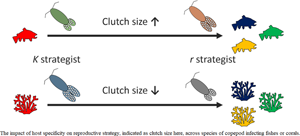
Introduction
Nature has produced, through natural selection, an astounding array of strategies that improve the fitness of animals. These strategies, which include innate life history traits and learned behaviours, stem from the trade-off between energy available in the immediate environment and the phenotypic constraints imposed by evolution (Roff, Reference Roff2002). Such trade-offs directly impact the allocation of energy towards reproductive output, which is usually measured in terms of associated trait values (Stearns, Reference Stearns1992), e.g. the number and size of offspring. The balance between the impact of past selective pressures and current resource availability ultimately shapes our description of species-wide reproductive traits (de Queiroz, Reference de Queiroz2007). A classical model that characterizes these traits between species is the r/K selection theory (Reznick et al., Reference Reznick, Bryant and Bashey2002), which relates to the selection of traits that regulate population density, also known as density-dependent selection (Bertram and Masel, Reference Bertram and Masel2019). Conceptually, a continuum can be drawn between r-selected species that have a capacity for rapid increase in population size and K-selected species that have populations close to environmental carrying capacity (MacArthur and Wilson, Reference MacArthur and Wilson1967), i.e. low and high density-dependent selection, respectively. Therefore, r-selected species are predicted to minimize self-preservation and invest more energy towards reproductive output, while K-selected species are predicted to maximize self-preservation and invest less energy towards reproductive output (Pianka, Reference Pianka1970). Other traits such as growth rate and longevity also differ between r- and K-selected species, e.g. r-selected species tend to have faster growth rates and are relatively short-lived. Albeit a simplified view of natural selection (Stearns, Reference Stearns1977; Parry, Reference Parry1981), one should be able to quantify the effects of density-dependent selection on the reproductive strategies of species in an r/K continuum (Mueller et al., Reference Mueller, Guo and Ayala1991; Reznick et al., Reference Reznick, Bryant and Bashey2002).
For instance, density-dependent selection impacts many ecological traits, including habitat specialization and the life history traits associated with reproductive strategy (Morris, Reference Morris1987; Bonte et al., Reference Bonte, Van Dyck, Bullock, Coulon, Delgado, Gibbs, Lehouck, Matthysen, Mustin, Saastamoinen, Schtickzelle, Stevens, Vandewoestijne, Baguette, Barton, Benton, Chaput-Bardy, Clobert, Dytham, Hovestadt, Meier, Palmer, Turlure and Travis2012; van Beest et al., Reference van Beest, Uzal, Vander Wal, Laforge, Contasti, Colville and McLoughlin2014; Büchi and Vuilleumier, Reference Büchi and Vuilleumier2016), i.e. the number and size of offspring. It has been shown that habitat specialists exhibit reproductive traits akin to what is expected in K-selected species, e.g. low fecundity and greater energetic investment per offspring. Conversely, habitat generalists appear to show r-selected species traits (McKinney, Reference McKinney1997; Quadros et al., Reference Quadros, Caubet and Araujo2009), e.g. low-energetic investment per offspring and greater fecundity. Studies relating habitat specialization to reproductive strategy have focused mainly on free-living species (Espirito-Santo et al., Reference Espirito-Santo, Rodriguez and Zuanon2013; Patrick and Weimerskirch, Reference Patrick and Weimerskirch2017) and little attention has been directed towards parasitic organisms (Esch et al., Reference Esch, Hazen, Aho and Esch1977; Koprivnikar and Randhawa, Reference Koprivnikar and Randhawa2013). For a parasite, the level of habitat specialization can be measured by the number of host species it infects at a particular life stage, i.e. host specificity (Poulin and Mouillot, Reference Poulin and Mouillot2003; Agosta et al., Reference Agosta, Janz and Brooks2010). Therefore, a parasite that infects only one or a few host species is highly specialized, whereas a parasite capable of infecting a great number of host species is a generalist. If host specificity evolved under similar density-dependent selective pressures as for habitat specialization in free-living species, one would expect a similar r/K-style continuum between highly host-specific and generalist parasites. In this case, specialist parasites are expected to invest relatively little energy in reproductive output (K-selected), while generalist parasites are expected to invest in it heavily (r-selected). For oviparous parasites, one could therefore predict that K-selected parasites typically invest in the quality of eggs (a relatively small number of large, energy-rich eggs), while r-selected parasites are expected to invest in the quantity of eggs (a relatively large number of small, energy-poor eggs).
In line with the idea that a stable environment over evolutionary time can select for habitat specialists with narrow physiological tolerances (Futuyma and Moreno, Reference Futuyma and Moreno1988), host specificity in parasites could have evolved under similar governing principles (Krasnov et al., Reference Krasnov, Mouillot, Shenbrot, Khokhlova and Poulin2011; Strona, Reference Strona2015). In order for a host–parasite association to be established, both antagonistic species would have to persist within the same environment over an evolutionary timescale and the host would have to be physiologically compatible for the parasite to develop (Combes, Reference Combes, Toft, Aeschlimann and Bolis1991). If new potential host species immigrate to a particular environment, selection may favour one of two likely outcomes. On the one hand, parasites may evolve traits for locating and infecting only certain compatible host species (high host specificity). On the other, selection may favour parasites capable of exploiting many different hosts, akin to a ‘jack of all trades and master of none’ (low host specificity) (Poulin, Reference Poulin2007). Therefore, the stability of the environment through time, which dictates the availability of compatible host species, should ultimately shape the host specificity of a parasite. Here, we define environmental stability as the local turnover rate of hosts within an ecosystem, which can be measured as the change of host richness per unit of time (Poulin et al., Reference Poulin, Krasnov and Mouillot2011). Accordingly, low environmental stability with high potential for host species turnover in an ecological time scale should favour low host specificity (generalist parasites), whereas high environmental stability with little potential for host species turnover should favour high host specificity (specialist parasites) (Fig. 1).
