INTRODUCTION
Protozoan parasites of the genus Leishmania are the aetiological agents of leishmaniasis, a group of diseases ranging in severity from cutaneous (CL) to mucocutaneous (MCL) and visceral (VL) forms (Murray et al. 2005). Worldwide, 2 million new cases of symptomatic disease occur each year (Desjeux, 2004). The different forms of the disease are produced by different species of Leishmania. CL is mainly caused by L. tropica, L. major and L. aethiopica in the Old World and by members of the L. mexicana (L. mexicana and L. amazonensis), L. braziliensis (L. braziliensis and L. peruviana), and L. guyanensis (L. guyanensis and L. panamensis) complexes in the New World (Anonymous, 1990). Cutaneous infection causes lesions that are normally resolved after a few months, although, depending on the causative agent, they can disseminate to mucosal surfaces. MCL is characterized by the dissemination of parasites from the skin to the naso-oropharyngeal mucosa, and is caused by infection with species of the L. braziliensis complex, especially L. braziliensis, but also by species of the L. guyanensis complex. VL is mainly caused by members of the L. donovani complex (L. donovani and L. infantum in the Old World, and L. chagasi in the New World). It is generally accepted that the clinical spectrum of leishmaniasis is governed by both parasite and host factors, but these remain poorly understood (Pearson and de Queiroz Sousa, 1996; Murray et al. 2005).
Leishmania parasites exhibit a digenetic life-cycle. In the sandfly, they exist as extracellular promastigotes that are transferred to the mammalian host when the sandfly takes a bloodmeal. The parasite is phagocytosed by macrophages and, inside the acidic phagolysosomes, promastigotes differentiate to amastigotes. The mechanisms implicated in this transformation are not completely understood, although it is known that environmental factors such as pH and temperature are triggering factors of this process (Garlapati et al. 1999; Zilberstein and Shapira, 1994). The heat shock response is considered to be of vital importance for the stage-specific differentiation of Leishmania parasites, and the genes encoding heat shock proteins (HSPs) in Leishmania have been extensively studied not only for their importance in the differentiation process, but also as a gene models to study gene expression in this parasite (Lee et al. 1988; Quijada et al. 1997; Clos and Krobitsch, 1999; Zilka et al. 2001).
Among HSPs, HSP70 is the most highly conserved in both sequence and function; it assists a wide range of cellular processes, such as folding of new synthesized proteins and refolding of misfolded and aggregated proteins (Hartl and Hayer-Hartl, 2002). Genes coding for HSP70 were among the first genes to be studied in Leishmania, in particular in L. major (Lee et al. 1988). In this species, 5 HSP70 genes were identified: 4 of them tandemly arranged and a fifth located at a separate locus, but on the same chromosome. Restriction and sequence analyses indicated that the coding regions of the 5 genes must be highly conserved. However, 5 nucleotides beyond the TAA termination codon, the sequence of the fifth gene was found to be divergent from that found in the tandemly linked HSP70 genes. In addition, it was observed that transcripts derived from the tandemly linked HSP70 genes increased in response to a temperature shift from 25 °C to 37 °C, whereas the transcripts derived from the fifth gene, being very abundant, are unaffected by temperature shifts (Lee et al. 1988). In L. donovani, a preliminary study showed the existence of 12–15 HSP70 genes tandemly arranged on a single chromosome (MacFarlane et al. 1990). In addition, the genomic organization of HSP70 genes in L. amazonensis has been examined (Bock and Langer, 1993). In this species, the existence of 7 HSP70 genes organized in tandem was deduced and an eighth HSP70 gene located at a distant site but on the same chromosome. Sequence analysis of cDNAs suggested the existence of 2 types of HSP70 differing in their 3′UTRs. Recently, in L. braziliensis, the sequence of the HSP70 coding region has been determined and the HSP70 genomic organization has been analysed (Zurita et al. 2003). The authors suggested the existence of a single gene, but they showed only preliminary data.
