INTRODUCTION
The Apicomplexan protozoan parasites belonging to the genus Cryptosporidium infect a wide range of vertebrate hosts. Cryptosporidium sp. infection is a major cause of diarrhoeal disease in humans worldwide and has been recognized as the predominant cause of waterborne protozoan diseases (Gargala, Reference Gargala2008; Ethelberg et al. Reference Ethelberg, Lisby, Vestergaard, Enemark, Olsen, Stensvold, Nielsen, Porsbo, Plesner and Mølbak2009; Plutzer and Karanis, Reference Plutzer and Karanis2009). Furthermore, the effects of cryptosporidiosis can go beyond acute diarrhoeal disease and lead to significant health sequelae (Hunter et al. Reference Hunter, Hughes, Woodhouse, Raj, Syed, Chalmers, Verlander and Goodacre2004). Immunocompromised hosts usually develop persistent progressive infection of greater severity, which may spread to extra-intestinal sites (Mor and Tzipori, Reference Mor and Tzipori2008).
Regarding treatment, although azithromycin and nitazoxanide have been shown to be somewhat effective in immunocompetent persons, there are no effective anti-cryptosporidial agents for immunocompromised persons (Cacciò et al. Reference Cacciò, Pinter, Fantini, Mezzaroma and Pozio2002; Amadi et al. Reference Amadi, Mwiya, Sianongo, Payne, Watuka, Katubulushi and Kelly2009). Several new potential drug targets and active agents have been identified, but there are no data on their use in treating the human disease (Rossignol, Reference Rossignol2010). In patients with cryptosporidiosis, HAART has been shown to resolve infection in some cases (Ives et al. Reference Ives, Gazzard and Easterbrook2001; Hommer et al. Reference Hommer, Eichholz and Petry2003; Pozio and Gómez-Morales, Reference Pozio and Gómez-Morales2005; Zardi et al. Reference Zardi, Picardi and Afeltra2005; Battegay et al. Reference Battegay, Fehr, Flückiger and Elzi2008; Violari et al. Reference Violari, Cotton, Gibb, Babiker, Steyn, Madhi, Jean-Philippe and McIntyre2008), even in the absence of recovery of host immunity (Bobin et al. Reference Bobin, Bouhouere, Dumpt, Boibieux, Grault and Peyrnamond1998; Miao et al. Reference Miao, Awad-El-Kariem, Franzen, Ellis, Müller, Counihan, Hayes and Gazzard2000). In particular, there is evidence of the ability of Indinavir (IND) to reduce Cryptosporidium parvum infection in both in vitro and in vivo models (Bobin et al. Reference Bobin, Bouhouere, Dumpt, Boibieux, Grault and Peyrnamond1998; Miao et al. Reference Miao, Awad-El-Kariem, Franzen, Ellis, Müller, Counihan, Hayes and Gazzard2000; Hommer et al. Reference Hommer, Eichholz and Petry2003; Mele et al. Reference Mele, Gómez-Morales, Tosini and Pozio2003; Zardi et al. Reference Zardi, Picardi and Afeltra2005; Violari et al. Reference Violari, Cotton, Gibb, Babiker, Steyn, Madhi, Jean-Philippe and McIntyre2008) yet there are limitations to its use, given its renal toxicity and the high rate of metabolism and degradation, which leads to the loss of activity in a short time (Jao and Wyatt, Reference Jao and Wyatt2010). To increase the anti-retroviral efficacy of IND, several attempts have been made to encapsulate it in solid lipid or phospholipid nanocapsules (Pereira de Oliveira et al. Reference Pereira de Oliveira, Garcion, Venisse, Benoit, Couet and Olivier2005; Dou et al. Reference Dou, Destache, Morehead, Mosley, Boska, Kingsley, Gorantla, Poluektova, Nelson, Chaubal, Werling, Kipp, Rabinow and Gendelman2006). The use of nanoparticles (Np) as a drug delivery device has been widely studied to achieve protection of the embedded drug (Torchilin, Reference Torchilin2006; Sanvivens and Pilar, Reference Sanvinvens and Pilar2008; Tosi et al. Reference Tosi, Costantino, Ruozi, Forni and Vandelli2008). However, the encapsulation of drugs in nanocarriers does not allow them to be selectively delivered to specific tissues, cells or organs. For this reason, several modification strategies have been developed to conjugate appropriate ligands on the surface of Np (Gref et al. Reference Gref, Couvreur, Barratt and Mysiakine2003); the most effective ligands may be polyclonal or monoclonal antibodies (Ab), given the high selectivity of Ab recognition of the relevant antigen. This approach has been applied in the treatment of several health disorders including cancer (Huwlyer et al. Reference Huwlyer, Wu and Pardridge1996; Cho et al. Reference Cho, Wang, Nie, Chen and Shin2008; Dimitrov et al. Reference Dimitrov, Feng and Prabakaran2008; Wang et al. Reference Wang, Gu, Zhang, Chang, Radovic-Moreno, Shaikh and Farokhzad2008). For IND, the use of an immune liposome in mice greatly increased drug accumulation in the lymph nodes, demonstrating that the drug had been selectively delivered to the primary HIV-1 reservoir (Gagné et al. Reference Gagné, Désormeaux, Perron, Tremblay and Bergeron2002).
