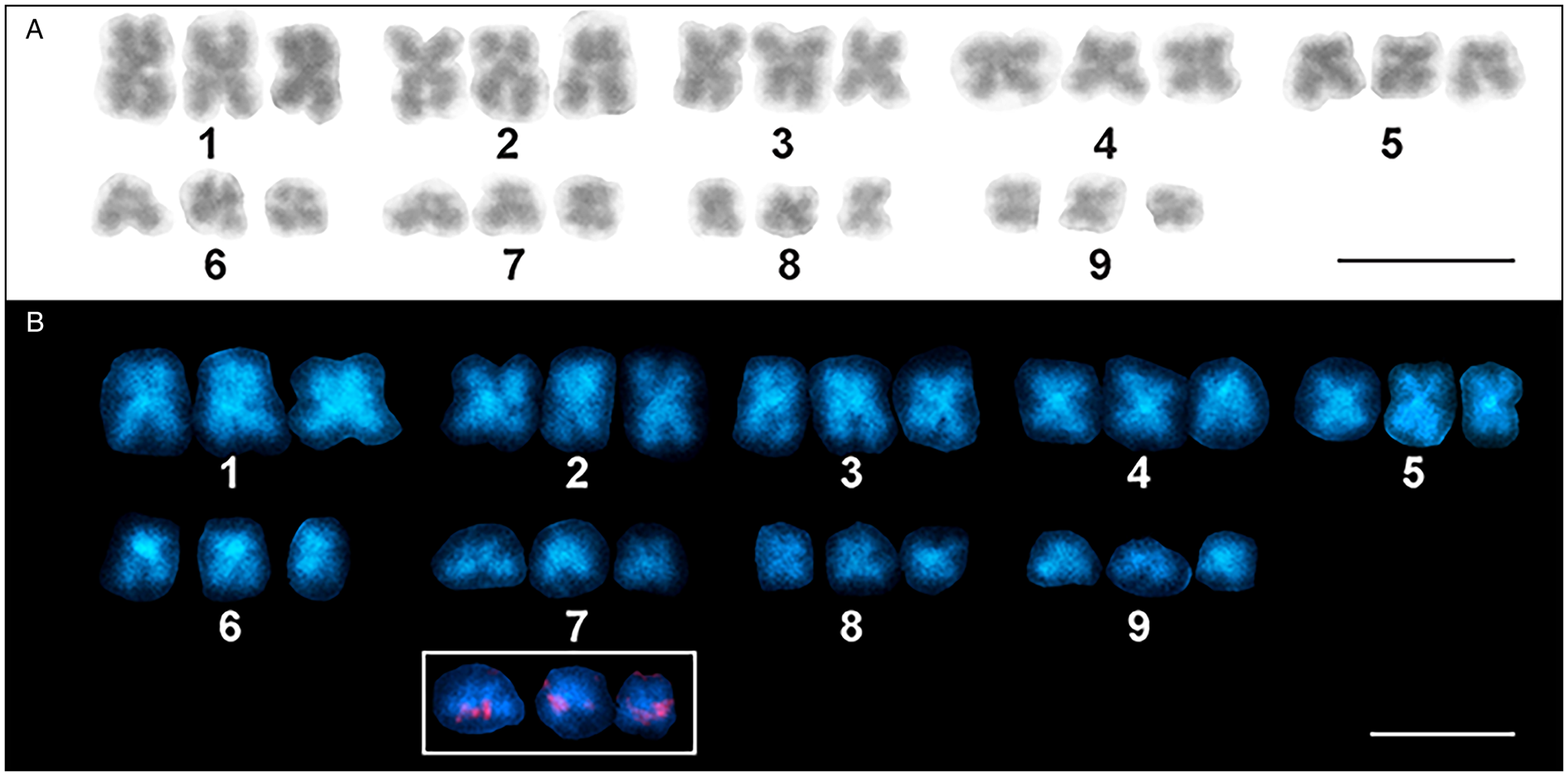
Fig. 2. Karyotype derived from mitotic metaphase cells of Dibothriocephalus latus (3n = 27 m). (A) Giemsa staining. (B) DAPI staining showing AT-rich bands. The small inset shows a detail of chromosomes of the triplet No. 7 with distinct 18S rDNA [nucleolar organizer region (NOR)] loci located on each individual chromosome (scale bar = 10 μm).
Table 1. Chromosome numbers in mitotic cells of Dibothriocephalus latus
Twenty mitotic metaphases from 2 individuals were karyotyped and the data obtained are summarized in Table 2. The chromosomes were relatively small; the largest and the smallest chromosomes measured 5.63 and 2.82 μm, respectively. The standard karyotype is symmetric; it is composed of 27 metacentric chromosomes divided into 9 triplets of homologous chromosomes of gradually decreasing size (Fig. 2A). Triplets nos. 1–4 were quite easily distinguishable by length, while triplets 5–9 were more difficult to distinguish due to their high similarity in length. No secondary constriction was seen on the Giemsa-stained chromosomes. The karyotype formula can be summarized as 3n = 27; n = 9 m and the fundamental number of arms is FN = 54. The total length of the haploid complement reached 36.53 μm on average.