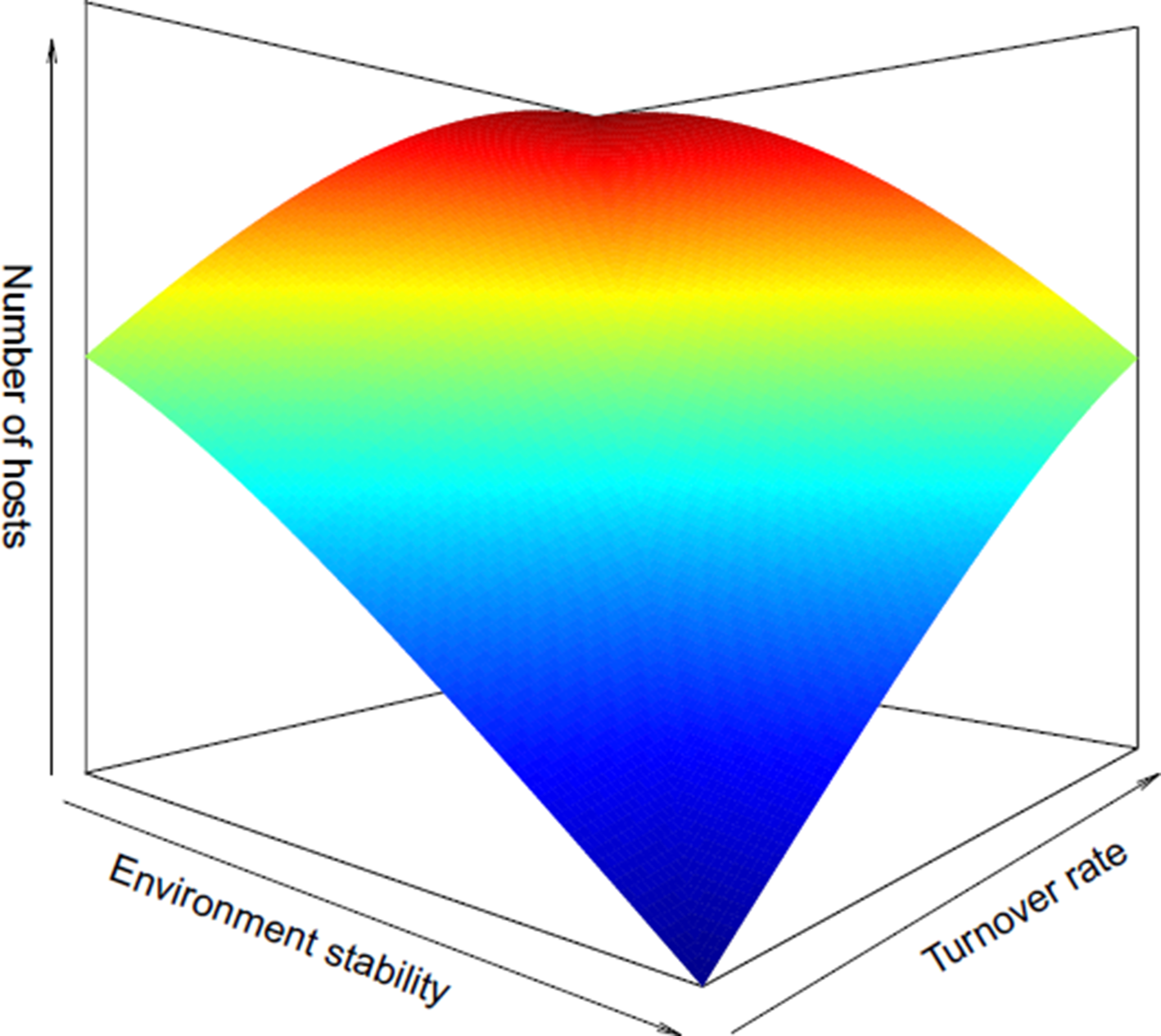
Fig. 1. Theoretical continuum of the potential main processes influencing parasitic copepod reproductive strategies in relation to host specificity. High environmental stability promotes low turnover rates of potential hosts, likely resulting in high host specificity. These conditions should select for r-selected traits in parasites, e.g. high fecundity and lower self-maintenance. In contrast, low environmental stability equals high turnover rates of potential hosts, which could select for parasites with low host specificity. These conditions should select for K-selected traits in parasites, e.g. low fecundity and higher self-maintenance.
General life history traits, including those associated with reproductive strategy, have been studied only in a few parasitic groups (Poulin, Reference Poulin1995, Reference Poulin, Baker, Muller and Rollinson1996; Morand, Reference Morand1996; Poulin and Hamilton, Reference Poulin and Hamilton1997; Whittington, Reference Whittington1997). These comparative analyses looked at interspecific variations in egg production of both endo- and ectoparasites. While such comparisons highlight the differences in reproductive strategy between free-living and parasitic species or between parasites that infect different host groups (e.g. vertebrate or invertebrate hosts) (Poulin, Reference Poulin1995), there is little information available on the impact of host specificity on the reproductive strategies of parasites. It has been suggested that endoparasites are simultaneously r- and K-selected, for they develop in an energy-rich environment (the host) that promotes the evolution of both self-maintenance and high reproductive output (Cavaleiro and Santos, Reference Cavaleiro and Santos2014). For instance, endoparasitic nematodes were found to have no particular trade-off between host specificity and the traits associated with reproductive strategy (Koprivnikar and Randhawa, Reference Koprivnikar and Randhawa2013). As for ectoparasites, which live on their host and are thus more exposed to the environment, it has been argued that selection favours the evolution of traits related to K-selected species, implying low reproductive output (Jennings and Calow, Reference Jennings and Calow1975). Such is the case for certain ectoparasitic ticks that appear to invest energy into a relatively small number of eggs (Schmidtmann, Reference Schmidtmann, Sonenshine and Mather1994). However, parasites vary widely in their life histories and transmission routes and the limited scope, both taxonomically and geographically, of previous studies makes it difficult to draw sound conclusions on the relation between host specificity and parasite reproductive strategies.