Since biodegradable poly(D,L-lactide-co-glycolide) Np (PLGA Np) are able to protect the drug, and thus decrease its degradation, we aimed to encapsulate IND in biodegradable PLGA Np and then to engineer their surface by conjugation with a specific anti-Cryptosporidium polyclonal Ab (IgG), in order to create a specialized system able to deliver the drug selectively against all stages of the parasite.
MATERIALS AND METHODS
Drug, polymers and polyclonal antibody
IND was obtained synthetically (Askin, Reference Askin1998); poly(D,L-lactide-co-glycolide) (PLGA, RG503H, Boehringer-Ingelheim) was labelled with tetramethylrhodamine (TMR-PLGA) (Costantino et al. Reference Costantino, Gandolfi, Bossy-Nobs, Tosi, Gurny, Rivasi, Vandelli and Forni2006). Pluronic F68 was used as a surface-active agent to stabilize the Np (Sigma-Aldrich). 2-(N-morpholino) ethanesulfonic acid (MES), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide-hydrochloride (EDC), and N-hydroxysuccinimide (NHS) (Sigma-Aldrich) were used for modification of the Np surface. As polyclonal Ab, a rabbit anti-C. parvum IgG, that recognizes all stages of C. parvum, was used (Ranucci et al. Reference Ranucci, Muller, La Rosa, Reckman, Gomez Morales, Spano, Pozio and Crisanti1993). All other chemicals were of analytical grade.
Preparation of Indinavir-loaded Np and conjugation with antibody
Np were obtained by the nanoprecipitation procedure (Fessi et al. Reference Fessi, Puisieux, Devissaguet, Ammoury and Benita1989), and the final volume of the suspension was adjusted to 10 ml with analytical grade water. The same procedure was followed using a 10% p/p of TMR-PLGA and 90% p/p of PLGA to produce unloaded labelled Np (TMR-B-Np) and loaded labelled Np (TMR-IND-Np). Np were purified by gel-filtration chromatography (160 ml Sepharose CL 4B gel, column 50×2 cm) using water as the mobile phase, and then freeze-dried (Lyovac GT-2, Leybold-Heraeus) without any cryoprotector. The antibody was then conjugated to loaded and unloaded TMR-Np (Nobs et al. Reference Nobs, Buchegger, Gurny and Allemann2003, Reference Nobs, Buchegger, Gurny and Allemann2004). Repeated centrifugations (28 300 g for 15 min) were applied to purify the samples. A summary of the samples prepared is shown in Table 1.
Table 1. Characterization of the nanoparticles (Np)

a O%=66·3; C%=33·7; O/C ratio=1·96.
b O%=37·6; C%=59·5; N%=2·8; O/C ratio=0·63; N/C ratio=0·04.
Determination of Indinavir loading
An exact amount (10 mg) of the Np loaded with IND (IND-Np, TMR-IND-Np, Ab-IND-Np, Ab-TMR-IND-Np) was solubilized in 1 ml of pure dichloromethane (DCM). An aqueous citrate buffer solution (pH 3) was then added and the reaction was placed under magnetic stirring for 3 h. The solution was then purified by centrifugation at 5500 g (freeze-ultracentrifuge, Sorvall Biofuge Stratos) and filtration by a 0·45 μm filter.
The solution was analysed by HPLC by injecting aliquots into a C18 column (Vydac C18, 4·6×250 mm, C18, 300 A pore size, 5 μm particle size). The mobile phase consisted of a solution of acetonitrile in sodium lauryl sulphate-triethylamine (pH 7), adjusted with ortho-phosphoric acid (flow rate at 1·0 ml/min) (Jancis et al. Reference Jao and Wyatt2005). The identification of IND was confirmed by absorbance at λ=214 nm, and the IND retention time (Rt) was 8 min. The loadings are expressed in mg of IND per 100 mg of particles of at least 3 tests.
Physical and physico-chemical characteristics of the antibody-conjugated Indinavir-loaded Np
A scanning electron microscope (SEM) (XL-40 Philips) (10 000×magnification) was used to evaluate the morphology of Np. Before the SEM analysis, the samples were coated under an argon atmosphere with a 10 nm layer of palladium gold (Emitech K550 Supper Coated, Emitech Ltd). Np in analytical grade water were analysed for particle size and zeta potential (z-p) by photon correlation spectroscopy and laser Doppler anemometry using a Zetasizer Nano ZS (Malvern, UK; Laser 4 mW He–Ne, 633 nm, Laser attenuator Automatic, transmission 100% to 0·0003%, Detector Avalanche photodiode, Q.E. >50% at 633 nm, T=25°C). The results were normalized with respect to a polystyrene standard solution.
To determine the Np surface composition during the modification procedure, Electron Spectroscopy for Chemical Analysis (ESCA) was performed on a 04-153 X-ray source analysis system (PHI, Uvalca-PHI) and an EA11 hemispherical electron analyzer (Leybold Optics), using MgKα1,2 radiations. The spectra were recorded in fixed retardation ratio (FAT) mode with 190 eV pass energy. The pressure in the sample analysis chamber was ca. 10−9mbar. The data were acquired and processed using the RBD AugerScan 2. 1H-HRMAS NMR spectra were recorded on a Bruker Avance 400 instrument; D2O was added to the sample which was then spun at 4000 Hz. All experiments were recorded at room temperature.
