INTRODUCTION
Ticks are obligate haematophagous mites parasitizing vertebrates. They act as vectors of a number of microbial agents that cause diseases in humans and animals. These pathogens include Borrelia spp., Coxiella burnetii, Francisella tularensis, as well as intracellular bacteria from the order Rickettsiales. Other tick-associated bacteria do not appear to be present in the salivary glands or to cause infections in humans and animals; some of these bacteria are transmitted to the progeny through the egg cytoplasm (transovarial transmission). These vertically transmitted microrganisms are usually referred to as symbionts, but their interaction with the host tick is generally not fully understood (for a general introduction to ticks, see Bowman and Nuttall, Reference Bowman and Nuttall2004).
The hard tick Ixodes ricinus (family Ixodidae) harbours an endosymbiont with the peculiar capacity to invade the mitochondria and multiply therein (first observation by Lewis, Reference Lewis1979). These mitochondrial bacteria found in I. ricinus have been identified based on 16S rRNA gene sequence data (Beninati et al. Reference Beninati, Lo, Sacchi, Bandi, Noda and Genchi2004), investigated at the ultrastructural level (Sacchi et al. Reference Sacchi, Bigliardi, Corona, Beninati, Lo and Franceschi2004), and described as a new genus and species, i.e. Midichloria mitochondrii (Sassera et al. Reference Sassera, Beninati, Bandi, Bouman, Sacchi, Fabbi and Lo2006). The biological role of M. mitochondrii in I. ricinus is still unknown. During oogenesis, the proportion of mitochondria invaded by M. mitochondrii is not negligible (e.g. see photos in Sacchi et al. Reference Sacchi, Bigliardi, Corona, Beninati, Lo and Franceschi2004), and this suggests that the bacterium could have a detrimental effect on the host tick. However, the fact that M. mitochondrii is apparently at fixation in females of I. ricinus (100% prevalence according to Lo et al. Reference Lo, Beninati, Sassera, Bouman, Santagati, Gern, Sambri, Masuzawa, Gray, Jaenson, Bouattour, Bitam, Kenny, Guner, Kharitonenkov and Bandi2006a) seems to indicate that its presence is somehow compatible with the survival and reproduction of the host tick (see discussion in Lo et al. Reference Lo, Beninati, Sassera, Bouman, Santagati, Gern, Sambri, Masuzawa, Gray, Jaenson, Bouattour, Bitam, Kenny, Guner, Kharitonenkov and Bandi2006a and Reference Lo, Beninati, Sacchi, Bandi, Bourtzis and Millerb).
While data have been acquired for M. mitochondrii in I. ricinus, which have permitted its description as a new genus and species, there is only circumstantial evidence for the presence of DNA sequences from related bacteria in other tick species (see Discussion section). The aim of this work was thus to obtain a more detailed picture of the distribution of bacteria related to Midichloria in hard ticks (family Ixodidae). The discovery of other tick species infected by bacteria related to M. mitochondrii could enhance investigations aimed at uncovering its biological role. For instance, a tick species could be found in which the prevalence of Midichloria-related symbionts is far from 100%, or in which different populations show different infection levels. This might permit both a comparison of the biology, fitness parameters and vector competence of infected versus uninfected ticks, and experiments on possible reproductive parasitism phenomena (Bandi et al. Reference Bandi, Dunn, Hurst and Rigaud2001). Furthermore, I. ricinus is the sole animal thus far shown to harbour an intra-mitochondrial bacterium and M. mitochondrii is the unique intra-mitochondrial bacterium thus far described. Determining whether intra-mitochondrial bacterial presence is an exception or a more widespread phenomenon was the main goal of this work.