Parasitic copepods are ideal subjects for addressing the impacts of host specificity on the reproductive strategies of parasites because these small crustaceans can be found infecting a great number of aquatic organisms including fishes, corals, starfishes and other invertebrates (Kabata, Reference Kabata1981; Stock, Reference Stock1988). In addition, there are both ecto- and endoparasitic species of copepods, and they can also be either be very host specific or generalist, infecting anywhere from one host species to several dozens (Horton et al., Reference Horton, Kroh, Ahyong, Bailly, Boyko, Brandão, Gofas, Hooper, Hernandez, Holovachov, Mees, Molodtsova, Paulay, Decock, Dekeyzer, Lanssens, Vandepitte, Vanhoorne, Adlard, Adriaens, Agatha, Ahn, Akkari, Alvarez, Anderberg, Anderson, Angel, Antic, Arango, Artois, Atkinson, Baldwin, Bank, Barber, Barbosa, Bartsch, Bellan-Santini, Bergh, Bernot, Berta, Bezerra, Bieler, Blanco, Blasco-Costa, Blazewicz, Bock, Bonifacino de León, Böttger-Schnack, Bouchet, Boury-Esnault, Boxshall, Bray, Bruce, Cairns, Calvo Casas, Carballo, Cárdenas, Carstens, Chan, Chan, Cheng, Churchill, Coleman, Collins, Collins, Corbari, Cordeiro, Cornils, Coste, Costello, Crandall, Cremonte, Cribb, Cutmore, Dahdouh-Guebas, Daly, Daneliya, Dauvin, Davie, De Broyer, De Grave, de Mazancourt, de Voogd, Decker, Decraemer, Defaye, d'Hondt, Dippenaar, Dohrmann, Dolan, Domning, Downey, Ector, Eisendle-Flöckner, Eitel, Encarnação, Enghoff, Epler, Ewers-Saucedo, Faber, Feist, Figueroa, Finn, Fišer, Fordyce, Foster, Frank, Fransen, Freire, Furuya, Galea, Gao, Garcia-Alvarez, Garcia-Jacas, Garic, Garnett, Gasca, Gaviria-Melo, Gerken, Gibson, Gibson, Gil, Gittenberger, Glasby, Glover, Gómez-Noguera, González-Solís, Gordon, Gostel, Grabowski, Gravili, Guerra-García, Guidetti, Guiry, Gutierrez, Hadfield, Hajdu, Hallermann, Hayward, Heiden, Hendrycks, Herbert, Herrera Bachiller, Ho, Hodda, Høeg, Hoeksema, Houart, Hughes, Hyžný, Iniesta, Iseto, Ivanenko, Iwataki, Janssen, Jarms, Jaume, Jazdzewski, Jersabek, Jóźwiak, Kabat, Kantor, Karanovic, Karthick, Katinas, Kim, King, Kirk, Klautau, Kociolek, Köhler, Kolb, Kotov, Kremenetskaia, Kristensen, Kulikovskiy, Kullander, Lambert, Lazarus, Le Coze, LeCroy, Leduc, Lefkowitz, Lemaitre, Liu, Loeuille, Lörz, Lowry, Ludwig, Lundholm, Macpherson, Madin, Mah, Mamo, Mamos, Manconi, Mapstone, Marck, Marek, Marshall, Marshall, Martin, Mast, McFadden, McInnes, Meidla, Meland, Merrin, Messing, Miljutin, Mills, Moestrup, Mokievsky, Monniot, Mooi, Morandini, Moreira da Rocha, Morrow, Mortelmans, Mortimer, Musco, Nesom, Neubauer, Neubert, Neuhaus, Ng, Nguyen, Nguyen Thi My, Nielsen, Nishikawa, Norenburg, O'Hara, Opresko, Osawa, Osigus, Ota, Páll-Gergely, Panero, Pasini, Patterson, Paxton, Pelser, Peña-Santiago, Perrier, Perrin, Petrescu, Picton, Pilger, Pisera, Polhemus, Poore, Potapova, Pugh, Read, Reich, Reimer, Reip, Reuscher, Reynolds, Richling, Rimet, Ríos, Rius, Rogers, Roque, Rosenberg, Rützler, Sabbe, Saiz-Salinas, Sala, Santagata, Santos, Sar, Satoh, Saucède, Schatz, Schierwater, Schilling, Schmidt-Rhaesa, Schneider, Schönberg, Schuchert, Senna, Serejo, Shaik, Shamsi, Sharma, Shear, Shenkar, Shinn, Short, Sicinski, Sierwald, Simmons, Sinniger, Sivell, Sket, Smit, Smit, Smol, Souza-Filho, Spelda, Sterrer, Stienen, Stoev, Stöhr, Strand, Suárez-Morales, Summers, Suppan, Susanna, Suttle, Swalla, Taiti, Tanaka, Tandberg, Tang, Tasker, Taylor, Taylor, Tchesunov, ten Hove, ter Poorten, Thomas, Thuesen, Thurston, Thuy, Timi, Timm, Todaro, Turon, Tyler, Uetz, Uribe-Palomino, Urtubey, Utevsky, Vacelet, Vachard, Vader, Väinölä, Valls Domedel, Van de Vijver, van der Meij, van Haaren, van Soest, Vanreusel, Venekey, Vinarski, Vonk, Vos, Walker-Smith, Walter, Watling, Wayland, Wesener, Wetzel, Whipps, White, Wieneke, Williams, Williams, Wilson, Witkowski, Witkowski, Wyatt, Wylezich, Xu, Zanol, Zeidler and Zhao2020). Finally, there exists an abundance of original species descriptions of parasitic copepods that include mean life history trait values such as female body size, clutch size and egg size. As opposed to other parasites with internal brood pouches, copepod eggs can be easily collected for counting and measuring from the external egg sacs produced by females (Kabata, Reference Kabata1981). Therefore, such descriptions can provide a number of species-wide reproductive trait values for a large number of parasitic species.