The drug release was determined by dispersing an exact weight (10–12 mg) of Np in 25 ml of pH 7·4 phosphate buffer solution (PBS). The bulk solution (1 ml) was placed in an acetate-cellulose membrane dialysis tube (molecular weight cut off=12 000–14 000 distribution; Spectrum Laboratory Inc.). The dialysis tube was placed in 25 ml of PBS and gently shaken with a magnetic stirrer in a water bath at 37±0·1°C. Samples of 0·2 ml were taken at fixed intervals from the medium outside the dialysis tube and the medium was immediately rinsed with the same volume of buffer. The IND concentration was determined by HPLC analysis. The result was expressed as the mean of at least 5 tests. As an additional control, in the same experimental conditions, an exact amount of IND was dissolved in PBS, and the diffusion of IND across the acetate-cellulose dialysis membrane was performed in 25 ml of a PBS solution.
To quantify the loss of IND during the steps of surface modification, the IND release was measured in MES buffer over a period of 7 h, which corresponds to the time needed for all of the modification steps (Nobs et al. Reference Nobs, Buchegger, Gurny and Allemann2003, Reference Nobs, Buchegger, Gurny and Allemann2004).
In vitro culture of C. parvum
Cryptosporidium parvum oocysts (isolate code ISSC6) were obtained from experimentally infected calves after feces purification by sucrose and Percoll density gradients (Rossi et al. Reference Rossi, Pozio, Besse, Gómez-Morales and La Rosa1990). For excystation, oocysts were resuspended in 10 mm HCl and incubated at 37°C for 10 min (Gut and Nelson, Reference Gut and Nelson1999).
A human ileocecaladenocarcinoma tumor cell line (HCT8) was cultured in RPMI 1640 (Gibco) supplemented with 5% fetal calf serum (FCS) (Gibco), 200 nm L-glutamine (Sigma), 1% sodium pyruvate (Sigma), 5% penicillin, and 5% streptomycin. The cells were maintained in tissue-culture flasks in a 5% CO2 atmosphere at 37°C and 85% humidity (Upton et al. Reference Upton, Tilley and Brillhart1995). Before infection with excysted oocysts, the HCT-8 cells were plated in Petri dishes at 1×106 cells/78 cm2 and on tissue-culture glass slides (Falcon). The FCS concentration was increased to 10%, and cells were grown to 80–90% confluence.
Cells were infected with 3 excysted oocysts/cell for 24 h (Slifko et al. Reference Slifko, Hoffman and Rose1999). The plated cells were then detached, fixed in 4% formaldehyde in PBS and treated with 1% Triton X-100 in PBS for 10 min. For evaluation of infection in control infected, non-treated cultures, a rabbit anti-C. parvum IgG, which recognizes all stages of C. parvum (Ranucci et al. 1993) was used as primary antibody. As secondary antibody, an FITC-labelled goat anti-rabbit IgG polyclonal was used. For the flow cytometry analysis, cells were centrifuged at 500 g for 10 min and then washed 3 times and re-suspended in 500 μl of PBS (Motta et al. Reference Motta, Gissot, Kanellopoulos and Ojcius2002). The instrument (FACS Calibur, Becton Dickinson) was set up to measure the forward-angle light scatter (FSC-H), side-angle light scatter (SSC-H) and the fluorescence intensities of FITC (on FL1 detector). The samples were analysed using an acquisition gate based on FL-1 and a FSC-H in order to select only the infected HCT-8 population. Four replicates of each experiment were performed. Results were obtained by comparison of the percentages of both non-treated and treated infected cells at each studied time-point (Mele et al. Reference Mele, Gómez-Morales, Tosini and Pozio2003).
In vitro model
Before treating the infected cells, 1·5 mg of the Ab-TMR- IND-Np or Ab-TMR-B-Np in analytical grade water were sonicated 6 times for 4 min each in an ultrasonic bath. Then 50 μl of each suspension were added to each ml of infected culture, the final concentration of IND was 50 μ m (Mele et al. Reference Mele, Gómez-Morales, Tosini and Pozio2003).
To determine whether or not Ab-TMR-IND-Np had an effect on the parasite attachment to, or invasion of, the HCT-8 cells, it was added to the cultures at the same time as the excysted oocysts at the concentration of 50 μ m (maximum DMSO content 0·5%). To determine whether or not Ab-TMR-IND-Np had an effect on established C. parvum infection, HCT-8 cells were infected with excysted oocysts at a ratio of 3 oocysts/cell at day 0; Ab-TMR-IND-Np at a concentration of 50 μ m was then added to the cultures every 24 h for 4 days. The infection level was then evaluated at the 2nd, 3rd, 4th and 5th day post-infection (p.i.).
To determine the ability of the Ab-TMR-IND-Np to target the parasite, glass slides from C. parvum-infected HCT-8 cells treated with Ab-TMR-IND-Np were stained for 10 min with 50 μl of a 4′-6-diamidino-2-phenylindole (DAPI LabVision Corporation) solution (125 ng/ml), which is known to form fluorescent complexes with natural double-stranded DNA (Xu et al. Reference Xu, Pilch, Srinivasan, Olson, Geacintov and Breslauer1997). The slides were observed under a fluorescence microscope (Axiophot, Zeiss) with triple excitation bands for DAPI, FITC and TMR using an emission filter set for fluorescence imaging ( 20–40–100 lenses were used in order to obtain a ×250–500–1250 magnification, e.g. 100×12·5).