Table 2. Measurement (mean ± s.d.) and classification of chromosomes of Dibothriocephalus latus

a Mean absolute lengths were calculated from 20 mitotic metaphases.
Abbreviations: s.d., standard deviation; M, metacentric chromosome.
Number, activity and chromosomal location of rDNA clusters and telomeric FISH
FISH with 18S rDNA probe showed clear hybridization signals on both chromatids of 3 homologous chromosomes in mitotically dividing cells in all chromosome number variants (all cytotypes) (Fig. 2B, Fig. 3) and also in interphase nuclei (Fig. 3A). These signals were approximately having the same intensity in all 3 chromosomes. Based on the results, we concluded that D. latus has a single locus for rRNA genes (i.e. the nucleolar organizer region, NOR) per haploid genome, which is located interstitially, postcentromeric on the short arms of the small metacentric chromosome No. 7 (7p cen). In DAPI-counterstained FISH preparations, the constitutive heterochromatin pattern showed only weak markings in the centromeric regions of all metaphase chromosomes (Fig. 2B).
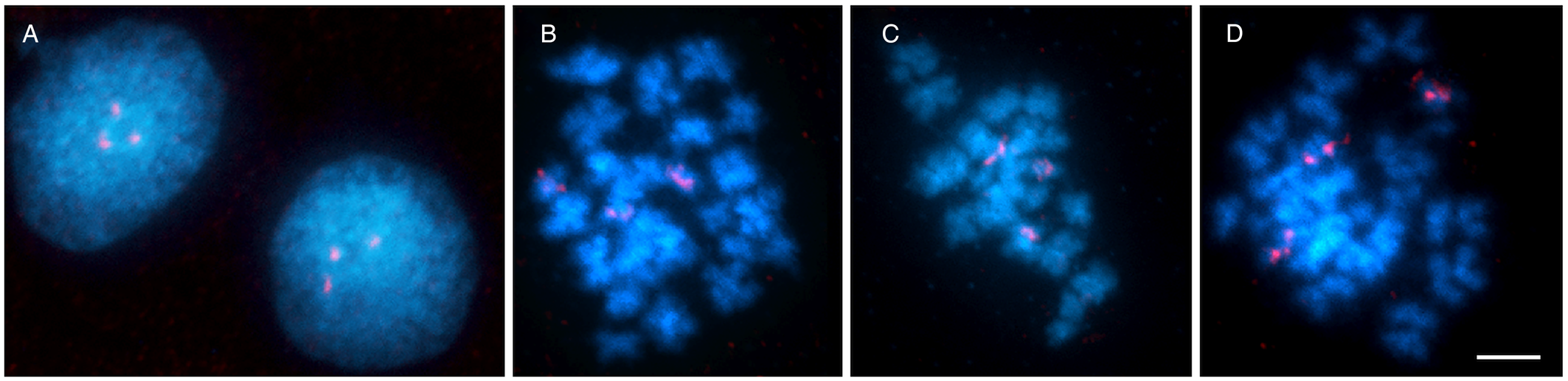
Fig. 3. Chromosomes of Dibothriocephalus latus stained with DAPI (blue) and 18S rDNA fluorescence in situ hybridization probe (red). (A) An interphase nucleus with 3 18S rDNA clusters. (B, C) Mitotic metaphases showing 3 chromosomes with hybridization signals. (D) Early mitotic anaphase showing early segregation of chromatids to opposite poles (scale bar = 10 μm).
FISH with telomeric (TTAGGG)n probe showed typical twin hybridization signals at the ends of both chromatids in all chromosomes (Fig. 4). Interstitial clusters of telomeric repeats (ITS) were not detected in D. latus chromosomes, although we used a more sensitive TSA-FISH method with this probe on long pachytene chromosomes (Fig. 4B and D).