Considering how little is known of the impacts that host specificity, a consequence of environmental stability, has on the reproductive strategies of parasites, here we address this issue using a worldwide dataset consisting of host species records for fish-infecting copepods [generally ectoparasitic (Kabata, Reference Kabata1981)] and coral-infecting copepods [generally endoparasitic (Humes, Reference Humes1985)]. If host specificity for parasites evolves under similar selective pressures as for habitat specialization in free-living species, we hypothesize that the traits associated with reproductive strategy (clutch size and egg size) in both these groups have evolved comparably. Using the r/K continuum usually associated with free-living species, we first predict that specialist parasites (high host specificity) have reproductive traits similar to those of K-selected species and that generalist parasites (low host specificity) have reproductive traits akin to those of r-selected species. Second, with respect to the type of parasitism (endo- and ectoparasitism), we predict that ectoparasitic copepods tend to have traits associated with K-selected species. This study provides deeper insight into the evolution of reproductive strategies in parasites under the selective constraints imposed by host specificity and, ultimately, the stability of the environment.
Materials and methods
Copepod species dataset
Parasitic copepod species reproductive traits, henceforth simplified to copepod traits, were compiled from a dataset consisting of various life history traits from original copepod species descriptions (Poulin, Reference Poulin1995). All fish-infecting and coral-infecting copepod species were considered, except for members of the family Pennellidae, which have an indirect life cycle as opposed to other parasitic copepods, i.e. they parasitize more than one host during their life (Kabata, Reference Kabata1981); removing pennellids thus eliminates a confounding factor in the analysis. Since the original dataset was created in 1995, we updated the accepted species names using the World Register of Marine Species (WoRMS) (Horton et al., Reference Horton, Kroh, Ahyong, Bailly, Boyko, Brandão, Gofas, Hooper, Hernandez, Holovachov, Mees, Molodtsova, Paulay, Decock, Dekeyzer, Lanssens, Vandepitte, Vanhoorne, Adlard, Adriaens, Agatha, Ahn, Akkari, Alvarez, Anderberg, Anderson, Angel, Antic, Arango, Artois, Atkinson, Baldwin, Bank, Barber, Barbosa, Bartsch, Bellan-Santini, Bergh, Bernot, Berta, Bezerra, Bieler, Blanco, Blasco-Costa, Blazewicz, Bock, Bonifacino de León, Böttger-Schnack, Bouchet, Boury-Esnault, Boxshall, Bray, Bruce, Cairns, Calvo Casas, Carballo, Cárdenas, Carstens, Chan, Chan, Cheng, Churchill, Coleman, Collins, Collins, Corbari, Cordeiro, Cornils, Coste, Costello, Crandall, Cremonte, Cribb, Cutmore, Dahdouh-Guebas, Daly, Daneliya, Dauvin, Davie, De Broyer, De Grave, de Mazancourt, de Voogd, Decker, Decraemer, Defaye, d'Hondt, Dippenaar, Dohrmann, Dolan, Domning, Downey, Ector, Eisendle-Flöckner, Eitel, Encarnação, Enghoff, Epler, Ewers-Saucedo, Faber, Feist, Figueroa, Finn, Fišer, Fordyce, Foster, Frank, Fransen, Freire, Furuya, Galea, Gao, Garcia-Alvarez, Garcia-Jacas, Garic, Garnett, Gasca, Gaviria-Melo, Gerken, Gibson, Gibson, Gil, Gittenberger, Glasby, Glover, Gómez-Noguera, González-Solís, Gordon, Gostel, Grabowski, Gravili, Guerra-García, Guidetti, Guiry, Gutierrez, Hadfield, Hajdu, Hallermann, Hayward, Heiden, Hendrycks, Herbert, Herrera Bachiller, Ho, Hodda, Høeg, Hoeksema, Houart, Hughes, Hyžný, Iniesta, Iseto, Ivanenko, Iwataki, Janssen, Jarms, Jaume, Jazdzewski, Jersabek, Jóźwiak, Kabat, Kantor, Karanovic, Karthick, Katinas, Kim, King, Kirk, Klautau, Kociolek, Köhler, Kolb, Kotov, Kremenetskaia, Kristensen, Kulikovskiy, Kullander, Lambert, Lazarus, Le Coze, LeCroy, Leduc, Lefkowitz, Lemaitre, Liu, Loeuille, Lörz, Lowry, Ludwig, Lundholm, Macpherson, Madin, Mah, Mamo, Mamos, Manconi, Mapstone, Marck, Marek, Marshall, Marshall, Martin, Mast, McFadden, McInnes, Meidla, Meland, Merrin, Messing, Miljutin, Mills, Moestrup, Mokievsky, Monniot, Mooi, Morandini, Moreira da Rocha, Morrow, Mortelmans, Mortimer, Musco, Nesom, Neubauer, Neubert, Neuhaus, Ng, Nguyen, Nguyen Thi My, Nielsen, Nishikawa, Norenburg, O'Hara, Opresko, Osawa, Osigus, Ota, Páll-Gergely, Panero, Pasini, Patterson, Paxton, Pelser, Peña-Santiago, Perrier, Perrin, Petrescu, Picton, Pilger, Pisera, Polhemus, Poore, Potapova, Pugh, Read, Reich, Reimer, Reip, Reuscher, Reynolds, Richling, Rimet, Ríos, Rius, Rogers, Roque, Rosenberg, Rützler, Sabbe, Saiz-Salinas, Sala, Santagata, Santos, Sar, Satoh, Saucède, Schatz, Schierwater, Schilling, Schmidt-Rhaesa, Schneider, Schönberg, Schuchert, Senna, Serejo, Shaik, Shamsi, Sharma, Shear, Shenkar, Shinn, Short, Sicinski, Sierwald, Simmons, Sinniger, Sivell, Sket, Smit, Smit, Smol, Souza-Filho, Spelda, Sterrer, Stienen, Stoev, Stöhr, Strand, Suárez-Morales, Summers, Suppan, Susanna, Suttle, Swalla, Taiti, Tanaka, Tandberg, Tang, Tasker, Taylor, Taylor, Tchesunov, ten Hove, ter Poorten, Thomas, Thuesen, Thurston, Thuy, Timi, Timm, Todaro, Turon, Tyler, Uetz, Uribe-Palomino, Urtubey, Utevsky, Vacelet, Vachard, Vader, Väinölä, Valls Domedel, Van de Vijver, van der Meij, van Haaren, van Soest, Vanreusel, Venekey, Vinarski, Vonk, Vos, Walker-Smith, Walter, Watling, Wayland, Wesener, Wetzel, Whipps, White, Wieneke, Williams, Williams, Wilson, Witkowski, Witkowski, Wyatt, Wylezich, Xu, Zanol, Zeidler and Zhao2020), visited in July 2019. All searches were performed using the ‘Advance Search’ option, which allows for the inclusion of both freshwater and marine species. Synonyms for each species found in the database, if any, were also added to the dataset. Within the updated dataset, some species had more than one entry with different copepod trait values, due either to multiple species descriptions or synonymy. For these cases, we calculated the median values of clutch size, egg size and female body size to obtain one entry per copepod species. The updated copepod species dataset can be found in the Supplementary material.