To verify whether Ab-TMR-IND-Np or the DMSO had a direct effect on HCT-8 cells, influencing their ability to support C. parvum infection, HCT-8 cells were pre-treated with Ab-TMR-IND-Np or with 0·5% DMSO for 24 h before the infection. No significant variation in the infection level was observed between C. parvum-infected cells and C. parvum-infected cells pre-treated with Ab- TMR-IND-Np or with 0·5% DMSO. Moreover, to eliminate any direct effect of the DMSO or of the engineered nanoparticles on the infected cells,C. parvum-infected HCT-8 cells treated with 0·5% DMSO, and C. parvum-infected HCT-8 cells treated with Ab-TMR-B-Np, were used as controls; no significant variation was observed in the infection level between C. parvum-infected cells and C. parvum-infected cells treated with Ab-TMR-B-Np or with 0·5% DMSO (data not shown).
Statistical analysis
A level of P< 0·01 was considered to be statistically significant by Student's t-test that was performed to compare the percentages of reduction in infected cells from experimental groups (C. parvum-infected HCT-8 cells treated with 50 μ m IND in 0·5% DMSO and C. parvum-infected HCT-8 cells treated with 50 μ m of Ab-TMR-IND-Np).
RESULTS
Nanoparticle characterization
Table 1 shows the dimensional range (size), zeta potential (z-p), polydispersity index (PDI) and drug loading of the Np. The encapsulation of IND affected neither the dimensional values (180–200 nm) nor the surface charges (z-p from −46·8 to −34·5 mV). Encapsulation resulted only in a decreased homogeneity of the Np population, expressed as PDI, which slightly increased from 0·09 to 0·2. No remarkable difference in the IND loading was found between IND-Np and TMR-IND-Np. As expected, the size of the antibody-engineered Np (234–287 nm) was slightly greater than that of the unmodified Np (184–201 nm), but the homogeneity of the antibody-engineered Np did not change (PDI: 0·12–0·18) when compared to unmodified Np. The most marked differences were observed for z-p, with values of surface charge still negative from −15·6 to −9·2 mV, but strongly decreased in comparison with the unmodified Np from −46·8 to −35·9 mV. Moreover, the average amount of IND in Ab-Np samples was lower than that in the unmodified Np (2·38–2·45 mg/100 mg of Np), demonstrating a loss of loaded IND over the modification procedures. By SEM all of the Np exhibited a compact shape and an intact surface (not shown), confirming that the antibody-conjugation did not affect the Np morphology.
Surface analysis of Np
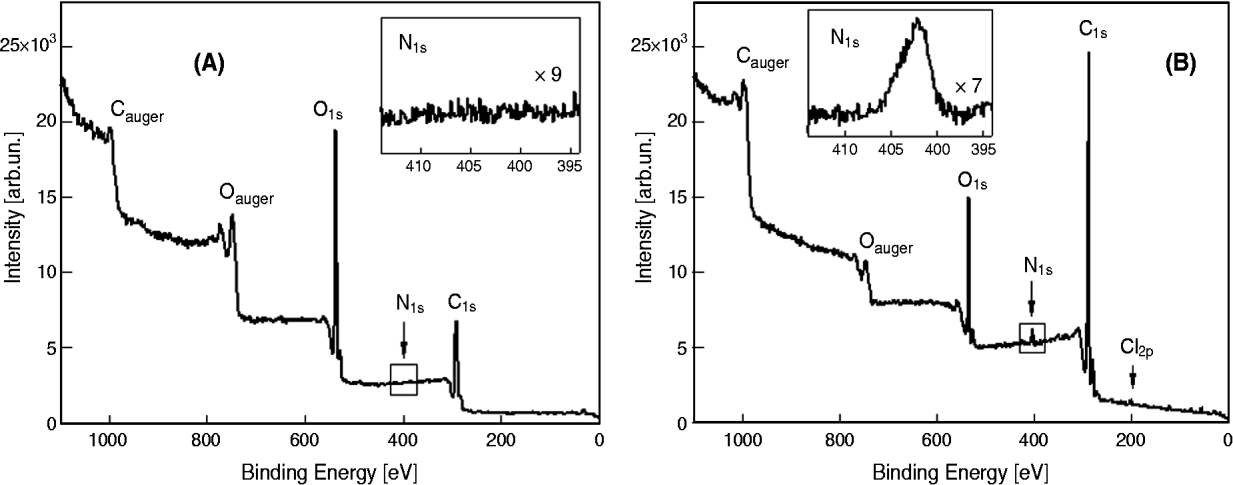
The surface changes of the modified Np were analysed using ESCA. The Np labelled with TMR were excluded from the analysis because of the presence of the rhodamine molecule, which contains N and thus creates a false positive. The surface of PLGA Np (B-Np, Fig. 1A) was characterized by the presence of carbon (C) and oxygen (O) (along with hydrogen, not detectable by ESCA). By contrast, the modified Np (Ab-B-Np, Fig. 1B) showed an increase in the signal of nitrogen (N), which was related to the surface modification resulting from the antibody linkage, demonstrating the success of the modification procedures. Table 1 shows the ratios of O to C and of N to C, which have been demonstrated to be good indicators of the rate of derivatization (Costantino et al. Reference Costantino, Gandolfi, Bossy-Nobs, Tosi, Gurny, Rivasi, Vandelli and Forni2006).