The screening for Midichloria-related bacteria reported here focused on hard ticks. Most of the ~900 species of ticks thus far described (suborder Ixodida in the order Acarina) are assigned to 2 main families: the Ixodidae (hard ticks) and the Argasidae (soft ticks). The Ixodidae are subdivided into 2 main lineages, the Prostriata and Metastriata (Hoogstraal and Aeschlimann, Reference Hoogstraal and Aeschlimann1982). The Prostriata encompass the subfamily Ixodinae (over 200 species in the single genus Ixodes). The Metastriata group comprises 370 species in 4 subfamilies: the Amblyomminae, Haemaphysalinae, Hyalomminae and Rhipicephalinae (Barker and Murrel, Reference Barker and Murrel2004). Our screening included representatives of all of the subfamilies of the Ixodidae and led to the detection of bacteria in tick specimens belonging to 7 different species (in addition to the previously investigated I. ricinus). 16S rRNA sequence data generated for these bacteria revealed a very close phylogenetic relationship with M. mitochondrii. Nucleotide identities between the 16S rRNA from M. mitochondrii of I. ricinus and the sequences from the other tick species were above 99%. We have thus referred to all of these tick bacteria using the species name M. mitochondrii (Sassera et al. Reference Sassera, Beninati, Bandi, Bouman, Sacchi, Fabbi and Lo2006).
MATERIALS AND METHODS
Tick species screened for bacteria related to Midichloria mitochondrii
Tick samples representing a total of 21 species of the Ixodidae were examined for the presence of bacteria related to M. mitochondrii. The Metastriata were represented by the genera Rhipicephalus (4 species), Dermacentor (4 species), Hyalomma (2 species), Amblyomma (3 species) and Haemaphysalis (3 species). The Prostriata were represented by 5 species of the genus Ixodes.
A total of 197 tick specimens were collected in Italy (Lazio, Sicily and Lombardia), Iceland (Grimsey) and USA (New Jersey, Florida, Missouri, South Carolina, Oregon, California and Georgia). Of these specimens, 125 were ethanol-preserved before DNA extraction, while the other 72 were processed immediately after collection. Ticks were collected at different developmental stages (larvae, nymphs, adults, and engorged females). Forty-two of the samples were pooled prior to DNA extraction. For the remaining 155 samples DNA was extracted from individual specimens. See Table 1 for a complete list of the specimens. All ticks were identified using standard taxonomic keys; in addition, for confirmation of tick identification, a fragment of the small subunit mitochondrial ribosomal RNA gene (12S rRNA) was sequenced for a subset of the examined specimens/pools of specimens.
Table 1. List of taxa, geographical origin, Accession numbers for bacterial 16S rRNA and tick 12S rRNA sequences, and screening results

1 DNA extracted from living specimens.
2 DNA extracted from samples preserved in ethanol.
3 Nymphs or larvae.
MSNPV, Museo di Storia Naturale, Università di Pavia; RML, U.S. National Tick Collection.
N.A., Sample Accession no. not available, or sequencing not performed because the samples were negative for midichlorians.
DNA extraction and polymerase chain reaction (PCR) amplification of tick 12S rRNA
DNA was extracted using Qiagen tissue kit (Qiagen, Hilden, Germany). Ethanol-preserved ticks were washed and dried for 45 min before DNA extraction. Afterwards, ticks were triturated into a solution containing 180 μl of ATL lysis buffer (Qiagen) and 200 ng/μl proteinase K (Sigma Aldrich, St Louis, USA). The lysis was carried out overnight with shaking at 56°C. The elution step was performed by 2 additions of 25 μl of hot (72°C) deionized water in the columns; the remaining extraction steps were performed according to the manufacturer's instructions. DNA was stored at −20°C.
PCR for amplification of 12S rRNA was performed as described by Beati and Keirans (Reference Beati and Keirans2001). PCR products were gel-purified using the QIAquick™ Gel Extraction Kit (Qiagen) according to the manufacturer's protocols, resuspended in 30 μl of deionized water and sequenced with PCR primers using the ABI PRISM BigDye Terminator Cycle Sequencing Reaction Kit, version 3.1 (Applera Europe, Warrington, UK), and run on an automated sequencer (ABI Prism 310 DNA sequencer, Applied Biosystems, Foster City, CA).