Copepod host species records
Using the updated copepod species dataset described above, we first searched for host species records in WoRMS using the current accepted copepod species name in the ‘Advance Search’ option. We only considered host records with a full star, i.e. entries verified by a taxonomic editor. For a host record and its reference to be included in the dataset, it had to be an original record of a host–parasite association using new host material, therefore most catalogues and reviews were excluded. Once a list of host species and its references for a given copepod species was obtained from WoRMS, it was cross-checked with Web of Science (WoS). The search in WoS was performed using the accepted copepod species name and all of its synonyms. To capture the most relevant publications, we used quotation marks around each species name along with the Boolean operator ‘OR’ between each synonymous species name (e.g. ‘Parabrachiella mirifica’ OR ‘Neobrachiella mirifica’) using the Topic Field search option. From these search results, only original records of host–parasite associations were compiled, just as for the host records acquired from WoRMS. We updated the host species names with the current accepted names found in WoRMS. With the results from both databases, we added the number of host species and the accompanying number of references for each copepod species to the dataset.
Testing for phylogenetic signals in copepod traits
In order to verify if the copepod trait values of clutch size and egg size depend on their phylogenetic relationships, i.e. if there is a phylogenetic signal, we attempted to reconstruct an evolutionary tree. Approximately half of the copepod species considered in this study do not have nucleotide sequences deposited in GenBank, therefore we tried to create a phylogenetic backbone following methods previously used for copepods (Khodami et al., Reference Khodami, McArthur, Blanco-Bercial and Arbizu2017). With this backbone, a post-MrBayes (Huelsenbeck and Ronquist, Reference Huelsenbeck and Ronquist2001) assessment was performed in the R package ‘PASTIS’ (Thomas et al., Reference Thomas, Hartmann, Jetz, Joy, Mimoto and Mooers2013). This package enables the inclusion of any species lacking genetic data through taxonomic placement constraints imposed on the initial consensus tree (backbone) produced by MrBayes. Unfortunately, after a series of runs each with different conditions, it was not possible to obtain a sensible consensus tree. These trees all contained one large polytomy that included all species, which is practically useless for calculating a phylogenetic signal (Molina-Venegas and Rodriguez, Reference Molina-Venegas and Rodriguez2017) (see the Supplementary material for more details on the phylogenetic analyses). Thus, to circumvent this problem, and to reduce the amount of variance due to evolutionary differences between species, we added copepod family as a random effect in the models (see below).
Statistical analyses
Correcting host specificity for study effort
Estimating host specificity is inherently difficult because of researcher biases and the variation in infection prevalence across communities and individual hosts, all of which can impact the likelihood of finding a parasite associated with a particular host (Dallas et al., Reference Dallas, Huang, Nunn, Park and Drake2017). Therefore, this important bias had to be corrected in the models. Methods exist to calculate the corrected host specificity of parasites, including those that account for the phylogenetic distance between hosts, which is also independent from study effort (Poulin and Mouillot, Reference Poulin and Mouillot2003). However, considering that our study focuses on the impact of the number of hosts rather than the phylogenetic relatedness between host species, we chose to correct for host records in the models. To do so, we used Bayesian multilevel modelling [‘brms’ package (Bürkner, Reference Bürkner2017)] with a negative binomial distribution to predict the influence of the number of references on the number of host records found. This type of modelling is ideal to analyse structured data in a Bayesian framework. The negative binomial distribution accounts for the overdispersion of data, thus avoiding biased estimates (Zuur et al., Reference Zuur, Ieno, Walker, Saveliev and Smith2009, Reference Zuur, Ieno and Elphick2010). The predict function was then used to fit the number of hosts based on the prediction of the model, creating a new variable hereafter referred to as the corrected number of hosts, which we use as a proxy for host specificity in the following models.