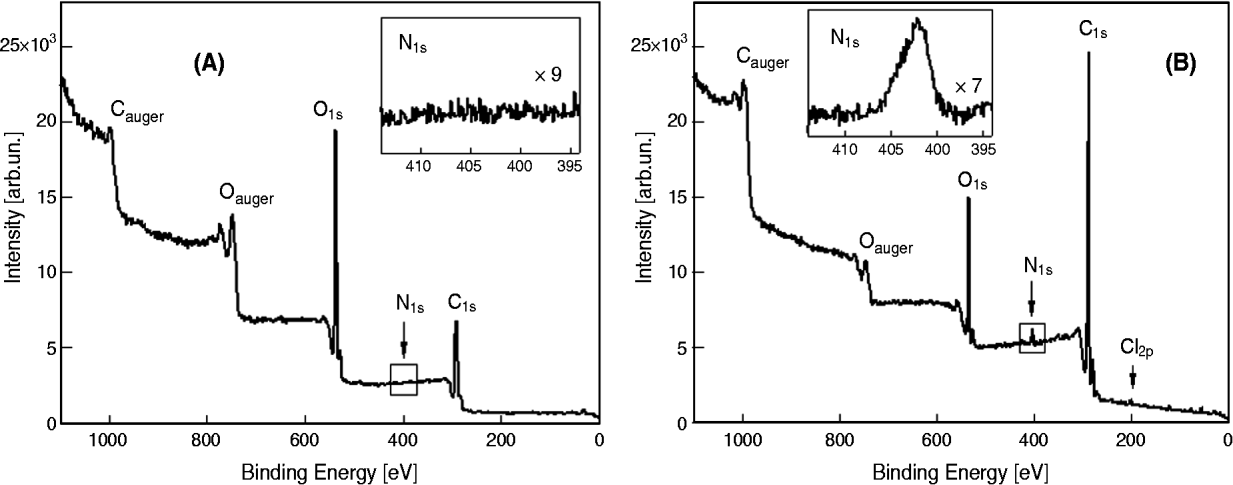
Fig. 1. Electron spectroscopy for chemical analysis spectrum of the unloaded nanoparticles (A) and the antibody-modified unloaded nanoparticles obtained (B). O1s and C1s signals represent the relative amount of, respectively, oxygen and carbon present in the surface layer analysed. Cauger and Oauger signals are referred to sample preparation. In the small box, N1s represents the relative amount intensity of nitrogen present in the considered surface layer.
In vitro study of Indinivar release
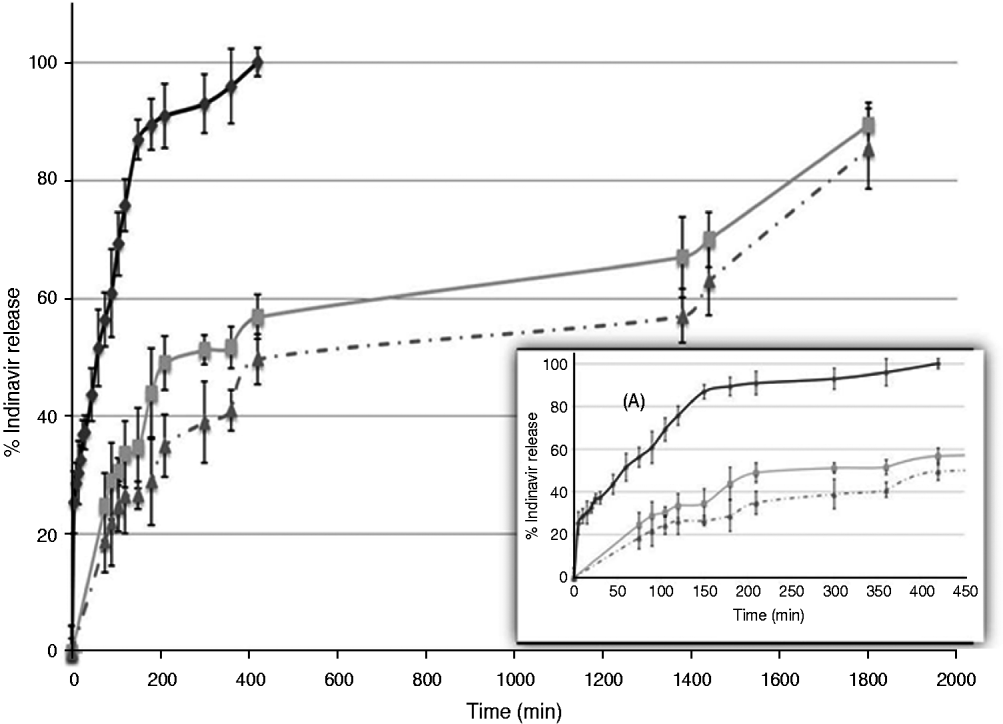
The IND-Np formulations produced a biphasic IND release profile, with an initial burst effect in which 24% of the total entrapped IND was released, over 75 min. In the second phase, a sustained controlled IND release was detected: 56% of the total IND was released after 7 h and 90% after 30 h. The free IND, used as control, was solubilized over 180 min (90%) (Fig. 2).