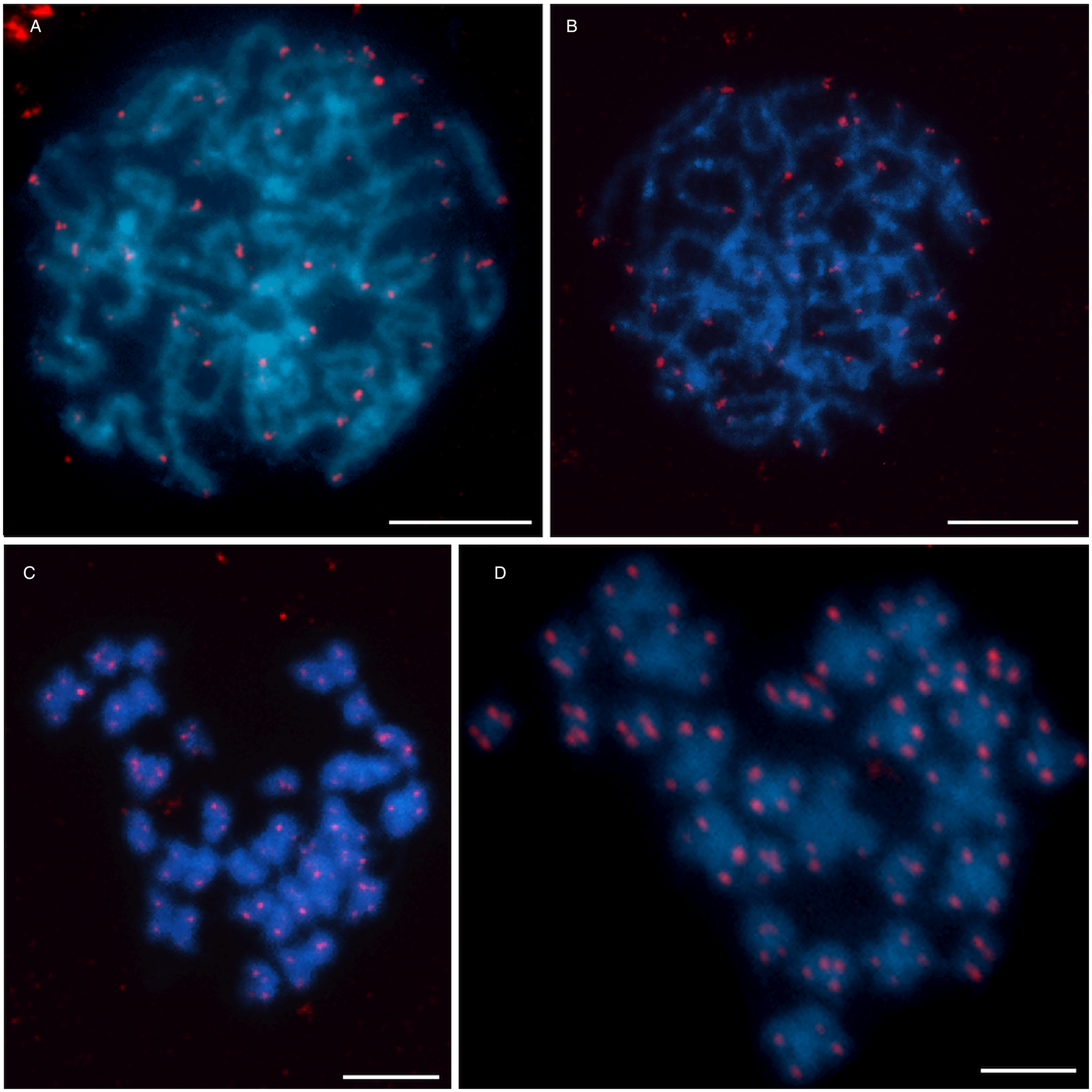
Fig. 4. Distribution of telomeric sequence (TTAGGG)n after FISH in Dibothriocephalus latus showing an exclusively telomeric pattern. (A) Pachytene nucleus and (C) mitotic metaphase after FISH with biotin-labeled telomeric probe. (B) Pachytene nucleus and (D) mitotic metaphase after TSA-FISH (scale bar = 10 μm). Abbreviations: FISH, fluorescence in situ hybridization; TSA-FISH, FISH with tyramide signal amplification.
AgNO3 staining (Ag-banding) of NOR-associated protein was used to demonstrate the activity of the rRNA genes. A variable number of 1–3 Ag-positive NORs were observed in pachytene nuclei of D. latus (Fig. 5) with 3 active nucleoli as the most common variant. A maximum of 2 nucleoli were observed in the interphase nuclei (Fig. 5C).
