Published online by Cambridge University Press: 01 November 2004
Dihydrofolate reductase-thymidylate synthase (DHFR-TS) from Plasmodium falciparum, a validated target for antifolate antimalarials, is a dimeric enzyme with interdomain interactions significantly mediated by the junction region as well as the Plasmodium-specific additional sequences (inserts) in the DHFR domain. The X-ray structures of both the wild-type and mutant enzymes associated with drug resistance, in complex with either a drug which lost, or which still retains, effectiveness for the mutants, reveal features which explain the basis of drug resistance resulting from mutations around the active site. Binding of rigid inhibitors like pyrimethamine and cycloguanil to the enzyme active site is affected by steric conflict with the side-chains of mutated residues 108 and 16, as well as by changes in the main chain configuration. The role of important residues on binding of inhibitors and substrates was further elucidated by site-directed and random mutagenesis studies. Guided by the active site structure and modes of inhibitor binding, new inhibitors with high affinity against both wild-type and mutant enzymes have been designed and synthesized, some of which have very potent antimalarial activities against drug-resistant P. falciparum bearing the mutant enzymes.
Drug resistance of the malaria parasites is one of the most important problems in malaria control. A major part of this problem is the resistance of the parasites to antifolates, inhibitors of dihydrofolate reductase (DHFR), a validated drug target which is a part of the bifunctional enzyme dihydrofolate reductase-thymidylate synthase (DHFR-TS) (Olliaro & Yuthavong, 1999; Hyde, 2002; Yuthavong, 2002). Not only have such inhibitors as pyrimethamine, cycloguanil and their derivatives been compromised by widespread resistance, but also their synergistic combinations with sulfa-drugs, which act on another enzyme, dihydropteroate synthase (DHPS) in the folate de novo synthesis pathway, are also under threat. Resistance to DHFR inhibitors is explained by the occurrence of point mutations in the enzyme, as shown by the changes in the base sequences of its gene, leading to changes in amino acids in certain positions of the enzyme (Cowman et al. 1988; Peterson, Walliker & Wellems, 1988). In order to develop new effective antifolates, it is important to know the molecular structure of DHFR-TS, how it interacts with inhibitors and substrates, and how mutations affect the interactions.
DHFR-TS from malarial parasites consists of the DHFR domain and the TS domain, joined through the junction region (Bzik et al. 1987). The bifunctional enzyme from Plasmodium falciparum (Pf) has 608 amino acid residues, with 231 residues of the DHFR domain at the N-terminus and 288 residues of the TS domain at the C-terminus joined by an 89-residue junction region. The amino acid identity of PfDHFR with DHFR from bacterial and mammalian species ranges from 24 to 42%, while the identity of PfTS with TS from the other species ranges from 42 to 63%. Significant sequence differences, especially around the active site regions, between DHFR of Plasmodium and the human host have allowed the development of differential inhibitors as antifolate drugs, in contrast with the more conserved nature of TS. However, in spite of the conserved sequence of PfTS, the fact that its activity depends on the integrity of the DHFR and the junction region (Shallom et al. 1999; Wattanarangsan et al. 2003) is a unique feature that may be exploited in the development of new differential TS inhibitors.
The recently solved crystal structures of the wild-type and mutant P. falciparum DHFR-TS complexed with pyrimethamine or WR99210, a dihydrotriazine derivative (Yuvaniyama et al. 2003) have given the molecular description of the antifolate target, and yielded insights into the mechanisms of resistance resulting from the mutations. In addition, the structures reveal some unique features of interactions among the 3 domains of the molecule that help to explain their kinetic properties, and may be exploited for development of new inhibitors. PfDHFR-TS is a dimeric enzyme, with 2 subunits of TS in extensive contact, together forming 2 active sites in the contact area. The overall structural features bear some similarities with those of Leishmania major DHFR-TS (LmDHFR-TS) (Knighton et al. 1994) and the recently solved DHFR-TS of Cryptosporidium hominis (ChDHFR-TS) (O'Neil et al. 2003), which are the only 2 other protozoal enzymes of known structure (Fig. 1). However, there are many differences between the overall features of the 3 enzymes, mainly resulting from the fact that PfDHFR-TS has additional interdomain contacts, due to the presence of the long junction region joining the 2 catalytic domains, and 2 additional sequences absent in other species (inserts) in the DHFR domain (Insert 1, residues 20–36 and Insert 2, residues 64–98). ChDHFR-TS has a junction region of 58 amino acids, while LmDHFR-TS has a very short junction of 2 amino acids, and has only 1 insert analogous to the second insert of the plasmodial enzyme. The junction region of each subunit of ChDHFR-TS has been shown to lead from one DHFR to the other DHFR and interact with it extensively through the ‘donated helix’ before returning to the TS domain (O'Neil et al. 2003). It was earlier shown (Yuvaniyama et al. 2003) that equivalent ‘donated helices’ are present in PfDHFR-TS, but it is still unclear, because of invisible electron density, whether for PfDHFR-TS each DHFR domain is linked to the TS domain underneath or to the other TS domain in a ‘domain swapping’ manner (Fig. 1B). The former arrangement would offer the possibility of control of the activity of one DHFR domain by the other subunit through the ‘donated helix’ of the junction region, as was proposed for ChDHFR-TS.