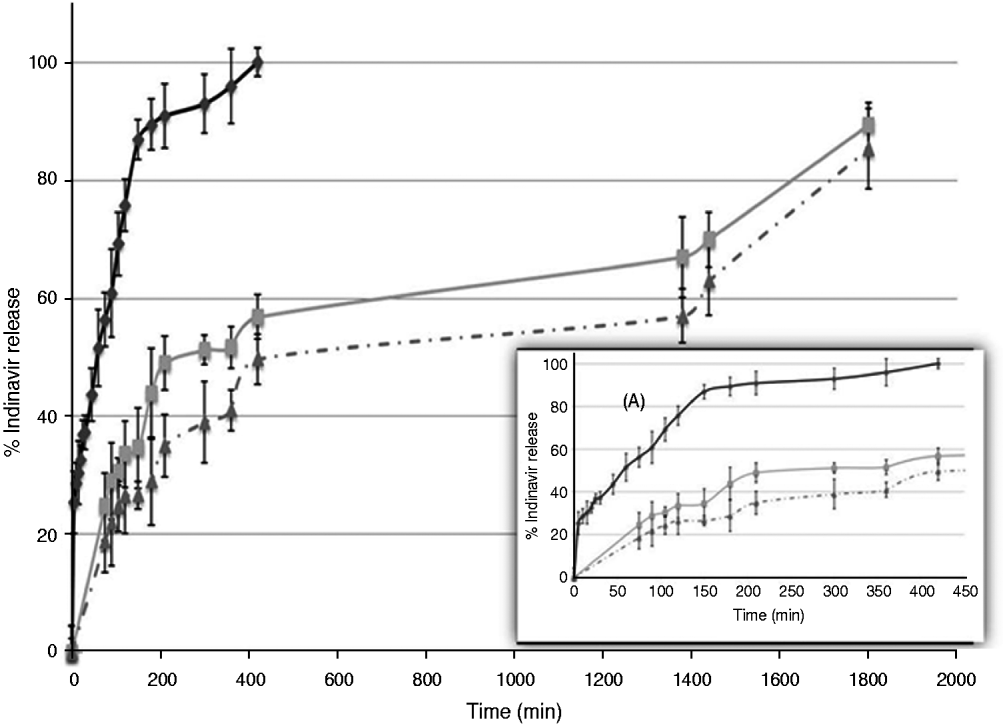
Fig. 2. In vitro Indinavir release from unmodified loaded nanoparticles (grey line), from antibody-modified drug-loaded nanoparticles (broken grey line) and free drug (black line). Box A, magnification of the same in vitro release, considering the first 420 min. The results are expressed as the mean of at least 5 experiments.
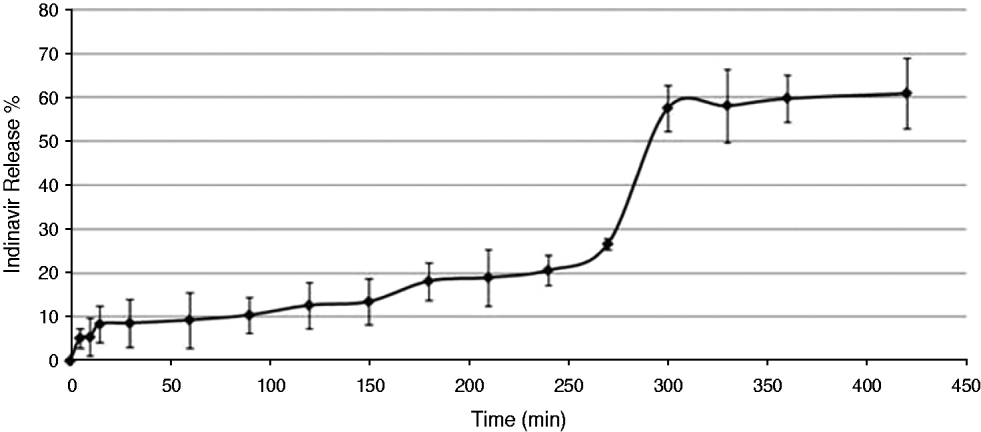
The release of IND during the 7 h required for antibody modification was measured in MES buffer (Fig. 3). Almost 60% of the IND was released from Np over this time, a finding that was confirmed by the loading content experiments, which showed that the IND content in the modified Np was half of the content in the unmodified Np.

Fig. 3. In vitro release of Indinavir (IND) from IND-loaded nanoparticles in 2-(N-morpholino) ethanesulfonic acid (MES buffer). The results are expressed as the mean of at least 3 experiments.
Affinity of the TMR-Ab- B-Np
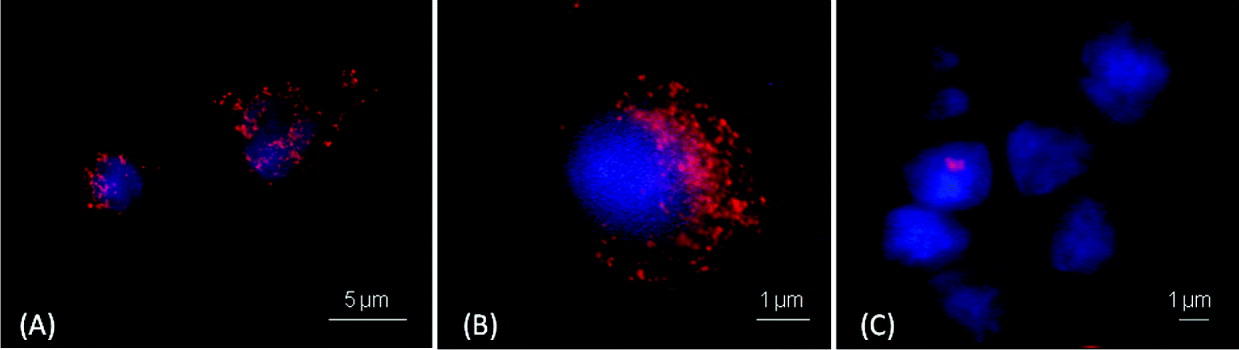
The results of the immunofluorescence assays regarding the affinity of the TMR-Ab-B-Np and TMR-B-Np to the target-C. parvum are shown in Fig. 4. TMR-Ab-B-Np covered part of the cell cytoplasm (Fig. 4A and B), whereas the Np that was not engineered by Ab (TMR-B-Np) did not exhibit the same affinity (Fig. 4C).