PCR screening for bacteria related to Midichloria mitochondrii
PCR screening for bacteria related to M. mitochondrii was performed using 2 sets of primers: Midi-F (5′-GTACATGGGAATCTACCTTGC-3′) and Midi-R (5′-CAGGTCGCCCTATTGCTTCTTT-3′); Midi-F2 (5′-CAACGAGCGCAACCCT TAT-3′) and MidiR2 (5′-CAGTCGTCAACCTT ACCGT-3′). All samples were tested with both primer copies, in order to double check the results obtained. These primers, targeted the 16S rRNA gene, amplify fragments of ~1100 (Midi-F –R) and ~350 bp (Midi-F2 –R2) and were designed to be conserved between the sequence of M. mitochondrii from I. ricinus (AJ566640) and the closely related sequences available in the data bases. In particular, an alignment was generated with the aim of representing all of the main lineages of the order Rickettsiales and the sequences already available that cluster as a monophyletic group with M. mitochondrii (see phylogenetic trees in Beninati et al. Reference Beninati, Lo, Sacchi, Bandi, Noda and Genchi2004; Lo et al. Reference Lo, Beninati, Sassera, Bouman, Santagati, Gern, Sambri, Masuzawa, Gray, Jaenson, Bouattour, Bitam, Kenny, Guner, Kharitonenkov and Bandi2006a; Sassera et al. Reference Sassera, Beninati, Bandi, Bouman, Sacchi, Fabbi and Lo2006). Primers were then designed to be conserved and specific for bacteria in the M. mitochondrii cluster. Primer specificity was then checked by BLAST searches and by PCR on biological samples positive for bacteria of the genera Rickettsia, Ehrlichia, Wolbachia and Anaplasma.
Amplifications were performed, with both primer sets, in 20 μl of buffer (10 mm Tris-HCl (pH 8·3), 50 mm KCl, 1·5 mm MgCl2, 0·001% gelatine) with 0·2 mm each deoxynucleoside triphosphate, 10 pmol of each primer, 0·5 U of Taq Polymerase (Eppendorf) and 2 μl of DNA sample. The thermal profile was: 2 min at 95°C; 40 cycles of 95°C for 30 sec, 56°C for 30 sec and 68°C for 45 sec; the elongation was completed at 10 min at 68°C. PCR products obtained with primer combination MidiF and MidiR were purified and sequenced as previously described using primers Midi-F and Midi-R and the internal primers P3b (5′-CTGTTTGCTCCCCACGCTTTC-3′) and P2b (5′-GATATTAGGAGGAACACCGC-3′).
Phylogenetic analyses
The bacterial 16S rRNA sequences obtained from ticks were subjected to BLAST analysis. These sequences were then aligned using as a mask an alignment downloaded from the Ribosomal Database Project (Cole et al. Reference Cole, Chai, Marsh, Farris, Wang, Kulam, Chandra, McGarrell, Schmidt, Garrity and Tiedje2003). The alignment was generated considering 16S rRNA secondary structures and included the sequence of M. mitochondrii from I. ricinus and the homologous Midichloria-related sequences available in the data bases (excluding sequences below 1000 bp). Representatives of the main lineages of the order Rickettsiales were also included. Phylogenetic analyses were performed using the programmes Mega 3.1 (Kumar et al. Reference Kumar, Tamura and Nei2004) and MrBayes (Huelsenbeck and Ronquist, Reference Huelsenbeck and Ronquist2001). Trees were generated using Neighbour Joining, Maximum Parsimony and Bayesian Inference methods. For Bayesian Inference, parameters for the model of substitution (General Time Reversible with Gamma correction for among-site heterogeneity) were estimated from the data. A total of 10 000 trees were obtained (ngen=1 000 000, samplefreq=100), and the first 3000 of these were considered as the ‘burn in’ and discarded. A 50% majority-rule consensus tree of the remaining trees, including branch lengths (sumt) was produced. For macro-taxonomic purposes, analyses were performed on alignments including representatives of the Rickettsiales (see tree in Fig. 1), while for investigating the relationships at a lower taxonomic level, sequences from Rickettsiales which are not closely related with M. mitochondrii were excluded (see tree in Fig. 2). The sequences obtained have been deposited in the EMBL data library (see Accession numbers in Table 1).