Fig. 1. Ribbon diagram comparison of protein fold among three DHFR-TS from (A) Leishmania major (Knighton et al. 1994), (B) Plasmodium falciparum (Yuvaniyama et al. 2003), and (C) Cryptosporidium hominis (O'Neil et al. 2003). DHFR and TS domains are coloured red and blue, respectively, where the junction regions are in dark green. The ‘donated helices’ within the junction regions in P. falciparum and C. hominis enzymes are drawn in yellowish green. The different shades denote individual subunits, although the actual connectivity of the P. falciparum DHFR-TS is still unknown owing to invisible electron density. The grey dashed curves represent possible linkages based on intermolecular space in crystal packing. Ligands bound in active sites are drawn in various colours: dUMP in magenta, TS inhibitor CB3 in orange, NADPH in cyan, whereas MTX (in L. major), WR99210 (in P. falciparum), and folate (in C. hominis) are in yellow. Coordinates of L. major and C. hominis DHFR-TS were kindly provided by D. Matthews and A. Anderson, respectively.
In addition, compared with human DHFR, LmDHFR-TS has 22 extra residues which are engaged in extensive contact with the TS domain, while the plasmodial enzyme has only 6 extra N-terminal residues, which are not in contact with the TS domain. It is notable, however, that deletion of only 5 amino acids on the N-terminus of PfDHFR-TS resulted in a protein with severely impaired DHFR function, and deletion of 15 amino acids resulted in non-functioning TS as well as DHFR (Wattanarangsan et al. 2003). This indicates that the N-terminal residues play an important role on both DHFR and TS functions, although the beginning of TS is 320 residues away from the N-terminus.
Based on comparison of the 3 known structures of protozoal DHFR-TS and the enzyme sequences of various protozoa, it was recently proposed that they comprise 2 structural families (O'Neil et al. 2003): PfDHFR-TS and ChDHFR-TS which are members of Alveolata are characterized by the presence of a long junction region, a part of which is used for DHFR-DHFR and DHFR-TS interdomain attachment, and LmDHFR-TS in the Euglenozoa which is characterized by a short junction, and use of the long N-terminal extension for DHFR-TS interaction.
The scaffold of the PfDHFR domain is similar to the core structures of other DHFRs, with 8 central β-strands (βA–βH) sandwiched by 4 α-helices (αB, αC, αE, αF). In addition, there are 3 extra short α-helical regions in the main DHFR domain, αA, a short 310-helix in Insert 1
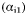
, and a long α-helix in Insert 2
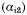
. While Insert 1 extends away from the domain surface and does not interfere with the core DHFR structure, a part of it interacts with the TS domain and helps to stabilize the interdomain attachment. The junction region, notably the

helix (or the ‘donated helix’) linking a DHFR domain with a TS domain interacts extensively with the other DHFR domain on the opposite side, including a part of Insert 2, explaining the closer proximity of the 2 DHFR domains than that observed in LmDHFR-TS (Yuvaniyama et al. 2003).
Comparison with DHFRs from bacterial and mammalian sources gives rise to the prediction that a number of residues in the active site of plasmodial DHFR may be essential for its activity. The prediction is confirmed by the revealed structures of the enzyme-inhibitor complexes (Yuvaniyama et al. 2003) and that of the enzyme-substrate complex (Chitnumsub et al. unpublished observations), as well as by mutagenesis studies. Several residues at the active site are engaged in H-bonding networks and other interactions to hold different parts of the substrate molecule in an orientation suitable for catalysis, including Asp54, Trp48 and Phe58 (Fig. 2), equivalent to Asp 27, Trp22 and Phe31 respectively in E. coli DHFR. Ile14, Ile164, and Thr185, equivalent to Ile5, Ile94, and Thr113 respectively in the E. coli enzyme, are also involved in the interactions. Knowledge of the role of these residues in the catalysis is important in understanding the mechanism of PfDHFR action and the design of new specific inhibitors.