In L. infantum, the genomic organization and expression of HSP70 genes have extensively studied by our group (Quijada et al. 1997, 2000; Folgueira et al. 2005). The L. infantum genome contains 6 HSP70 genes that are located in a single locus and arranged tandemly in a head-to-tail manner (Quijada et al. 1997). All the genes are highly conserved in sequence with the exception of the 3′-UTR of gene 6 (located at the 3′-end of the cluster) that is absolutely divergent relative to the 3′-UTR shared by the other 5 genes. Hence, for simplicity, genes 1 to 5 are referred as HSP70-I genes and gene 6 as HSP70-II gene (Folgueira et al. 2005). Remarkably, transcripts derived from gene 6 are more abundant than transcripts derived from all the other HSP70 genes put together (Quijada et al. 1997). However, only mRNAs derived from HSP70-I genes accumulate in response to heat shock treatments, while the HSP70-II mRNAs remain unaffected. Subsequently, it was shown that the temperature-dependent accumulation of HSP70-I transcripts is associated with the presence of a cis-acting sequence in their 3′-UTR (Quijada et al. 2000). In a recent work, we have reported that HSP70-II mRNAs are preferentially translated at heat shock temperatures, but not at 26 °C, whereas HSP70-I mRNAs are bound to polysomes at 26 and 37 °C (Folgueira et al. 2005).
Because a relationship between gene organization and expression seems to exist in Leishmania HSP70 genes, we considered it of interest to determine the extent of similarities and variations of the HSP70 loci within the spectrum of Leishmania species. Here we describe the genomic organization of the HSP70 genes in 6 Leishmania species. Our results are discussed in the light of the recently completed sequence of the L. major genome (Ivens et al. 2005). We have demonstrated the existence of 2 types of HSP70 genes, previously reported in L. major and L. infantum, in all the species tested with the exception of L. braziliensis. The study was completed by analysing the expression of HSP70 genes in representative Leishmania species at the levels of mRNA and protein synthesis.
MATERIALS AND METHODS
Parasites and treatments
Promastigotes of L. infantum (MCAN/ES/96/BCN150), L. tropica (MHOM/SU/74/K-27), L. mexicana (MNYC/BZ/62/M-379), L. amazonensis (IFLA/BR/67/PH-8), L. braziliensis (MHOM/BR/75/M-2904) and L. major (MHOM/IL/80/Friedlin) were cultured in vitro at 26 °C in RPMI 1640 medium (Sigma), supplemented with 10% heat-inactivated foetal calf serum (Sigma). The L. major Friedlin strain (clone V1) was generously provided by Dr Davis Sacks (NIAID, National Institutes of Health, Bethesda, USA). Logarithmic phase cultures (5–9×106 promastigotes/ml) were used in the experiments. For heat shock experiments, aliquots of cultured promastigotes were incubated at 37 °C for different periods of time. Immediately after heat shock, parasites were spun down and harvested for the analysis of the steady-state mRNA levels or de novo protein synthesis.
Southern blot and Northern blot analyses
Genomic DNA was isolated as described previously (Requena et al. 1988). One μg of total DNA was digested with a variety of restriction endonucleases, electrophoresed on 0·8% agarose gels, and transferred to nylon membranes (Hybond-N, Amersham Corp.) by standard methods (Sambrook et al. 1989). RNA from promastigotes was isolated using the Total Quick RNA Cells and Tissues kit (Talent, Trieste, Italy). Four μg per line of total RNA were size separated on 1% (w/v) agarose-formaldehyde gels and electrophoresed to nylon membranes using the Transfer Power Lid System (Hoefer, San Francisco, CA). DNA probes, either for DNA and RNA analysis, were labelled with [α-32P]dCTP by nick translation (Sambrook et al. 1989). Hybridizations were performed as reported earlier (Quijada et al. 1997). For reuse, blots were treated with 0·1% SDS for 30 min at 95 °C to remove previously hybridized probes.
DNA probes
The 3′-UTR I probe corresponds to the 3′-UTR of L. infantum HSP70-I genes. The DNA fragment containing this region was obtained by BamHI cleavage of clone pB3′UTRIc (Folgueira et al. 2005). The 3′-UTR II probe corresponds to the 3′-UTR of L. infantum HSP70-II gene; the corresponding DNA fragment was obtained by HindIII+SacI double digestion of clone pTC6 (Folgueira et al. 2005). The CDS probe contains a region of the coding region, common to both types of L. infantum HSP70 genes. The DNA fragment was obtained by EcoRI cleavage of clone pLd70.2, which contains the L. infantum HSP70 gene region corresponding to positions 1701-2138 of the EBI Data Bank entry with Accession number Y08020.