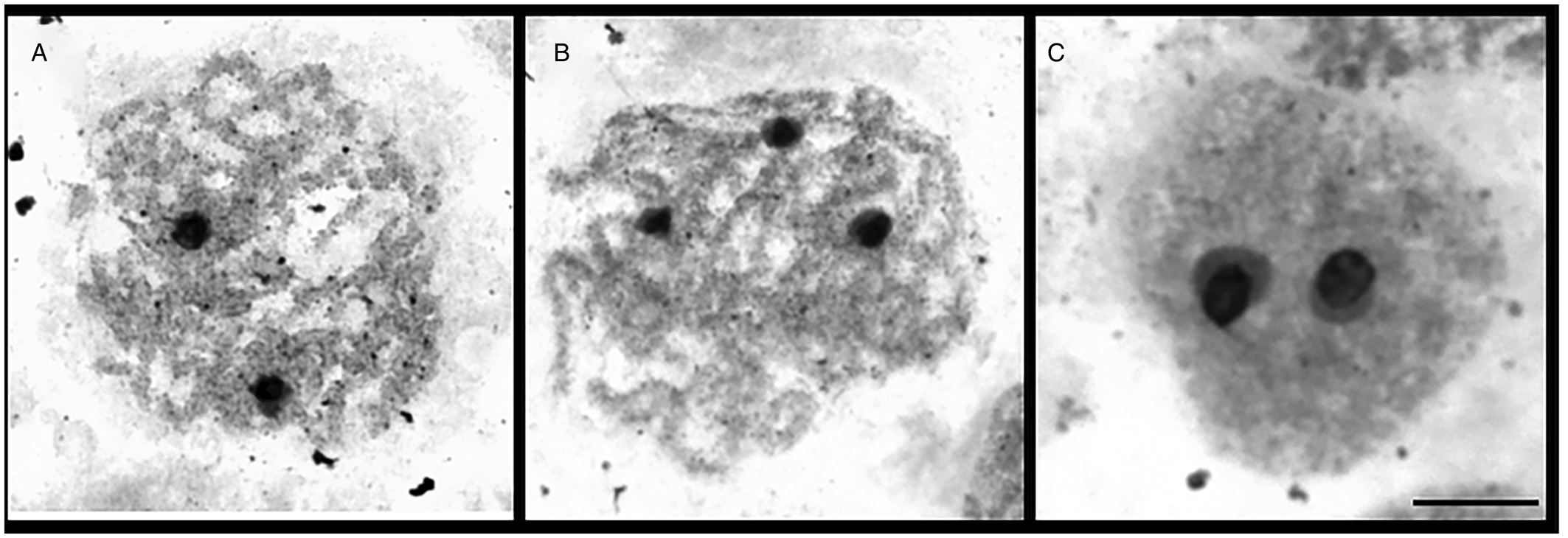
Fig. 5. Visualization of nucleoli (N) in meiotic spermatocytes of Dibothriocephalus latus after AgNO3 staining (scale bar = 10 μm).
The course of meiosis
During the slide preparation from the fixed material, we processed the whole worm, piece by piece from scolex to the last proglottid. However, we did not record all stages of meiotic division. When the worm was divided into 3 parts, numerous mitotic metaphases were observed in the first proximal third of the strobila, while few mitotic metaphases and selected stages of spermatocyte division (early and late pachytene nuclei) were found in the middle part of the body. In the last third of the strobila, mitotic chromosomes were not observed and meiotic figures (pachytene and late metaphase II nuclei) occurred only very sporadically; this part of strobila often contained proglottids full of eggs.
As shown by 18S rDNA FISH (Fig. 6), both univalent and bivalent of the triplet No. 7 might be present during meiotic prophase. Three homologues of triplet No.7, carrying ribosomal genes, did not pair and remained as 3 univalents in most spermatocyte prophases (Fig. 6E–G) or formed 1 bivalent plus 1 univalent (Fig. 6C and D). The subsequent stages of meiosis I (metaphase I and anaphase I) were not found on the preparations. However, there are 2 lines of evidence that meiosis II occurred, though rarely. First, we observed metaphase II nuclei with unbalanced chromosome numbers (Fig. 6H) due to failure of chromosome segregation during meiosis I. Second, we found early stages of spermiogenesis (developing spermatozoa) on slides from both specimens examined (Fig. 6I). However, fully developed sperm were missing.
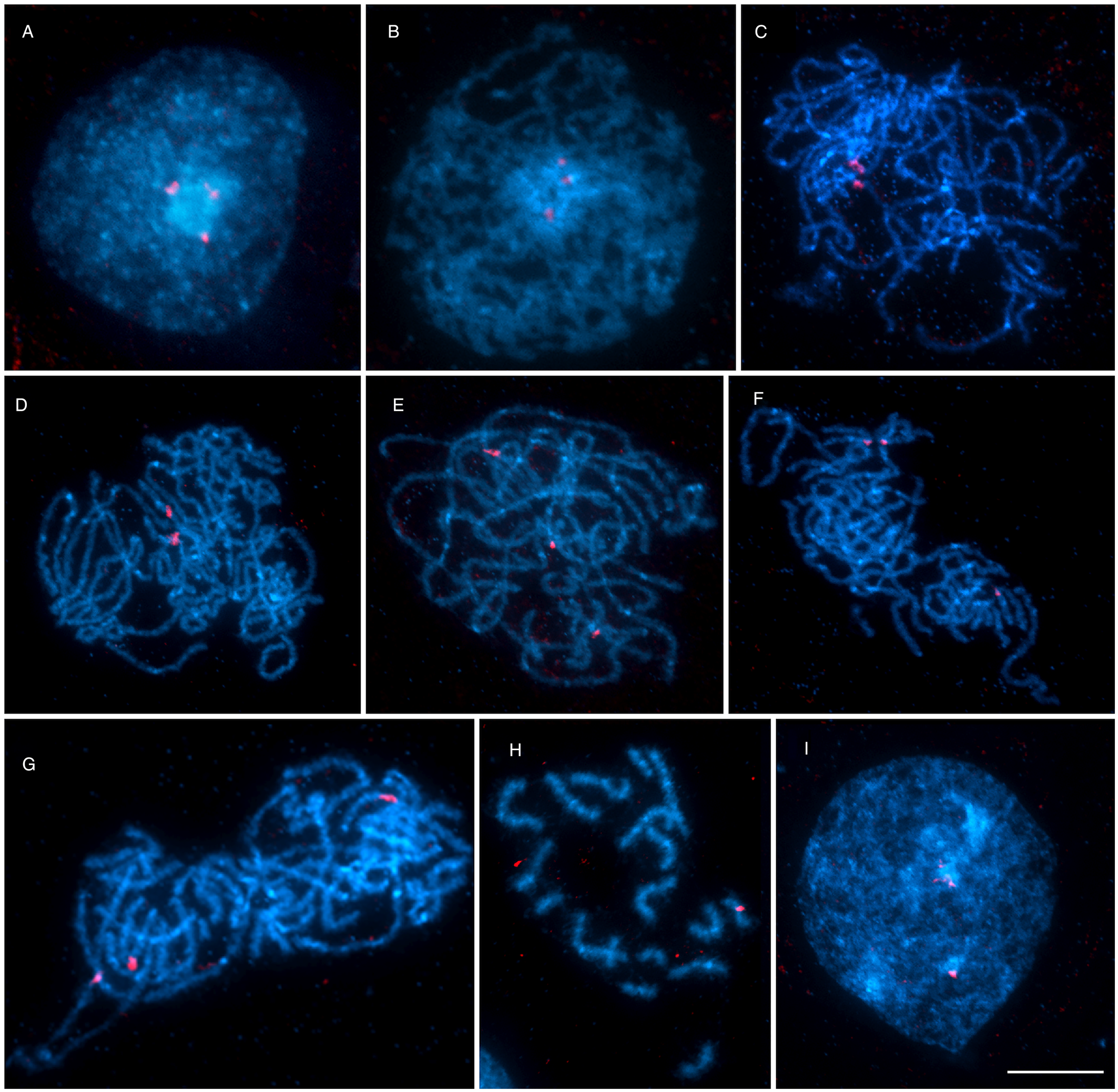