Fig. 2. (A) Enzyme–substrate interactions at the active site of the wild-type PfDHFR-TS. Dihydrofolate substrate (DHF) is coloured magenta at carbon atoms. Two water molecules (W1 and W2) important in binding of the ligand are drawn as red spheres. Four amino acid residues that are mutated in the pyrimethamine-resistant mutant are denoted with red labels. (B) Diagram of the interactions. Dashed lines indicate hydrogen bonds, while a squiggled line denotes ring-stacking interaction. The four residues responsible for pyrimethamine-resistance are bolded and underlined while other residues important in other interactions are also shown. The positions of some atoms of the substrate are assigned as numbers.
The carboxyl oxygens of Asp54 of PfDHFR, form hydrogen bonds with the 3-N, 2-amino and 4-carboxyl O of the pteridine ring of dihydrofolate and the 1-N and 2-amino of 2,4-diaminopyrimidines and equivalent groups of dihydrotriazine inhibitors (Figs 2 and 3). Up to the present, no single Asp54 mutant enzyme has been identified in the field, supporting its essential role in catalysis, although some D54N mutants created from random mutagenesis were identified from screening for resistance to WR99210 (a cycloguanil analogue highly effective against known resistant mutants) in a bacterial surrogate system carrying the PfDHFR (Chusacultanachai et al. 2002). However, as found similarly for E. coli DHFR, PfDHFR carrying mutations at Ala16, Asp54, Ser108 and Phe223 (A16V+D54N+S108T+F223S) has been identified in P. falciparum subjected to chemical mutagenesis and pyrimethamine pressure (Tanaka et al. 1990). This raises the possibility that compensating mutations can revive the enzyme activity, and may make it bind poorly with antimalarial inhibitors, explaining the evolution of drug resistance. It was found (Sirawaraporn et al. 2002) that while the A16V+D54E+S108T mutant had very low enzyme activity, the A16V+D54E+S108T+F223S mutant had greatly enhanced activity, while binding very poorly with pyrimethamine, cycloguanil or WR99210. The molecular explanation for restoration of the enzyme activity can be seen from the model of the enzyme, which shows that Ser223, with a shorter side-chain, restores the ability of Glu54 to form hydrogen bonds with the substrate. This adjustment slightly widens the active-site cavity which reduces the binding affinity of small inhibitors, while interactions with the substrate which is bigger can still be tolerated. It can be concluded that multiple mutations with compensating effects on PfDHFR activity can lead to the evolution of resistance.

Fig. 3. Comparisons of enzyme–inhibitor interactions at the active site: (A) between wild-type–WR99210 and double mutant–pyrimethamine complexes, and (B) between wild-type–WR99210 and quadruple mutant–WR99210 complexes. The complexes of the double mutant (A) and quadruple mutant (B) are drawn in full colour, while the wild-type complex is shown in thinner drawing with carbon atoms in black. In the mutant complexes, carbon atoms of pyrimethamine (A) and WR99210 (B) are coloured magenta, while those of the NADPH are in cyan. The two water molecules, resembling those in Fig. 2A, are drawn in red and black spheres for the mutant and wild-type complexes, respectively. The four residues responsible for pyrimethamine-resistance are labelled in red. Comparing with the wild-type complex, the double mutant-pyrimethamine complex shows significant dislocation of the nicotinamide ring of NADPH while the protein conformation is generally the same (A). Notably, the quadruple mutant shows large dislocation of the backbone conformation at residues 48–51 while the ligand binding modes are largely preserved except a shift at the trichlorophenyl ring of WR99210 (B). (C–E) Diagrams of the interactions in the double mutant (C), wild-type (D), and quadruple mutant (E) complexes use the same notations for symbols as in Fig. 2B. The positions of some atoms of the inhibitors are assigned as numbers.
In order to find out the role of Asp54 in the catalysis and binding to substrates and inhibitors of PfDHFR, this residue was mutated to all other possible 19 amino acids. It was found that the Glu54 (D54E) mutant has significant enzyme activity, albeit only about 2% that of the wild-type enzyme (Sirawaraporn et al. 2002). The reduction in activity was due to both decrease in the catalytic constant and increase in Km values for dihydrofolate and NADPH. Seven mutants had only 0·03–0·2% of the activity of the wild-type enzyme, and the 11 remaining mutants had undetectable DHFR activity. The D54E mutant enzyme binds poorly with pyrimethamine, and very poorly with cycloguanil. The results indicate that precise orientation of the carboxyl oxygen atoms is important in the binding of substrates and inhibitors.