Testing the impact of host specificity on copepod reproductive strategies
Fish-infecting and coral-infecting copepods were analysed separately. Bayesian multilevel modelling was used to investigate the impact of host specificity on the reproductive strategies of copepods, i.e. clutch size and egg size. This method was selected because it is very efficient at sampling posterior distributions using the Hamiltonian Monte Carlo algorithm with the Stan sampling algorithm (a probabilistic programming language used in statistics). All models were constructed using a Gamma family distribution with a log link function as response variables were strictly positive and continuous, since some species trait values were calculated as medians from multiple synonymous species descriptions. We then built a series of candidate models for each response variable and each host type. As we were dealing with reproductive traits, every model was corrected for species average female body size, which was log transformed and scaled. The fixed effects with non-normal distributions were normalized with log transformations.
Model building and selection
Each model was built with priors (prior probability distribution) obtained from the get_prior function in the ‘brms’ package using four chains of 4000 iterations each (2000 for warmup and 2000 for sampling). The adapt_delta function was increased to 0.99 to lower the number of divergent transitions after warmup. We considered that the corrected number of hosts (the ‘fixed’ effect in the models) had an effect on a reproductive trait if the 95% credible interval did not overlap with zero. We ensured that every parameter in the model converged by verifying the potential scale reduction factor on split chains with the $\hat{R}$![]() convergence diagnostic (at convergence, $\hat{R}$
convergence diagnostic (at convergence, $\hat{R}$![]() is equal to one). Afterwards, stacking and pseudo-BMA (Bayesian model averaging) weights (used for model selection in Bayesian statistics) were computed with the PSIS-loo criterion using the ‘loo’ package (Vehtari et al., Reference Vehtari, Gelman and Gabry2017) to verify if the corrected number of hosts was better at fitting the posterior distribution than the model with female body size alone (hereafter referred to as the null model). All statistical analyses were performed in R version 4.0.0 (an annotated example of the statistical analysis is provided in the Supplementary material).
is equal to one). Afterwards, stacking and pseudo-BMA (Bayesian model averaging) weights (used for model selection in Bayesian statistics) were computed with the PSIS-loo criterion using the ‘loo’ package (Vehtari et al., Reference Vehtari, Gelman and Gabry2017) to verify if the corrected number of hosts was better at fitting the posterior distribution than the model with female body size alone (hereafter referred to as the null model). All statistical analyses were performed in R version 4.0.0 (an annotated example of the statistical analysis is provided in the Supplementary material).
Results
Copepod species dataset
A total of 100 fish-infecting copepod species and 50 coral-infecting copepod species were included in the dataset. Host species records obtained from the literature range from 1 to 52 in fish-infecting copepods and from 1 to 11 in coral-infecting copepods. Fish- and coral-infecting copepods are both found in the orders Cyclopoida and Siphonostomatoida, whereas only two species of coral-infecting copepods are found in the order Harpacticoida. For more details on the dataset compiled for this study, including references, see full datasets in the Supplementary material.
Fish-infecting copepods
For clutch size, model averaging using stacking and pseudo-BMA weights indicate that the most complex model, including female body size and the corrected number of hosts (posterior estimate = 0.140, estimate error = 0.070, 95% credible interval = 0.010–0.290), is better than the null model. Both selection methods produce values that are close to the recommended threshold of 0.5 (Vehtari et al., Reference Vehtari, Gelman and Gabry2017), however, as both stacking and pseudo-BMA indicators point in the same direction, we chose to present this output. According to the selected model, host specificity has a positive effect on clutch size (Fig. 2). As for egg size, host specificity also has a positive effect (posterior estimate = 0.078, estimate error = 0.047, 95% credible interval = −0.009 to 0.173). However, since the lower credible interval crosses zero and both stacking and pseudo-BMA methods add more weight to the null model, this indicates that host specificity adds little predictive value to copepod egg size (Fig. 2).