Fig. 6. The course of meiotic division in Dibothriocephalus latus after FISH with 18S rDNA probe (red) counterstained with DAPI (blue). (A) Interphase nucleus with a large nucleolus and 3 18S rDNA clusters. (B) Leptotene. (C–G) Pachytene nuclei showing irregular pairing of spermatocyte chromosomes carrying rDNA genes; (C, D) pachytene with NOR-bearing triplet No. 7 in form of 1 univalent and 1 bivalent or (E–G) 3 univalents. (H) Metaphase II. (I) Secondary spermatocyte, the early stage of spermiogenesis (scale bar = 10 μm). Abbreviations: FISH, fluorescence in situ hybridization; NOR, nucleolar organizer region
Discussion
Chromosome numbers, karyotype structure and interspecific chromosome variations
Currently, there is very limited knowledge about the karyotypes of diphyllobothridean species. Karyological data have so far been obtained for only 7 species out of 58 valid taxa of the family Diphyllobothriidae (Špakulová et al., Reference Špakulová, Orosová and Mackiewicz2011 ; Scholz et al., Reference Scholz, Kuchta and Brabec2019). These data showed that the basic chromosome number is x = 9 with a predominance of diploid species (2n = 2x = 18), except for 2 triploids with 3n = 3x = 27 (Sasada, Reference Sasada1978; Liu and He , Reference Liu and He1988; Okino et al., Reference Okino, Ushirogawa, Matoba, Nishimatsu and Saito2017). Published karyotypes are composed exclusively of metacentric and submetacentric chromosomes; the obvious exception is S. solidus, which is diploid and has 5 metacentric and 4 acrocentric chromosome pairs (Petkevičiūtė, Reference Petkevičiūtė1996).
As found in this study, D. latus from the isolated Alpine Lake Iseo shares the basic set of chromosomes (x = 9) with other diphyllobothridean species, as well as the overall gross morphology of chromosomes (i.e. all metacentric). Based on our detailed analysis of karyotype architecture and morphometric data, we can conclude that D. latus tapeworms from the isolated Alpine Lake Iseo population are triploids (3n = 3x = 27 m). In addition to the triploid number of 27, our study revealed a surprisingly high level of aneuploidy with 5 different chromosome numbers, namely 18, 20, 22, 24 and 26.