Trp48, equivalent to Trp22 in the E. coli enzyme and Trp24 of the human enzyme, is involved in hydrogen bonding with a water molecule (W1 in Figs 2 and 3), which in turn hydrogen-bonds to Asp54 and 4-O of the substrate. It is also in van der Waals contact with both inhibitor and substrate. Trp48 of PfDHFR is expected to be an essential or an important residue, based on analogy with the enzyme from other species (Beard et al. 1991; Warren et al. 1991). It has been found (Kamchonwongpaisan et al. unpublished observations) that all plasmodial DHFR mutants of Trp48 except one do not have any significant catalytic activity. The Tyr48 (W48Y) mutant is the only one with significant enzyme activity, with 7% activity of that of the wild-type enzyme. Both the Km values for dihydrofolate and NADPH, and the Ki values for pyrimethamine and cycloguanil of the W48Y mutant DHFR are moderately increased. Trp48 can therefore be considered an important residue for PfDHFR, similarly to mammalian and bacterial DHFRs.
Phe58, equivalent to Phe31 of E. coli DHFR, is also important for both substrate and inhibitor binding. In other DHFRs, this conserved residue participates in the network of coupled motions in the catalytic cycle, pressing against the substrate and facilitating hydride transfer (Benkovic & Hammes-Schiffer, 2003). In PfDHFR-TS, Phe58 is on αB helix close to the αj1-helix of the junction region, a situation similar to that in ChDHFR-TS. As pointed out for ChDHFR-TS (O'Neil et al. 2003), this indicates a possible signaling mechanism for regulation of catalysis from one subunit to another. No Phe58 mutant parasite has been identified from field isolates to date, although random mutagenesis of PfDHFR, followed by screening in a bacterial surrogate system under drug pressure generated F58L and F58C mutants of PfDHFR-TS, which were found to have very poor enzyme activities (Chusacultanachai et al. 2002). The equivalent F57L mutant of P. vivax DHFR also had very poor enzyme activity (Leartsakulpanich et al. 2002).
The structures of the wild-type and mutant enzymes bound with inhibitors give insight into the significance of the S108N mutation in reducing the binding affinity of pyrimethamine (Yuvaniyama et al. 2003). The modes of inhibitor binding as found from the X-ray structures generally confirm earlier predictions from modelling studies (McKie et al. 1998; Warhurst, 1998; Lemcke et al. 1999; Rastelli et al. 2000; Warhurst, 2002; Sardarian et al. 2003). Inhibitors like pyrimethamine and cycloguanil have a rigid p-chlorophenyl group in which the Cl atom lies very close to the side-chain of residue 108. The crystal structures show that changing the side-chain from that of Ser to Asn would create steric constraint to the binding of the p-chlorophenyl group, as well as dislocation of the nicotinamide ring of NADPH, which in turn affects the binding of the inhibitor (Fig. 3A and C). This prediction was supported by the finding that a pyrimethamine analogue with Cl in the m- rather than the p-position binds much better with the mutant enzymes carrying the S108N mutation, and indeed has much better antimalarial activity against the mutant parasites than pyrimethamine (McKie et al. 1998; Tarnchompoo et al. 2002; Sardarian et al. 2003; Kamchonwongpaisan et al. 2004). It had also been earlier noted that the reduced sensitivity to pyrimethamine and cycloguanil of DHFR residue-108 mutants (Asn, Thr, Gln, Cys) tends to be correlated with the side-chain length and bulk (i.e. molecular weight, molar refractivity, volume and surface area), while the charge on the non-H atom distal to the backbone and the lipophilicity of the side-chain apparently have no impact (Warhurst, 1998).