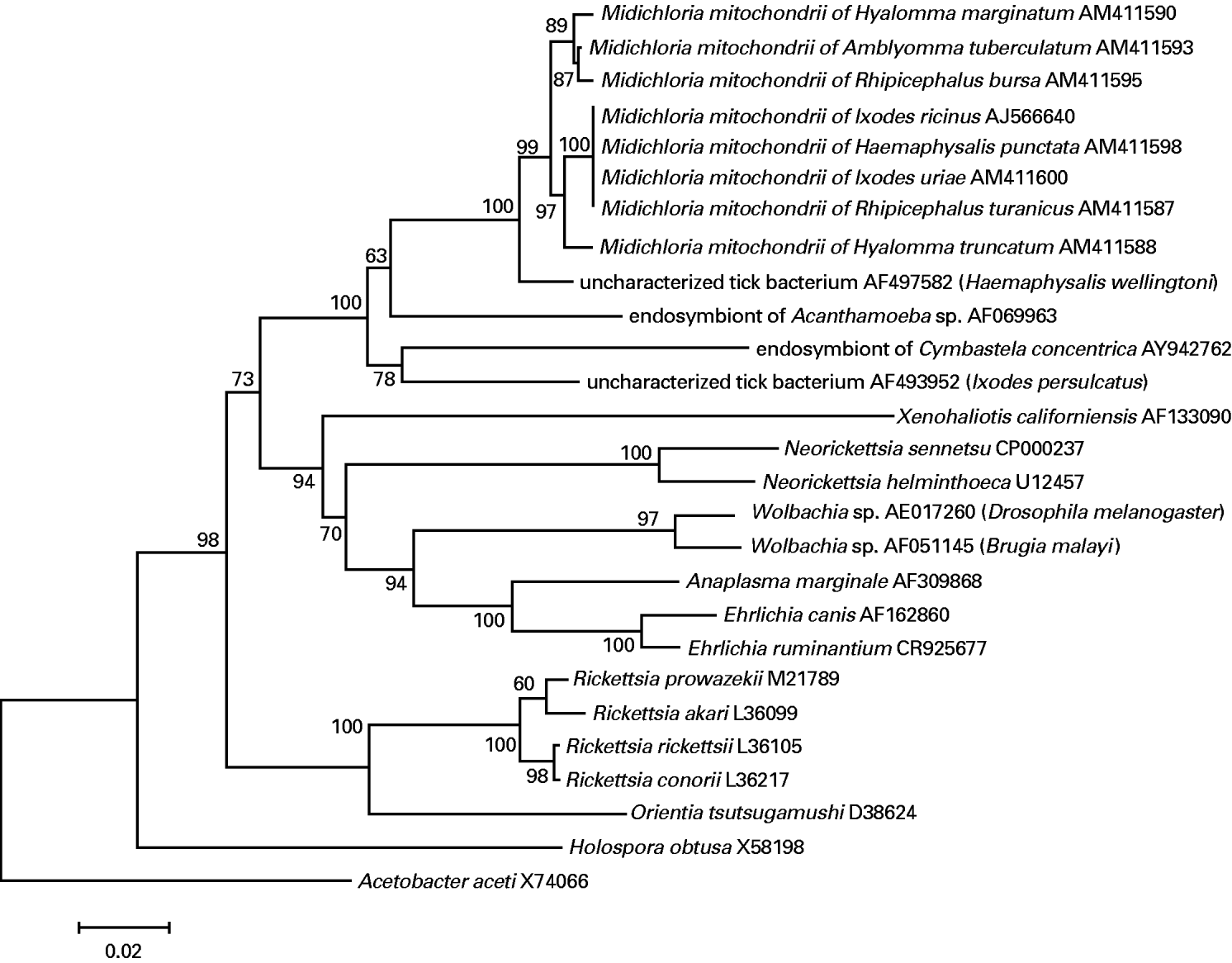
Fig. 1. Example of tree based on 16S rRNA gene sequences showing the position of Midichloria mitochondrii from different ticks relative to representatives of the order Rickettsiales. The tree was generated using MEGA (Neighbour Joining; Kimura correction). The Accession number for each sequence is indicated. Numbers adjacent to each node represent the bootstrap percentages (5000 repetitions). Acetobacter aceti (Rhodospirillales) was included as an outgroup. Additional analyses in which other Alphaproteobacteria were included as outgroups, or using other tree-building approaches, generated trees showing the same overall topology. Bar: 0·02 inferred substitutions per site.