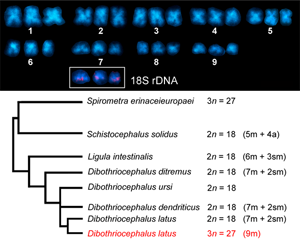
A former cytogenetic study on 3 congeneric diphyllobothridean species from Finland, including our model species D. latus (syn. Diphylobothrium latum), D. ditremus (syn. Diphylobothrium osmeri) and D. dendriticus (syn. Diphylobothrium dendriticum), revealed similar variations in chromosome numbers from 8 to 28, although to a lesser extent compared to this work (Wikgren and Gustafsson, Reference Wikgren and Gustafsson1965). All 3 analyzed species then belonged to the genus Diphyllobothrium at that time and were characterized as diploids. The authors attributed these variations to either preparation artifacts or the occurrence of somatic aneuploidy. Regarding chromosome morphology, they did not find any differences between the congeners. Haploid complements included 7 metacentric and 2 submetacentric chromosomes, which is slightly different from our Italian triploid population of D. latus with 9 metacentric chromosomes. However, it should be noted that chromosome morphometries in those works have been documented on squashed or section slides without indicating centromeric indexes and should therefore be treated with caution. Petkevičiūtė (Reference Petkevičiūtė1992) reported the same chromosome number and ploidy level (2n = 18) and only minor morphological differences between chromosomes for 2 species of this family, D. ditremus (syn. Diphylobothrium ditremum) from a geographically distant population in Chukotka, Russia and Ligula intestinalis from a population in Lithuania. Apart from ploidy in the population of D. latus from Italy, a comparison of the chromosome sets of D. ditremus, D. dendriticus, L. intestinalis and D. latus population from Finland indicates only small karyotype diversity, with no apparent structural and numerical variations. As the fundamental number of arms is also identical in these 4 species, small interspecific variations in chromosome morphology can be best explained by intrachromosomal changes, i.e. small deletions or duplications. These cytogenetic similarities also reflect well their close phylogenetic relationships (see Fig. 3 in Waeschenbach et al., Reference Waeschenbach, Brabec, Scholz, Littlewood and Kuchta2017 and Fig. 7 in this study). In the current phylogenetic tree, S. solidus was placed in ‘earlier diverging position together with Spirometra spp.’ (p. 837 in Waeschenbach et al., Reference Waeschenbach, Brabec, Scholz, Littlewood and Kuchta2017). This is in line with known karyological data, as S. solidus has a less symmetrical complement, with nearly half of the chromosome pairs being acrocentric (Petkevičiūtė, Reference Petkevičiūtė1996), and therefore can be considered as evolutionary older species in this group. Its karyotype may represent an ancestral condition in Diphyllobothriidea compared to other more derived chromosome forms and taking into account that pericentric inversion contributed to the karyotype divergence of some species (Petkevičiūtė, Reference Petkevičiūtė1996). However, traditional karyological methods alone are not sufficient to elucidate the evolutionary history of these tapeworms, which points to the need of molecular cytogenetic markers. Moreover, mapping of cytogenetic markers can also determine a level of ploidy by identification of homologous chromosome.

Fig. 7. Summary of available data on the diploid/triploid chromosome number (2n/3n) and chromosome morphology in karyologically studied diphyllobothridean species along with their latest phylogenetic relationships based on Waeschenbach et al. (Reference Waeschenbach, Brabec, Scholz, Littlewood and Kuchta2017).
Chromosomal location of the 18s ribosomal RNA and telomere constitution
The Italian D. latus population is the first investigated for the chromosomal location of ribosomal RNA genes in the order Diphyllobothriidea. Using FISH with 18S rDNA probe, we identified a single major rDNA site on all 3 chromosomes of triplet No. 7, clearly demonstrating the triploid nature of the studied population. Although data on rDNA in related diphyllobothrideans are not available, 18S rDNA in D. latus mapped to a pericentromeric position on small metacentric chromosomes, as in all other previously analyzed tapeworm species (Orosová et al., Reference Orosová, Provazníková, Xi and Oros2019). Our results are thus in line with the hypothesis of a generally constant position of rDNA loci in different families of Cestoda (Orosová et al., Reference Orosová, Provazníková, Xi and Oros2019).
The detection of vertebrate telomere repeat motif (TTAGGG)n at all chromosome end in D. latus population is consistent with results obtained in mitotic and meiotic chromosomes of other tapeworm species. Exclusively telomeric signals were reported in 4 tapeworm species, namely Caryophyllaeus laticeps, Caryophyllaeides fennica and Khawia abbottinae (Caryophyllidea), and Nippotaenia mogurndae (Nippotaeniidea) (Bombarová et al., Reference Bombarová, Vítková, Špakulová and Koubková2009; Orosová et al., Reference Orosová, Provazníková, Xi and Oros2019). Only the karyotype of Atractolytocestus huronensis (Caryophyllidea) exhibited also hybridization signals for interstitial telomeric sequences (ITSs), in addition to the telomeric signals, but always only in 1 chromosome of 3 of 5 triplets (Špakulová et al., Reference Špakulová, Bombarová, Miklisová, Nechybová and Langrová2019). Our findings together with results obtained so far indicate that this hexanucleotide repetitive sequence is a conserved feature of cestode telomeres. The absence of ITS in D. latus karyotype suggests that this species has undergone no structural arrangements of chromosomes during its evolution.