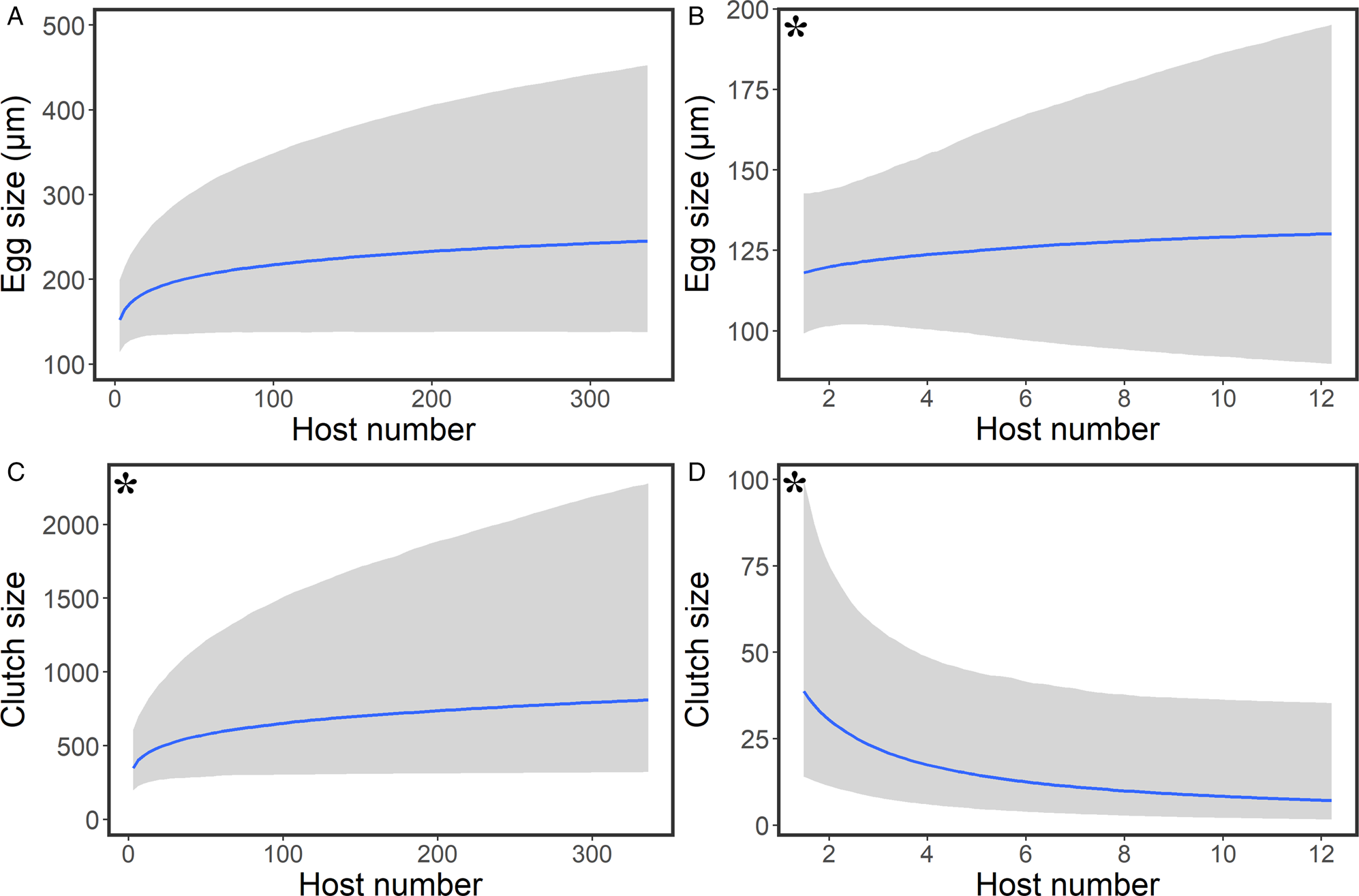
Fig. 2. Interspecific relationship (blue line with 95% credible intervals; shaded area) between host specificity and egg size for (a) fish-infecting copepods and (b) coral-infecting copepods and between host specificity and clutch size for (c) fish-infecting copepods and (d) coral-infecting copepods. An asterisk indicates the best possible model was selected using stacking and pseudo-BMA weights.
Coral-infecting copepods
For this set of models, female body size has no effect on the response variables. Therefore, this parameter was not included alongside the main fixed effects. Host specificity has a negative impact on clutch size (posterior estimate = −0.369, estimate error = 0.159, 95% credible interval = −0.664 to −0.033), and this model was selected by both stacking and pseudo-BMA weights (Fig. 2). In contrast, we found no effect of host specificity on egg size (posterior estimate = 0.023, estimate error = 0.047, 95% credible interval = −0.069 to 0.116) (Fig. 2).
Discussion
In this study, we show that host specificity impacts the reproductive strategies of both endo- and ectoparasitic copepods. We predicted that parasites, with respect to habitat specialization (host specificity), evolve to invest resources towards reproductive output just like free-living species do. We found that fish-infecting copepods have larger clutch sizes (r-selected trait) as host specificity decreases across species, controlling for copepod phylogeny with family as a random effect. This trend compares with free-living species; habitat generalists tend to exhibit r-selected traits (high reproductive output) as opposed to habitat specialists that tend to exhibit K-selected traits (low reproductive output). In contrast, coral-infecting copepods show the opposite pattern; species with low host specificity have relatively smaller clutch sizes (K-selected trait), and species with high host specificity have relatively larger clutch sizes (r-selected trait).
Copepods infecting fishes produce larger clutch sizes and egg sizes than copepods infecting corals. In this respect, ectoparasitic copepods of fishes do not appear to exhibit typical K-selected traits, which goes against our initial prediction. However, this does fall in line with the idea that larger parasites tend to produce more eggs of a larger size (Poulin, Reference Poulin, Baker, Muller and Rollinson1996, Reference Poulin2007), as many fish-infecting copepod species have larger body sizes compared to coral-infecting ones. This difference in size may be due to the limited space occupied by coral-infecting copepods living within the gastrovascular cavity of polyps (Humes, Reference Humes1985; Cheng and Dai, Reference Cheng and Dai2009). While this alone could explain why copepods of fish, which are larger and more resource-rich hosts in comparison with coral polyps, tend to produce more eggs and larger eggs, it has been suggested that host mobility plays a role in shaping the reproductive traits of parasites (Poulin, Reference Poulin2007). A mobile host, such as a fish, is likely more difficult to infect, simply because the copepod has to locate the fish and attach itself to it (Poulin, Reference Poulin1995). Coral-infecting copepods, however, likely experience a higher probability of transmission because corals form large colonies of immobile individuals. The selective pressures imposed by host mobility may favour higher fecundity (r-selected trait) in parasites of mobile hosts and lower fecundity (K-selected trait) in parasites of immobile hosts. This difference has already been discussed in previous studies on marine parasites (Bauer, Reference Bauer1994; Poulin, Reference Poulin1995). More recently, population genetics studies have shown that host traits such as mobility can impact the spatial genetic structure of parasite populations (Blasco-Costa et al., Reference Blasco-Costa, Waters and Poulin2012; Blasco-Costa and Poulin, Reference Blasco-Costa and Poulin2013), highlighting the potential evolutionary restrictions that hosts impose on parasite evolution in general and reproductive strategy in particular.