Fig. 2. Comparison of the phylogenies of Midichloria mitochondrii and ixodid ticks. Nucleotide sequence Accession numbers for ixodid ticks and for the different ‘strains’ of M. mitochondrii are reported in Table 1. Accession numbers for outgroup sequences are indicated. (A) Example of tree based on 16S rRNA gene sequences showing the inferred phylogeny of M. mitochondrii. Names at the terminal nodes are those of the host ticks (for full-length genus names, see (B). The tree was generated using MEGA (Neighbour Joining; Kimura correction). Numbers adjacent to each node represent the bootstrap values (5000 repetitions). An endosymbiont of Acanthamoeba sp. was used as outgroup. Additional analyses in which other Alphaproteobacteria were included as outgroups, or using other tree-building approaches, generated trees showing the same overall topology. Bar: 0·001 inferred substitutions per site. (B) Example of phylogenetic tree based on partial 12S rRNA gene sequences showing the relationships of the examined ticks. The + symbol indicates tick species that were found to be positive for M. mitochondrii. The tree was generated using MEGA (Neighbour Joining; Kimura correction). Numbers adjacent to each node represent the bootstrap percentages (5000 repetitions). Nodes with bootstrap values below 50% are shown as unresolved. Analyses using other tree-building approaches did not lead to a better resolution of these nodes. The argasid tick Argas persicus was included as outgroup. Bar: 0·02 inferred substitutions per site.
The 12S mitochondrial rRNA sequences obtained were subjected to BLAST analysis (http://www.ncbi.nlm.nih.gov/blast) and compared to the sequences available in the data bases in order to confirm morphology-based species identification. The sequences were aligned with ClustalX (Thompson et al. Reference Thompson, Gibson, Jeanmougin and Higgins1997) and manually adjusted taking into account secondary structures (Hickson et al. Reference Hickson, Simon, Cooper, Spicer, Sullivan and Penny1996). Phylogenetic analyses were then performed as described for 16S rRNA sequences. These sequences have been deposited in the EMBL data library (see Accession numbers in Table 1).
Electron microscopy
Specimens of the tick species Rhipicephalus bursa were found positive for M. mitochondrii during the PCR screening; this tick species was thus chosen for transmission electron microscopy (TEM) examination. Eleven adult engorged females were collected from the field (Lazio, Italy), brought to the lab within 48 h and immediately dissected. Half of the ovary from each female specimen was fixed for TEM examination, while the remaining half was used for DNA extraction and PCR. The 4 ovarian specimens that were found to be PCR positive for M. mitochondrii as well as a negative ovarian specimen were then subjected to TEM examination. For TEM examination, ovaries were fixed in 0·1 m cacodylate buffer (pH 7·2) containing 2·5% glutaraldehyde for 3 h at 4°C. The samples were then washed in the same buffer and post-fixed in 1% OsO4 in the same buffer for 1·5 h at 4°C. Successively all samples were dehydrated in ethanol and embedded in Epon 812. The semithin sections (1 μm) for light microscopy were stained with 0·5% toluidine blue; thin sections (80 nm) were stained with uranyl acetate and lead citrate and examined under an EM900 transmission electron microscope (Zeiss).
RESULTS
All of the ticks examined have been identified using morphological keys. In addition, all specimens were examined by PCR amplification of mitochondrial 12S rRNA to check for the quality of the DNA. A fragment of about 350 bp of the 12S rRNA was sequenced for a subset of samples, in order to confirm morphological identification. The fragment was also sequenced for all samples positive for M. mitochondrii. In total, forty 12S rRNA sequences were generated. BLAST search of 12S rRNA sequences confirmed morphological identifications in all cases. The 12S rRNA sequences obtained have been deposited in the EMBL data library (see Accession numbers in Table 1).
The 2 PCR protocols designed to be specific for the 16S rRNA of M. mitochondrii and related bacteria gave amplifications of the expected size from tick specimens belonging to 8 species out of the 21 species included in this study. In total, 15 tick specimens were found positive out of the 155 individual ticks examined. In addition, out of 42 pooled tick samples, 7 were found to be positive for M. mitochondrii. Table 1 summarizes the results obtained.
The ~1100 bp PCR-amplified 16S rRNA gene fragments from positive specimens were then sequenced, and the sequences were, in all cases, unambiguous. The sequences obtained were run against the databases and gave the highest similarity scores with the 16S rRNA sequences of M. mitochondrii from I. ricinus and with those sequences which have already been shown to form a monophyletic group with this bacterium (see Fig. 1 and Sassera et al. Reference Sassera, Beninati, Bandi, Bouman, Sacchi, Fabbi and Lo2006).