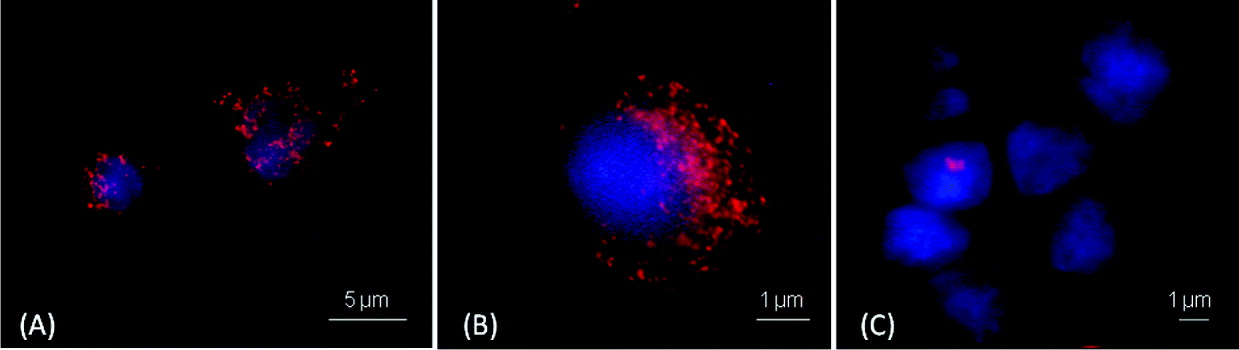
Fig. 4. Immunofluorescence assays demonstrating the affinity of the tetramethylrhodamine (TMR)-labelled antibody-engineered nanoparticles (A and B) and TMR-labelled non-antibody engineered nanoparticles (C) for the target-Cryptosporidium parvum-infected HCT-8 cells. Red spots represent the nanoparticles labelled with TMR. Nuclei of the target-C. parvum-infected HCT-8 cells are stained blue.
Anti-C. parvum activity of Ab-TMR-IND-Np in HCT-8 cells
The treatment with 50 μM Ab-TMR-IND-Np, added to the culture at the same time as the excysted oocysts, resulted in complete inhibition of the infection. The parasites were no longer able to infect the cells; in fact, the histograms of the intensity of fluorescence for FITC were similar in non-infected cells and Ab-TMR-IND-Np-treated infected cells. The same results were obtained when IND alone was used (data not shown).
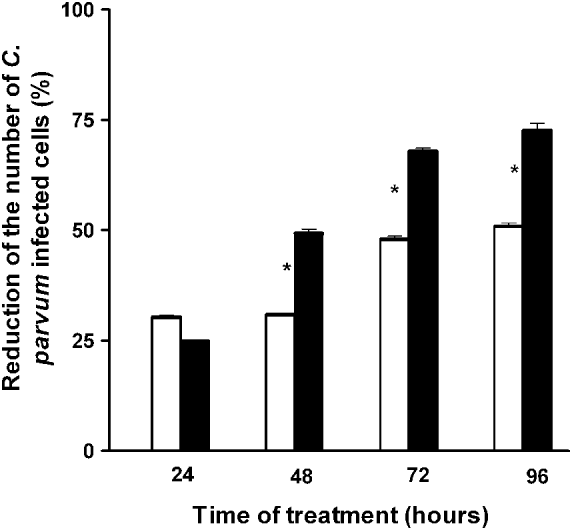
When IND was added to the C. parvum-infected cells, the extent to which the infection decreased was found to be dependent on the duration of treatment. Treatment with Ab-TMR-IND-Np or with free IND for up to 24 h (i.e., 48 h of culture) induced a non-significant reduction in the percentage of infected cells (25% and 30%, respectively). After 48 h of treatment, there was a 51% reduction in the population of infected cells treated with Ab-TMR-IND-Np and a 31% reduction in cells treated with IND alone; that is, the percentage of infected cells treated with Ab-TMR-IND-Np was significantly lower than the percentage of infected cells treated with free IND (P<0·01). After 72 h, there was a 67% reduction in infected cells treated with Ab-TMR-IND-Np and a 48% reduction in the infected cells treated with IND alone. After 96 h, the percentages were 70% and 50%, respectively (Fig. 5).
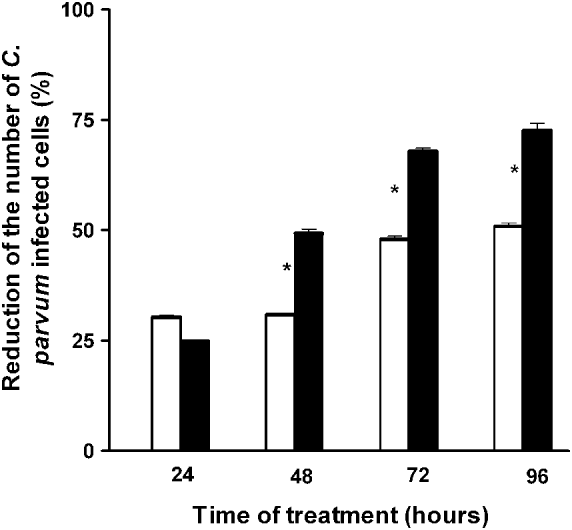
Fig. 5. Histogram of the reduction (percentage) of the number of Cryptosporidium parvum-infected HCT-8 cells after treatment with 50 μ m Indinavir (open bars) or 50 μ m antibody-modified loaded Indinavir nanoparticles (solid bars) for 24, 48, 72 and 96 h. * P<0·01.