The general validity of this conclusion was further supported by more recent extensive binding data for these plus other mutants with bulky side-chains (Tarnchompoo et al. 2002; Sardarian et al. 2003; Kamchonwongpaisan et al. 2004). The S108N mutant also binds poorly with other pyrimethamine derivatives with bulky groups in place of the p-Cl, and the binding was generally progressively poorer for the double (S108N+C59R) mutant. Such cumulative effects on inhibitor binding are seen more clearly with the triple (S108N+N51I+C59R) and the quadruple (S108N+N51I+C59R+I164L) mutants, leading to increasingly poorer antimalarial effects of pyrimethamine and cycloguanil (Kamchonwongpaisan et al. 2004). Structural explanation of this cumulative effect is given by the observation that the N51I mutation causes a substantial main-chain movement of residues 48–51 by 0·5–2·2 Å, with a 48–49 peptide flip, and the I164L mutation causes a minor shift (0·3–0·5 Å) of residues 164–167 (Fig. 3B), which together open up the active site gap and weaken the binding of small inhibitors like pyrimethamine (Yuvaniyama et al. 2003).
Removal of the p-Cl or replacement with m-Cl led to better binding with the mutant DHFRs. A number of other inhibitors with flexible groups which can avert the potential steric constraint around the side-chain of residue 108 also proved to have better binding affinities with mutant enzymes. These include WR99210 and other cycloguanil analogues, and some trimethoprim analogues, which show good antimalarial activities with the mutant parasites, reflecting the binding affinities with the mutant enzymes which they carry (Kamchonwongpaisan et al. 2004; Sirichaiwat et al. 2004). In contrast to rigid inhibitors like pyrimethamine and cycloguanil, WR99210 with a flexible side-chain can adopt a conformation that fits well in the active site of the mutant enzymes, explaining its effectiveness as an antimalarial against resistant parasites. The structure of WR99210 bound with the quadruple mutant as compared with the wild-type DHFR-TS (Fig. 3B) shows that its side-chain can avert steric hindrance caused by the S108N and subsequent mutations, and can further interact with other residues in the active site, explaining its retention of its binding affinity (Yuvaniyama et al. 2003). Fig. 3D and E show diagrams of important residues of the wild-type and the quadruple (S108N+N51I+C59R+I164L) mutant enzyme respectively, within the vicinity of and interacting with the inhibitor.
The importance of the side-chain of residue 108 in inhibitor binding was assessed by site-specific mutation (Sirawaraporn et al. 1997a; Tarnchompoo et al. 2002). Significant correlations were found between the Ki values for pyrimethamine and cycloguanil and the length and bulk of the side-chain of this residue. Furthermore, pyrimethamine derivatives with bulky groups in place of the p-Cl bind even more poorly with the S108N and multiple mutant enzymes in the series, indicating that the binding affinities are reduced with increased steric interference (Tarnchompoo et al. 2002; Sardarian et al. 2003; Kamchonwongpaisan et al. 2004). Removal of the p-Cl or replacement with m-Cl led to better binding with the mutant enzymes as expected.
Modelling of binding of cycloguanil and its derivatives has helped to understand the specific reduction in its binding with another mutant DHFR, namely, the A16V+S108T mutant (Rastelli et al. 2000). Cycloguanil carries two methyl groups at the 2-position, in contrast to pyrimethamine that has only one ethyl group in the equivalent position. The binding model shows that one of the two methyl groups is in steric conflict with the enlarged side-chain of Val16. In support of this model, it was found that 2-desmethyl cycloguanil and other derivatives with only one substituent in the 2-position generally have better binding to the mutant relative to the wild-type enzyme than the disubstituted derivatives (Yuthavong et al. 2000). A similar finding has also been made with variation of the side-chain of residue 16 in the series of A16X+S108T mutants (Kamchonwongpaisan et al. unpublished observations). Increased resistance to cycloguanil can be observed in mutants with increasing size of the A16X side-chains, from Ala, Cys, Ser, Val, and Thr, respectively.
Although the crystal structures of the complex of cycloguanil with the enzyme and its mutants are not known, that of the cycloguanil analogue WR99210 has been solved (Yuvaniyama et al. 2003), and found to be similar to the modelled structure (Rastelli et al. 2000), although there are significant differences. Modelling of new analogues into the active site should therefore help in designing effective inhibitors against resistant parasites.