The course of meiosis
The course of the meiotic division of spermatogonial cells of D. latus was aberrant. We did not observe all stages of spermatogenesis, which could be to some extent associated with the colchicine treatment, as it blocks the segregation of chromosomes or chromatids so that cell division is interrupted in metaphase. However, the main cause of abnormal meiosis is undoubtedly triploidy. During the first meiotic prophase, in pachytene nuclei, 2 different arrangements of 3 homologous chromosomes No. 7 were easily identified in FISH preparations by 18S rDNA marker. We observed either a bivalent plus univalent or 3 unpaired univalents, but never a trivalent configuration that is probably unstable (see Marec, Reference Marec1996). Similar pairing behavior can also be expected in the other chromosome triplets. Interestingly, we did not observe any diplotene/diakinesis nuclei, although in the case of bivalent configurations, crossing-over could occur to some extent (Shaffer et al., Reference Shaffer, Brearley, Littlewood and Fink1971). We assume that chromosomes segregated randomly during anaphase I and also segregation of chromatids during anaphase II was irregular, resulting in aneuploid gametes with variable chromosomes numbers. Similar random segregation of multivalent chromosome associations and the production of aneuploid gametes were described in many taxa (Comai, Reference Comai2005; Chinone et al., Reference Chinone, Nodono and Matsumoto2014). Regarding the meiotic division of triploid tapeworm species, there are very few comparable data in the literature. Abnormal spermatogenesis and production of aneuploid gametes by random chromosome segregation were also observed in 2 triploid caryophyllidean species, A. huronensis and Glaridacris catostomi (Jones and Mackiewicz, Reference Jones and Mackiewicz1969; Grey and Mackiewicz, Reference Grey and Mackiewicz1980; Bruňanská et al., Reference Bruňanská, Drobníková and Oros2009; Špakulová et al., Reference Špakulová, Bombarová, Miklisová, Nechybová and Langrová2019). As with D. latus, no mature functional sperm was detected.
Origin of polyploidy
Polyploidy is a well-tolerated widespread phenomenon in the eukaryotic world and plays an important role in the evolution and development of species (Storchova and Pellman, Reference Storchova and Pellman2004). The most interesting result of the present study is the revealed difference in the ploidy (2n vs 3n) of 2 geographically distant populations of the same species D. latus. Such interpopulation differences are not uncommon and have been observed in many plants, and, to a lesser extent, occurs in animals (Park et al., Reference Park, Yong, Im and Chung2000; Giuliano-Caetano et al., Reference Giuliano-Caetano, Jorge, Moreira-Filho and Bertollo2001; Gassner et al., Reference Gassner, Dejaco, Schönswetter, Marec, Arthofer, Schlick-Steiner and Steiner2014; Winterfeld et al., Reference Winterfeld, Ley, Hoffmann, Paule and Röser2019) and flatworms and oligochaetes as well (Fujino and Ishii, Reference Fujino and Ishii1982; Rhee et al., Reference Rhee, Eun and Lee1987; Agatsuma et al., Reference Agatsuma, Terasaki, Yang and Blair1994; Shen et al., Reference Shen, Tsai, Fang and Chen2011; Zadesenets et al., Reference Zadesenets, Vizoso, Schlatter, Konopatskaia, Berezikov, Schärer and Rubtsov2016). In fact, diploid/triploid populations are known even among the basal cestode orders (Grey, Reference Grey1979; Grey and Mackiewicz, Reference Grey and Mackiewicz1980; Petkevičiūtė and Kuperman, Reference Petkevičiūtė and Kuperman1992; Špakulová et al., Reference Špakulová, Orosová and Mackiewicz2011). Triploidy is frequently associated with parthenogenesis (Otto and Whitton, Reference Otto and Whitton2000), a phenomenon also known in parasitic flatworms. For instance, sexual worms tend to be diploid, whereas parthenogenetic worms are triploid and tetraploid (Benazzi, Reference Benazzi and Barigozzi1982). Among tapeworms, examples of such close association are A. huronensis (3n = 24) (Jones and Mackiewicz, Reference Jones and Mackiewicz1969) and G. catostomi (3n = 30) (Grey and Mackiewicz, Reference Grey and Mackiewicz1980). The difference between them is that A. huronensis is an exclusively triploid parthenogenetic species without an obvious diploid ancestor while only 1 geographically isolated triploid population was found in the generally diploid G. catostomi. The latter type has been found in several other parasitic platyhelminths, including representatives of Trematoda (e.g. Paragonimus westermani, Blair et al., Reference Blair, Nawa, Mitreva and Doanh2016).
Apparently, polyploidization events could arise by chance several times across a wide range of taxa. There are 2 different ways for parthenogens to amplify their genomes; first by autopolyploidy (additional haploid chromosome complement comes from the same species) or second by allopolyploidy (due to interspecific hybridization) (Otto, Reference Otto2007). In the case of A. huronensis, Špakulová et al. (Reference Špakulová, Bombarová, Miklisová, Nechybová and Langrová2019) hypothesized that ancestral interspecific hybridization with the diploid congener Atractolytocestus tenuicollis could be the cause of its triploidy. The origin of the polyploid nature of the G. catostomi population is a matter of speculation. Geographic parthenogenesis is a common phenomenon denoting ‘large variety of patterns where sexual and related asexual forms differ in their geographic distribution’ (p. 1 in Tilquin and Kokko, Reference Tilquin and Kokko2016). Grey and Mackiewicz (Reference Grey and Mackiewicz1980) mentioned a possible link between parthenogenesis and triploidy in G. catostomi population isolated due to the separation of their fish host (suckers) in the dam, while the other 5 populations were diploid.