DISCUSSION
According to the results of this study, IND embedded in an antibody-modified polymeric Np, compared to IND alone, exerts a higher inhibitory effect on the growth of C. parvum in HCT-8 infected cells, and the extent of this effect depends on the duration of treatment. Other studies have demonstrated that IND embedded in sterically stabilized immunoliposomes increases the drug's accumulation in lymph nodes up to 126 times, compared to free IND, increasing the efficiency of treatment for HIV infection (Gagné et al. Reference Gagné, Désormeaux, Perron, Tremblay and Bergeron2002). However, considering C. parvum targeting, neither active-strategies nor an antibody-conjugation have so far been applied as drug delivery approaches.
Based on the morphological and chemical-physical analyses, the antibody-engineered Np were found to be well formed, with nanometric size and negative surface charges; moreover, ESCA demonstrated that these Np have antibody on their surface. The most marked differences between surface-modified and unmodified Np were observed in z-p (−15·6 to −9·2 in modified Np versus −46·8 to −35·9 in unmodified Np); this difference could be explained by the different antibody surface coverage of antibody-engineered Np compared to unmodified Np, in which carboxyl groups are not-conjugated and thus exposed onto the Np surface, giving a negative surface charge effect. This finding could be considered as further evidence of the efficiency of Np surface modification with Ab. The encapsulation of IND in the Np proved to be highly efficient, allowing low doses of the drug to be administered, thus decreasing toxicity. The in vitro analysis of IND release in phosphate buffer confirmed the controlled release of the drug over time and an initial burst effect, which was probably associated with IND absorption onto the Np surface and/or embedded near the surface. It is important to note that a decreased initial burst effect was observed in association with the loss of adsorbed IND during the modification procedure (Cohen et al. Reference Cohen, Yoshioka, Lucarelli, Huang and Langer1991; Sah et al. Reference Sah, Toddywala and Chien1994; Uchida et al. Reference Uchida, Yagi, Oda and Goto1996; Batycky et al. Reference Batycky, Hanes, Langer and Edwards1997; Rafati et al. Reference Rafati, Coombes, Adler, Holland and Davis1997; Huang et al. Reference Huang, Chestang and Brazel2002). A similar trend of IND release was observed for Ab-IND-Np, confirming that the surface modification of the Np did not affect the IND release. The additional labelling of the Np with TMR allowed the Np to be localized in the cell cultures and showed the affinity of the antibody-engineered labelled Np (Ab-TMR-B-Np or Ab-TMR-IND-Np) for the oocysts, when added to the cell culture.
Other studies have demonstrated the effect of IND in decreasing the percentage of infected cells, whether alone (Mele et al. Reference Mele, Gómez-Morales, Tosini and Pozio2003) or in combination with paromomycin (Hommer et al. Reference Hommer, Eichholz and Petry2003), suggesting that IND mainly affects the free stages of the parasite, acting on the proteinase activity (Mele et al. Reference Mele, Gómez-Morales, Tosini and Pozio2003). In general, the Ab-TMR-IND-Np were, in our study, more effective against C. parvum in comparison with free IND. However, during the first 24 h of treatment, no statistical difference was observed between the percentage of infected cells treated with IND and those treated with Ab-TMR-IND-Np. This could be explained by the time needed for the release of the embedded drug, which at 30 h was still at 60%. By contrast, at 48, 72 and 96 h p.i., there was a greater reduction in infected cells after Ab-TMR-IND-Np treatment; at these times, the process of drug release had already been completed. Moreover, the reduction in infected cells could be explained by the ability of antibody-engineered Np to interact with high specificity with C. parvum, as a function of immune-recognition which was also demonstrated by the fluorescent images which clearly showed how antibody-engineered labelled Np ‘attack’ the target C. parvum-infected cells. Thus, once in contact with the target, the IND released from Np could be more effective than when it is free in the solution. Another hypothesis for explaining the high effectiveness of the antibody-engineered Np loaded with IND in decreasing the infection could be the possible entry of Np into the cells. Several studies (e.g., Harush-Frenkel et al. Reference Harush-Frenkel, Altschuler and Benita2008) have demonstrated that small nanoparticulate systems (polymeric and lipidic) can enter the cell by crossing the cell membranes.
The finding that antibody-engineered Np loaded with IND are able to target C. parvum in infected cells could represent a novel therapeutic strategy against Cryptosporidium sp. infection. Moreover, the Np as an IND delivery device, allows the development of a more appropriate dose formulation reducing the IND side effects.
ACKNOWLEDGMENTS
We are very grateful to Mr Daniele Tonanzi and Mr Marco Amati (Istituto Superiore di Sanità, Rome, Italy) for their invaluable technical assistance. We also thank Dr Andrea Tombesi and Cinzia Restani (Centro Interdipartimentale Grandi Strumenti, CIGS, University of Modena and Reggio Emilia, Italy) for their advice in fluorescence analysis.
FINANCIAL SUPPORT
This work was supported by the Istituto Superiore di Sanità.