Phylogenetic analyses were performed on 3 different data sets. The first included 16S rRNA sequences from bacteria of the main branches of the Rickettsiales and from M. mitochondrii and related bacteria (Fig. 1). The second alignment involved bacterial 16S rRNA from M. mitochondrii of I. ricinus and all closely related tick endosymbionts (Fig. 2A). The last alignment focused on mitochondrial 12S rRNA from ixodid ticks (Fig. 2B). The results of the analyses performed for each data set were similar irrespective of the phylogenetic method employed.
Figure 1 shows an example of 16S rRNA-based tree (Neighbour Joining) including representatives of the genera and families of the Rickettsiales. All of the sequences generated in this study form a monophyletic branch, which also includes the sequence of M. mitochondrii from I. ricinus, and the bacterial sequences that were already available in the data bases from the ticks Ixodes persulcatus (Mediannikov et al. Reference Mediannikov, Ivanov, Nishikawa, Saito, Sidel'nikov, Zdanovskaia, Mokretsova, Tarasevich and Suzuki2004) and Haemaphysalis wellingtoni (Parola et al. Reference Parola, Cornet, Sanogo, Miller, Thien, Gonzalez, Raoult, Telford and Wongsrichanalai2003), from the marine sponge Cymbastela concentrica (Taylor et al. Reference Taylor, Shupp, de Nys, Kjelleberg and Steinberg2005) and from the amoeba Achantamoeba sp. (Fritsche et al. Reference Fritsche, Horn, Seyedirashti, Gautom, Schleifer and Wagner1999). Support for this branch was very high in all of the analyses performed (Neighbour Joining; Maximum Parsimony; Bayesian Inference), with bootstrap or posterior probability values of 97–100%. Within this branch, molecular diversity at the nucleotide level is limited: with the exclusion of the sequences from Acanthamoeba sp., C. concentrica and from I. persulcatus, pairwise nucleotide differences ranged from 3 to 22 nt out of 1000.
Figure 2A is a 16S rRNA-based tree (Neighbour Joining) including the M. mitochondrii sequences generated in this study and the sequence from the endosymbiont of Acanthamoeba sp. (included as an outgroup). Sequences from M. mitochondrii are assigned to 3 main branches, supported by high bootstrap values. Figure 2B shows a 12S rRNA-based tree (Neighbour Joining) including the tick species examined in this study. The tree is not fully resolved, but the main branches correspond to the traditional systematic arrangement of hard tick genera into subfamilies and of the subfamilies into the groups Prostriata and Metastriata (Hoogstraal and Aeschlimann, Reference Hoogstraal and Aeschlimann1982), with the exception of Ixodes uriae whose positioning relative to the two groups is not resolved. The uncertain positioning of I. uriae is not surprising if we consider the results of total evidence phylogenetics of hard ticks, placing this and other species of the genus Ixodes in different positions depending on the methods of analysis (Klompen et al. Reference Klompen, Black, Keirans and Norris2000). Indeed, phylogenetic analyses of our tick data set performed using other algorithms (Maximum Parsimony and Bayesian Inference; see Materials and Methods section) did not lead to a better resolution. A detailed comparison of the host and symbiont phylogenies was therefore outside the scope of this work, considering that (i) tick phylogeny is still not fully resolved and (ii) the level of divergence here reported in the 16S rRNA of M. mitochondrii is not sufficient for the reconstruction of a robust phylogeny. Examining the symbiont (Fig. 2A) and host trees (e.g. the tree in Fig. 2B or the trees in Fig. 4 in Klompen et al. Reference Klompen, Black, Keirans and Norris2000) it is clear that the positioning of M. mitochondrii endosymbionts is not always congruent with the taxonomic/phylogenetic placements of the hosts. For example, endosymbionts from ticks of the Ixodes genus (the Prostriata of traditional taxonomic arrangements) cluster with endosymbionts from ticks of the Metastriata group. In some cases, tick species harbouring closely related M. mitochondrii are highly divergent at the level of their 12S rRNA gene. For example, M. mitochondrii from distantly related ticks of the genus Ixodes (I. ricinus and I. uriae) are identical to each other in their 16S rRNA, and are also identical to M. mitochondrii of the Metastriata ticks R. turanicus and Ha. punctata. There are also single-tick species in which different individuals harbour M. mitochondrii with different 16S rRNA sequences (Hy. truncatum, Hy. marginatum and R. bursa).