It is unclear whether stepwise mutations, giving rise to increasing resistance, occurred during or before introduction of antifolates. The former would represent evolution due to drug pressure, while the latter would be selection of pre-existing mutants. In either case, the generation of resistance can be studied from a combination of site-directed (Sirawaraporn et al. 1997b) and random mutagenesis (Ferlan et al. 2001; Hankins et al. 2001; Chusacultanachai et al. 2002), which yielded mutants both found and not found in nature for comparative studies. In the generation of the quadruple mutant (at codons 51, 59, 108, and 164), S108N was probably the first mutation, yielding moderately resistant P. falciparum, which could give rise to further mutations N51I, C59R and I164L with increasing levels of resistance (Sirawaraporn et al. 1997b). The reasons for absence in nature of other single mutants than Ser108 can be obtained from the study of these artificially created mutants, as either due to too low enzyme activity (A16V), or to no significant decrease in affinity for the inhibitors (single N51I, C59R, I164L mutations). The rationale for selection of the Asn mutant at codon 108 was provided by site-directed mutagenesis studies, in which all possible 19 other amino acids were substituted for Ser (Sirawaraporn et al. 1997a; Tarnchompoo et al. 2002). Except for Lys, all other possible mutants could be expressed from the heterologous system. Nine of the mutants showed no detectable enzyme activity. The relatively higher Ki values for pyrimethamine and cycloguanil of the mutants with bulky side-chains (Asn, Gln, Leu, Val and Met), compared with the Ki values of the wild-type DHFR confirm the validity of the interaction model proposed.
Generation of antifolate resistance can be studied by simulated, directed ‘evolution’, achieved through exposure of randomly mutated PfDHFR genes complementing growth of a micro-organism used in a suitable selection system. Sibley's group (Ferlan et al. 2001; Hankins et al. 2001) used Saccharomyces cereviseae with disrupted endogenous DHFR as a selection system for PfDHFR function. PCR mutagenesis of the PfDHFR followed by selection in this system against drug pressure produced PfDHFR mutants associated with high drug resistance. Mutations that increased resistance to pyrimethamine were identified in 3 regions of the DHFR domain, around codons 50, 188 and 213 (Ferlan et al. 2001). If the mutagenesis is done on the background of the triple mutant N51I+C59R+S108N, further mutations can be identified in the 3 clusters of codons 50–57, 187–193 and 213–214 (Hankins et al. 2001). Several mutations previously identified in field samples were also isolated, including codons 50 and 164. While the majority of the mutants identified were found to be resistant to chlorcycloguanil, most mutants were still sensitive to WR99210, prompting the prediction that this and related compounds will be clinically effective against new multiple mutants in the field.
Another selection system based on transformation of E. coli by the PfDHFR gene, previously subjected to PCR mutagenesis, has been developed in our laboratory (Chusacultanachai et al. 2002). In this system the endogenous bacterial DHFR activity is suppressed by trimethoprim, at a concentration which does not inhibit the PfDHFR. This system identified the single and multiple mutations originally found in the field correctly: all obtained pyrimethamine-resistant mutants possessed S108N mutation, in combination with common mutations of N51I, C59R and I164L. New resistant mutants with novel mutations not found in the field were also identified. Exposure of the randomly mutated PfDHFR libraries to WR99210 or SO3 (a pyrimethamine analogue, with m-Cl in place of p-Cl effective against multiple mutant DHFRs; McKie et al. 1998; Sardarian et al. 2003) resulted in selection of novel single and multiple mutants, which exhibited 2- to over 2000-fold increase in resistance against these two antifolates. It is of interest that, starting from the quadruple mutant gene with N51I+C59R+S108N+I164L, a number of genes with additional mutations conferring resistance to WR99210 were identified. Most of these WR99210-resistant mutants have F58L mutation, which implies that this residue is important for inhibitor binding, in line with the binding mode seen from the X-ray structures (Fig. 3). Importantly, most of the WR99210-resistant mutants contain the reversion mutation N108S or N108T, implying that in the background of other mutations the side-chain of Ser is important for the binding of WR99210, or that the side-chain of Asn now becomes deleterious to its binding. Alternatively, the reversions may be needed for enzyme activity in the presence of other mutations. It should also be noted that pyrimethamine and WR99210 have been shown to exert opposing selection on P. vivax DHFR (Hastings & Sibley, 2002; Ridley, 2002). In addition, the mutation studies including kinetic analysis of the mutants show that, apart from the active site residues that are crucial for DHFR activity, residues remote from the binding pocket also play essential roles in substrate and inhibitor binding.