Hermaphroditic tapeworms can reproduce by cross-fertilization as well as by selfing (Christen and Milinski, Reference Christen and Milinski2003). Dibothriocephalus latus is usually found as a single plerocercoid in the musculature of the second intermediate fish host or as a single worm in the intestine of the final host (Gustinelli et al., Reference Gustinelli, Menconi, Prearo, Caffara, Righetti, Scanzio, Raglio and Fioravanti2016; Radačovská et al., Reference Radačovská, Bazsalovicsová, Blasco Costa, Orosová, Gustinelli and Králová-Hromadová2019). This model has also been confirmed for the D. latus from the Lake Iseo, considering that a single plerocercoid from 1 fish develops into 1 adult worm in the definitive host and thus causes a low level of genetic polymorphism in this D. latus population (Bazsalovicsová et al., Reference Bazsalovicsová, Koleničová, Králová-Hromadová, Minárik, Šoltys, Kuchta and Štefka2018). This suggests self-fertilization as a likely way of reproduction of this tapeworm. However, sexual reproduction involves the fusion of an egg and sperm. Since functional mature sperm were not observed in our work and some steps of meiosis were apparently skipped in this species, parthenogenetic reproduction is evident. Parthenogens do not need to find a partner to mate and reproduce (Schiffer et al., Reference Schiffer, Danchin, Burnell, Creevey, Wong, Dix, O'Mahony, Culleton, Rancurel, Stier, Martínez-Salazar, Marconi, Trivedi, Kroiher, Thorne, Schierenberg, Wiehe and Blaxter2019). Reproduction without male sperm suppresses the ability to gain genetic variability in a species, as the process of meiotic recombination is responsible for maintaining genetic heterozygosity. It means that parthenogenetic reproduction in D. latus also leads to the loss of genetic variability and to an increase of homozygosity; microsatellite analysis of the same Italian D. latus population revealed a high level of monomorphic (72 out of 78) loci but not complete loss of heterozygosity (Bazsalovicsová et al., Reference Bazsalovicsová, Koleničová, Králová-Hromadová, Minárik, Šoltys, Kuchta and Štefka2018). It is very likely that crossing-over, in univalent/bivalent arrangements (discussed above), where 2 chromosomes pair, occurs between sister chromatids and a small amount of variations are maintained in the genome (Lutes et al., Reference Lutes, Neaves, Baumann, Wiegraabe and Baumann2010).
The karyotyped triploid D. latus population has 3n = 27 chromosomes, i.e. 3 haploid sets of chromosomes. No significant morphometric differences in chromosomes within the individual triplets were seen. Therefore, it is probable that the isolated D. latus population from Iseo Lake became parthenogenetic through autopolyploidy rather than by interspecific hybridization and that all 3 complements come from the same ancestral species. Parthenogenesis, or more specifically parthenogenetic races, is often associated with the occupancy of habitats that are at higher altitudes or latitudes and tend to be characterized by anthropogenic pollution compared with their sexual relatives (Gassner et al., Reference Gassner, Dejaco, Schönswetter, Marec, Arthofer, Schlick-Steiner and Steiner2014). Northern Italy is known for its heavily industrialized cities that surround several natural lakes, including Lake Iseo. Moreover, this lake is also polluted by trash and untreated sewage system; it means that the lake receives urban and industrial effluent (Gustinelli et al., Reference Gustinelli, Menconi, Prearo, Caffara, Righetti, Scanzio, Raglio and Fioravanti2016).
In summary, we assume that most likely all the facts discussed (intensity of infection – missing sexual partner, geographical isolation and to some extent also polluted environment) gave rise to the Italian triploid parthenogenetic population of D. latus and hypothetically, this phenomenon is not historically distant. Nevertheless, the question of the origin of triploidy and parthenogenesis remains open until further in-depth analysis of this unusual karyotype is performed and further studies on samples from different populations are available.
Author contribution
M.O. and F.M. conceived the study; M.O. and A.R. performed the field-collection, M.O. and Mi.O. examined the fishes, fixed the parasites and infected the experimental animals. Z.Č. was responsible for laboratory animals and their welfare; M.O., Mi.O., A.M. and I.P. performed experiments and data analysis. M.O. and F.M. prepared a draft of the manuscript. All authors discussed the results and commented on the manuscript. All authors approved the manuscript.
Financial support
This work was supported by Slovak Research and Development Agency (no. APVV-15-0004) and by Bilateral Mobility Project SAV-18-20.
Conflict of interest
The authors declare there are no conflicts of interest.
Ethical standards
The research related to animals has been complied with all the relevant national regulations and institutional policies for the care and use of animals.