Fish-infecting copepods follow the same reproductive trend typically seen in free-living species. Within this group, highly host-specific copepods produce fewer eggs, a K-selected trait seen in habitat specialists. The relatively low fecundity of specialist fish-infecting copepods could reflect historically stable environments, with lower host species turnover or a high abundance of a certain host species. Conversely, copepods with low host specificity produce more eggs, an r-selected trait seen in habitat generalists. The relatively high fecundity of generalist fish-infecting copepods could reflect historically unstable environments within which host species immigrate and emigrate frequently. Surveys of localized aquatic systems, such as reefs (Bohnsack, Reference Bohnsack1983) or river stretches (Haubrock et al., Reference Haubrock, Pilotto, Innocenti, Cianfanelli and Haase2020), show that fish turnover rates can be quite high, enough to completely change community composition within a few decades or centuries. Recently, it has been shown that the functional turnover rate of fishes, i.e. the replacement of species performing different ecological functions, was higher than expected in a marine environment (Vallee et al., Reference Vallee, Villanueva and Blanchard2019). Even though such studies suggest that fish community composition can change rapidly over relatively short periods of time, it is admittedly difficult to assess the historical turnover rates of species for many environments (Buckley and Jetz, Reference Buckley and Jetz2008). Generalist fish-infecting copepods evolved to invest more energy into the quantity of eggs, without any obvious trade-off with egg size, in order to maximize the chances of offspring infecting compatible host species. If these parasites evolved in an environment with relatively high turnover rates of mobile hosts that move around in three dimensions, releasing many eggs that have the potential to infect several host species would ensure parasite transmission, and thus, species survival. If environmental stability dictates the degree of host species turnover, based on our results, we posit that greater environmental stability (with more potential for high host specificity) selects for low reproductive output in parasites infecting mobile hosts, whereas an unstable environment selects for the opposite pattern.
Coral-infecting copepods with low host specificity produce fewer eggs than their highly host-specific counterparts. For this group, generalists invest less energy in the quantity of their eggs, a trait associated with K-selected organisms. Unlike fishes, it is possible to obtain the ecological history of coral communities using the fossil record: corals form resistant skeletons that accumulate in place, providing relative abundance data of past communities (Pandolfi and Jackson, Reference Pandolfi and Jackson2001). Studies have shown that coral community structures are highly ordered over large temporal scales and can persist for tens of thousands of years (Pandolfi, Reference Pandolfi2002; Pandolfi and Jackson, Reference Pandolfi and Jackson2006). These copepods have marked morphological adaptations, such as highly modified body forms, for living with or within corals (Humes, Reference Humes1985; Cheng et al., Reference Cheng, Mayfield, Meng, Dai and Huys2016). Parasites that evolved in such stable environments would likely invest in the quality of eggs rather than the quantity. According to our results, a generalist coral-infecting copepod should have a high transmission success rate in a stable patch of diverse corals, whereas a highly host-specific one may need to invest more into the quantity of eggs in order for offspring to locate one particular coral host amid several species. Therefore, in a stable environment where it is almost guaranteed to locate compatible host species, a generalist parasite would need to invest relatively little energy into reproduction.
Of course, there are some limitations to our study. First, host specificity is likely not the only factor impacting the reproductive strategies of parasitic copepods. According to life history theory (Stearns, Reference Stearns1992), the age-specific mortality rates that occur in a life cycle dictate the energy invested by females into reproductive output. Although this component may play an important role here, we cannot account for it in the models, as it is impossible to accurately determine the mortality rates in the early life stages of all copepod species before they locate and attach themselves successfully to a host. Second, there are large credible intervals associated with our prediction lines. Since this study includes multiple parasite species, each with unique evolutionary histories and life history traits, we should expect considerable variation across the dataset. Nevertheless, we can still conclude that host specificity can predict parasite reproductive traits in both groups tested here. Third, latitudinal gradient could have affected our results, which is thought to impact the host specificity of parasites (Vázquez and Stevens, Reference Vázquez and Stevens2004) since lower-latitude environments are generally more stable, although this concept has been met with some inconsistent evidence among parasite groups (Rohde, Reference Rohde1978; Krasnov et al., Reference Krasnov, Shenbrot, Khokhlova, Mouillot and Poulin2008). Finally, the current dataset excludes free-living copepods, which could account for differences in patterns observed here in relation to other studies (Poulin, Reference Poulin1995). Despite these limitations, we were able to extract complex evolutionary patterns of life history trait selection from parasitic copepods dictated by the environment.
Here, we show that host specificity plays an important role in shaping the reproductive traits of parasites. Like free-living species, reproductive strategies in parasites can be drawn in a continuum ranging from low reproductive input (traits usually related to K-selected species) to high reproductive input (traits usually related to r-selected species). From host species records compiled from the literature, we found that fish-infecting copepods produce smaller clutch sizes in highly host-specific species and produce larger clutch sizes in more generalist species. In contrast, the opposite pattern was observed in coral-infecting copepods. Both reproductive traits varied independently, suggesting little energetic trade-off between them. These differences may reflect the evolutionary history that both groups share with their hosts. It is possible that the environmental stability that dictates fish turnover rates varies widely across systems and geographical ranges, which could select for parasites that maximize reproductive efforts (relatively larger clutch sizes) to increase transmission success rates. Conversely, evidence shows that coral communities remain stable over large temporal scales, suggesting that parasites have higher transmission success rates and less selection pressure to invest heavily into reproductive output. These possible scenarios highlight the importance of environmental stability, which ultimately determines the host specificity of parasites, and its impact on parasite reproductive strategies. This study is an important step towards a better understanding of large-scale evolutionary and ecological patterns governing parasite life histories.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182021002122
Data
The full dataset and R code are provided in the Supplementary material.
Acknowledgements
We thank Eunji Park and Robert Poulin for feedback and constructive criticism on an earlier draft. We also acknowledge Fátima Jorge and Diogo Provete for their help with the phylogenetic analyses.
Author contributions
AE, JFD and MM conceived and designed the study; AE, JFD and MM compiled the data; AF led the data analysis; JFD led the writing with contributions to the methods and results from AF and critical input from AE, AF and MM. All authors gave final approval for publication.
Financial support
This study was supported by the University of Otago Doctoral Scholarships to JFD, MM and AF, and by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001 grant to AE (process number 88881.187634/2018-01). JFD was also supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) scholarship (PGSD3-530445-2019).
Conflict of interest
The authors declare there are no conflicts of interest.