Out of the 7 new tick species that were found positive for M. mitochondrii, we had the possibility to collect samples on 2 different occasions only for the species R. bursa. The number of specimens collected allowed us to obtain a first picture of the prevalence of M. mitochondrii in this tick species, with 7 positive females out of the 21 examined and 2 males out of 16. We emphasize that all positive specimens were from a single population, while the specimens from the other population examined were all negative. In general, for all of the species that were found positive for M. mitochondrii in this screening, not all of the specimens examined were positive (see Table 1), though only for R. bursa were we able to collect a number of specimens sufficient for concluding that prevalence is far from 100%.
Female specimens of R. bursa were prepared for both PCR and TEM examinations. TEM was performed on ovaries from 4 female specimens that were found positive for M. mitochondrii, and from 1 specimen that was found negative. TEM examinations of the ovaries from positive females revealed the presence of bacteria in the mitochondria of the oocytes (Fig. 3A). The few bacteria that were observed outside the mitochondria were free in the cytoplasm (i.e. without surrounding vacuoles). The size of these bacteria is ~1 μm in length and ~0·25 μm in diameter. The number of bacteria in the mitochondria ranged from a single bacterium to over 20 observed in a single section. Infected mitochondria in R. bursa do not present clear signs of reduction/degeneration of the matrix, even in cases where numerous bacteria infect the mitochondria (Fig. 3B).

Fig. 3. Electron micrographs of ovarian cells of Rhipicephalus bursa showing mitochondria harbouring bacteria (A). Mitochondrion containing several bacteria does not present clear signs of reduction or degeneration of the matrix (B).
DISCUSSION
The screening reported here shows that M. mitochondrii is commonly found in ticks. Eight species out of 21 were positive, and only in 1 of the 7 genera examined (Dermacentor) were positive individuals not detected. The prevalence of M. mitochondrii in the various tick species examined here differs markedly from that observed in I. ricinus, where 100% of females and 44% of males have been found positive (Lo et al. Reference Lo, Beninati, Sassera, Bouman, Santagati, Gern, Sambri, Masuzawa, Gray, Jaenson, Bouattour, Bitam, Kenny, Guner, Kharitonenkov and Bandi2006a). In general, the results here reported on the distribution of M. mitochondrii in ticks likely represent an underestimation. Firstly, for most of the species a limited number of specimens was examined, which implies that future screenings could lead to the detection of positive tick individuals from further tick species. Secondly, our screening included ethanol-preserved specimens. Partial degradation of the DNA cannot thus be excluded, even though control PCR for mitochondrial tick genes provided evidence for a good DNA quality.
Figure 2A shows a phylogenetic tree of M. mitochondrii; a tree representing a phylogeny of the ticks included in this study is also shown for comparison (Fig. 2B). The overall phylogeny of ticks is still unresolved (Barker and Murrel, Reference Barker and Murrel2004), and it was not the goal of this work to address this issue. However, an incomplete congruence in the phylogenies of ticks and their endosymbionts can be inferred, both comparing the M. mitochondrii tree with the host tree in Fig. 2B (or with trees in other studies; e.g. Black et al. Reference Black, Klompen and Keirans1997; Klompen et al. Reference Klompen, Black, Keirans and Norris2000), as well as considering that ticks belonging to different genera (Ixodes spp. and R. turanicus) harbour M. mitochondrii with identical 16S rRNA sequences. Considering the Midichloria-like 16S rRNA sequences already available in the data bases, we note that the endosymbiont of I. persulcatus belongs to the sister group of the main cluster that includes the sequences generated in this study plus those from Ha. wellingtoni and Acanthamoeba sp. In the tree in Fig. 1, the endosymbiont of I. persulcatus clusters with a bacterial sequence obtained from a sponge. The distance between this Ixodes bacterium and those found in other representatives of the genus Ixodes is relatively large.