Structural and mutagenesis studies of PfDHFR-TS have given valuable information on the development of resistance of the malaria parasites to antifolate drugs. The information can be used to help design new effective antifolates, either as single drugs or as combinations. While single drugs need to be designed to be effective against all different parasites bearing multiple mutants, drug combinations would consist of components, each of which is directed against some mutant enzymes, and all of which would be effective against all different mutant parasites. The main rationale for developing these single or combined drugs is that the parasite has limitation in their mutation possibilities, since the DHFR has to have a minimal activity in order to allow the parasite to survive. The fact that many mutant PfDHFRs conferring resistance to WR99210, selected from the background of the quadruple mutant, showed reversion of N108S or N108T (Chusacultanachai et al. 2002), is an indication that the parasite may have such limit in mutation possibilities. Some combinations such as A16V and S108N have never been found in the field, nor have mutants with more than 4 mutations. Lack of enzyme activity was also shown for many site-directed mutants, such as those at codons 108 and 164, which would be expected to have poor binding to antifolate inhibitors (Sirawaraporn et al. 1997a; Chusacultanachai et al. 2002) and would have led to resistant parasites had the enzymes been active.
Modelling of enzyme-inhibitor interaction, supported by the X-ray structures, led to the definition of essential features for effective inhibitors. The following features are noted (Yuthavong, 2002): (a) hydrogen bonding with both of the carboxyl oxygens of Asp54, (b) hydrogen bonding with backbone oxygens of Ile164 and Ile14, (c) optimum rigid length of the inhibitor e.g., from the 2-amino group of a 2,4-diaminopyrimidine to the distal end interacting with Ser108 and mutants with bulkier side-chains, preferably with stacking interaction with Phe58 and the bound NADPH, (d) preferably a flexible side-chain, as in WR99210, which can avoid steric clash with residue 108 of the resistant mutants, and can form hydrophobic and other interactions with various residues in the active site (Fig. 3), (e) free space close to Ala16 in order to avoid resistance as in the case of cycloguanil, where one of the 2,2-dimethyl groups clashes with the side-chain of mutated Val16, and (f) other parts of the molecule, the binding of which cannot be affected by permissible enzyme mutations.
A number of inhibitors were designed and synthesized, with general structure as shown in Fig. 4, based partly on the features defined above, and tested against wild-type and mutant PfDHFRs. Many of the inhibitors have been shown to have very good binding affinities with the mutant PfDHFRs (Fig. 5A) (Kamchonwongpaisan et al. 2004; Kamchonwongpaisan et al. unpublished observations). They have also been tested for anti-plasmodial activity in culture against wild-type and resistant P. falciparum carrying the mutant DHFRs (Fig. 5B), and in vivo against P. berghei in mice. Some of these new inhibitors show good anti-plasmodial activities against both the wild-type and mutant parasites, with relatively low inhibition of human DHFR and low toxicity against mammalian cell lines (Tarnchompoo et al. 2002; Kamchonwongpaisan et al. 2004). Selectivity to DHFR inhibitors also arises from the fact that the human host can respond to DHFR inhibitors by increasing DHFR expression, while the parasite cannot do so (Zhang & Rathod, 2002). These inhibitors are therefore potential leads to the design and synthesis of effective and selective antifolate antimalarials in the future.

Fig. 4. The general structures of effective inhibitors against multiple mutant PfDHFR-TS. The nucleus (light pink) is either a diaminopyrimidine or a diaminodihydrotriazine, with 2 or 3 N atoms in the ring respectively, while the side-chain is either a rigid aryl derivative (dark pink) or a flexible chain starting with a C or O atom. For diaminodihydrotriazines, the 2-position should be monosubstituted by a hydrophobic group.

Fig. 5. The ranges of inhibition constants (Ki) and in vitro IC50 values against Plasmodium falciparum for some cycloguanil inhibitors of wild-type and mutant PfDHFR-TSs with general structures as shown in Fig. 4, with p-Cl Ph (pink), m-Cl Ph (green), and flexible side-chains (yellow) at N-1 position. Data for cycloguanil and WR99210 are shown in deep red and deep blue respectively. (A) Ki values. (B) In vitro IC50 values against P. falciparum with the wild-type or mutant DHFR-TSs corresponding to (A), of cycloguanil derivatives with p-Cl Ph (pink), m-Cl Ph (green), and flexible side chains (yellow) at N-1 position. Data are from 12, 21 and 16 compounds in the p-Cl Ph, m-Cl Ph and flexible side chain categories respectively. Designations: P. falciparum carrying wt-DHFR-TS (TM4/8.2), C59R+S108N (K1CB1), N51I+C59R+S108N (W2), C59R+S108N+I164N (Csl-2), N51I+C59R+S108N+I164N (V1/S).
Our work is supported by the Medicines for Malaria Venture (MMV), EU INCO-DEV, the Wellcome Trust, the Thailand Tropical Diseases Research (T2) Programme, and the BIOTEC Center of Thailand.