Previous studies have provided microscopical and molecular evidence for vertical transmission of M. mitochondrii in I. ricinus (Sacchi et al. Reference Sacchi, Bigliardi, Corona, Beninati, Lo and Franceschi2004; Lo et al. Reference Lo, Beninati, Sassera, Bouman, Santagati, Gern, Sambri, Masuzawa, Gray, Jaenson, Bouattour, Bitam, Kenny, Guner, Kharitonenkov and Bandi2006a). The results reported here reveal that in some tick species different individuals harbour M. mitochondrii with identical gene sequences (i.e. in I. ricinus, A. tuberculatum, Ha. punctata). There is thus an overall consistency of information which indicates an effective role of vertical transmission in the diffusion of these bacteria. On the other hand, the results reported above also indicate a partial lack of congruence in the phylogenies of M. mitochondrii endosymbionts and their hosts, and suggest that horizontal transmission of the symbionts might have occurred. How horizontal transmission could take place is unknown. However, based on the presence of DNA from M. mitochondrii in the blood of roe deer (Skarphédinsson et al. Reference Skarphedinsson, Jensen and Kristiansen2005), and the possible detection of a related bacterium in samples from human patients (Mediannikov et al. Reference Mediannikov, Ivanov, Nishikawa, Saito, Sidel'nikov, Zdanovskaia, Mokretsova, Tarasevich and Suzuki2004), we might suggest that transmission to tick hosts could occur, and that blood could represent a potential source of infection for ticks. In any case, based on the above studies, the possibility that M. mitochondrii could infect humans and animals is worthy of further investigation.
In R. bursa, the species for which we could obtain the most samples, the prevalence of M. mitochondrii was 33% in females and 13% in males. The discovery of a tick species in which the prevalence of M. mitochondrii is far from fixation in both males and females will facilitate experiments aimed at uncovering the biological role of these endosymbionts. For example, fitness parameters could be compared in positive and negative individuals, as could the relative prevalence of tick-borne pathogens in positive and negative individuals. Experiments examining the possible role of M. mitochondrii as a reproductive parasite (e.g. cytoplasmic incompatibility, as in the case of Wolbachia; see Werren, Reference Werren1997) could also be envisioned.
TEM observations on the ovaries from R. bursa gave results comparable to those obtained in I. ricinus, in terms of the size, shape and overall appearance of the bacterial symbionts (Sacchi et al. Reference Sacchi, Bigliardi, Corona, Beninati, Lo and Franceschi2004). Evidence was also obtained that the bacteria reside in mitochondria. I. ricinus mitochondria harbouring numerous bacteria (i.e. over 10/20 per section) always present clear signs of degeneration of the matrix, in some cases resembling empty sacs; this has been interpreted as a consumption of the matrix by the bacteria (Sacchi et al. Reference Sacchi, Bigliardi, Corona, Beninati, Lo and Franceschi2004). Such degeneration was less evident in R. bursa, even in mitochondria containing up to 20 bacteria per section. These TEM observations, associated with the lower prevalence of M. mitochondrii in R. bursa, may indicate that the kind of interaction between the host tick and symbionts is different in R. bursa and in I. ricinus. We note that within the monophyletic branch of M. mitochondrii, the bacteria of these 2 tick species are assigned to 2 different branches.
The capacity to invade the host mitochondria is thus present in different lineages of M. mitochondrii, infecting different tick species belonging to different genera and subfamilies. We could therefore hypothesize that the tropism for the mitochondria could represent an ancestral trait of M. mitochondrii, and it is reasonable to expect that other tick species in addition to the two that we investigated at the microscopic level harbour bacteria with the capacity to invade the mitochondria. In summary, intra-mitochondrial symbiosis is probably not just a curiosity limited to I. ricinus, but could represent a more widespread biological phenomenon, at least in ticks of the family Ixodidae, a group of arthropods encompassing over 600 species, many of significant medical and veterinary importance. The fact that 16S rRNA gene fragments almost identical to those from M. mitochondrii have been found in samples from humans and deer (Mediannikov et al. Reference Mediannikov, Ivanov, Nishikawa, Saito, Sidel'nikov, Zdanovskaia, Mokretsova, Tarasevich and Suzuki2004; Skarphédinsson et al. Reference Skarphedinsson, Jensen and Kristiansen2005) opens the intriguing question of whether the invasion of mitochondria by these bacteria also occurs in mammals.
We thank the French Polar Institute (IPEV programme n°333) and A. Petersen for enabling collection of I. uriae in Iceland. N. L. is supported by an Australian Research Council Postdoctoral Fellowship. The authors are most grateful to George Lucas for his permission to derive the genus name Midichloria after the fictional creatures midichlorians (see Sassera et al. Reference Sassera, Beninati, Bandi, Bouman, Sacchi, Fabbi and Lo2006). Experimental lab work was supported by MIUR-PRIN grant to C. B.