Fig. 1. Ribbon diagram comparison of protein fold among three DHFR-TS from (A) Leishmania major (Knighton et al. 1994), (B) Plasmodium falciparum (Yuvaniyama et al. 2003), and (C) Cryptosporidium hominis (O'Neil et al. 2003). DHFR and TS domains are coloured red and blue, respectively, where the junction regions are in dark green. The ‘donated helices’ within the junction regions in P. falciparum and C. hominis enzymes are drawn in yellowish green. The different shades denote individual subunits, although the actual connectivity of the P. falciparum DHFR-TS is still unknown owing to invisible electron density. The grey dashed curves represent possible linkages based on intermolecular space in crystal packing. Ligands bound in active sites are drawn in various colours: dUMP in magenta, TS inhibitor CB3 in orange, NADPH in cyan, whereas MTX (in L. major), WR99210 (in P. falciparum), and folate (in C. hominis) are in yellow. Coordinates of L. major and C. hominis DHFR-TS were kindly provided by D. Matthews and A. Anderson, respectively.

Fig. 2. (A) Enzyme–substrate interactions at the active site of the wild-type PfDHFR-TS. Dihydrofolate substrate (DHF) is coloured magenta at carbon atoms. Two water molecules (W1 and W2) important in binding of the ligand are drawn as red spheres. Four amino acid residues that are mutated in the pyrimethamine-resistant mutant are denoted with red labels. (B) Diagram of the interactions. Dashed lines indicate hydrogen bonds, while a squiggled line denotes ring-stacking interaction. The four residues responsible for pyrimethamine-resistance are bolded and underlined while other residues important in other interactions are also shown. The positions of some atoms of the substrate are assigned as numbers.

Fig. 3. Comparisons of enzyme–inhibitor interactions at the active site: (A) between wild-type–WR99210 and double mutant–pyrimethamine complexes, and (B) between wild-type–WR99210 and quadruple mutant–WR99210 complexes. The complexes of the double mutant (A) and quadruple mutant (B) are drawn in full colour, while the wild-type complex is shown in thinner drawing with carbon atoms in black. In the mutant complexes, carbon atoms of pyrimethamine (A) and WR99210 (B) are coloured magenta, while those of the NADPH are in cyan. The two water molecules, resembling those in Fig. 2A, are drawn in red and black spheres for the mutant and wild-type complexes, respectively. The four residues responsible for pyrimethamine-resistance are labelled in red. Comparing with the wild-type complex, the double mutant-pyrimethamine complex shows significant dislocation of the nicotinamide ring of NADPH while the protein conformation is generally the same (A). Notably, the quadruple mutant shows large dislocation of the backbone conformation at residues 48–51 while the ligand binding modes are largely preserved except a shift at the trichlorophenyl ring of WR99210 (B). (C–E) Diagrams of the interactions in the double mutant (C), wild-type (D), and quadruple mutant (E) complexes use the same notations for symbols as in Fig. 2B. The positions of some atoms of the inhibitors are assigned as numbers.

Fig. 4. The general structures of effective inhibitors against multiple mutant PfDHFR-TS. The nucleus (light pink) is either a diaminopyrimidine or a diaminodihydrotriazine, with 2 or 3 N atoms in the ring respectively, while the side-chain is either a rigid aryl derivative (dark pink) or a flexible chain starting with a C or O atom. For diaminodihydrotriazines, the 2-position should be monosubstituted by a hydrophobic group.

Fig. 5. The ranges of inhibition constants (Ki) and in vitro IC50 values against Plasmodium falciparum for some cycloguanil inhibitors of wild-type and mutant PfDHFR-TSs with general structures as shown in Fig. 4, with p-Cl Ph (pink), m-Cl Ph (green), and flexible side-chains (yellow) at N-1 position. Data for cycloguanil and WR99210 are shown in deep red and deep blue respectively. (A) Ki values. (B) In vitro IC50 values against P. falciparum with the wild-type or mutant DHFR-TSs corresponding to (A), of cycloguanil derivatives with p-Cl Ph (pink), m-Cl Ph (green), and flexible side chains (yellow) at N-1 position. Data are from 12, 21 and 16 compounds in the p-Cl Ph, m-Cl Ph and flexible side chain categories respectively. Designations: P. falciparum carrying wt-DHFR-TS (TM4/8.2), C59R+S108N (K1CB1), N51I+C59R+S108N (W2), C59R+S108N+I164N (Csl-2), N51I+C59R+S108N+I164N (V1/S).