Metabolic labelling, Western blotting, and immunoprecipitation
For labelling, 6×107 promastigotes were collected and resuspended in 100 μl of Dulbecco's modified Eagle's medium without methionine and cysteine (Met-, Invitrogen), but supplemented with 10% heat-inactivated foetal calf serum. Proteins were labelled with 100 μCi of [35S]methionine/cysteine protein labelling mix (Redivue Pro-mix [L-35S], >1000 Ci/mol, Amersham Biosciences). After labelling, cells were harvested, washed twice in pre-chilled PBS, and incubated for 30 min with shaking in 100 μl of lysis buffer (50 mM Tris/HCl, pH 7·5, 150 mM NaCl, 1% (v/v) Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 8 μg/ml leupeptin, 4 μg/ml aprotinin and 4 μg/ml pepstatin). Lysates were sonicated for 10 min and centrifuged at 13000 g for 15 min at 4 °C. The soluble extract was mixed with 20 μl of rabbit anti-L. infantum HSP70 polyclonal serum (Rico et al. 1999), and incubated for 15 h at 4 °C. Agarose beads (15 μl), conjugated with Protein A (Sigma), were equilibrated in 50 μl of lysis buffer and added to the protein extract-HSP70 antiserum mixture. The mixture was incubated on a rotator for 2 h at 4 °C, after which the beads were collected by centrifugation, and washed 3 times with 0·5 ml of buffer A (10 mM Tris-HCl, pH 8·0, 30 mM NaCl, 2% (v/v) Triton X-100), twice in 0·5 ml of buffer B (10 mM Tris-HCl, pH 8·0, 50 mM NaCl, 0·1% (v/v) Triton X-100), and once in 0·5 ml of buffer C (10 mM Tris-HCl, pH 8·0, 0·05% (v/v) Triton X-100). Finally, the beads were resuspended in 60 μl of 2× Laemmli buffer. Immunoprecipitated proteins were resolved by SDS-PAGE (10%) and, after drying the gel, proteins were analysed by exposure to X-ray films. The total amount of immunoprecipitated HSP70 protein was analysed by Western blotting using the rabbit anti-L. infantum HSP70 serum (Rico et al. 1999).
Quantitative analysis
Autoradiographs were scanned with the GS-710 calibrated imaging densitometer (Quantity One version 3.0, Bio-Rad). Measurements were performed under conditions in which a linear correlation existed between the amount of proteins, DNA or RNA and the intensities of the bands.
RESULTS
Genomic organization of the Leishmania HSP70 genes
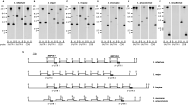
On the basis of the well-known genomic organization of HSP70 genes in L. infantum, we studied the genomic organization of the HSP70 genes in other Leishmania species by Southern blot analysis (Fig. 1). Hybridizations were performed using specific probes for the L. infantum HSP70-I and HSP70-II genes (3′-UTR I and 3′-UTR II, respectively). A probe common to both types of genes (CDS probe) was also used. Thus, in order to determine whether both types of HSP70 genes exist in other Leishmania species, Southern blot analyses were carried out. Genomic DNA of L. infantum, L. major, L. amazonensis, L. tropica, L. mexicana and L. braziliensis was digested with restriction enzymes, transferred onto nylon membranes, and hybridized with probes corresponding to the 3′-UTR I, 3′-UTR II, and the coding region of L. infantum HSP70 genes (Fig. 1). The results showed that the genomic organization for HSP70 genes in L. major (Fig. 1B) and L. tropica (Fig. 1C) genomes is very similar to that present in L. infantum (Fig. 1A). In these species, several HSP70 genes exist arranged in a head-to-tail manner. The 3·8-kb hybridization band, present in lanes containing the DNA digested with SalI or BamHI, corresponds to the size of the repetition unit of the tandem. The repetition unit is observed after probing with either the coding region (CDS) or the 3′-UTR I, suggesting that most of the HSP70 genes in L. major (Fig. 1B) and L. tropica (Fig. 1C) are HSP70-I type genes as occurs in L. infantum (Fig. 1A; Quijada et al. 1997). However, the 3′-UTR II probe hybridized with a 5·7-kb SalI band, but not with the 3·8-kb repetition unit, indicating the existence of a single HSP70-II gene in these species, located at the 3′-end of the cluster. The densitometric analysis of the CDS-hybridization bands (in lanes with DNA digested with either BamHI or SalI), assuming that the largest bands contain the HSP70 gene located at the 5′-end of the cluster, indicated that L. major haploid genome must contain 5 HSP70-I genes whereas L. tropica should have 6 HSP70-I genes. Densitometric analysis carried out in parallel with L. infantum DNA samples confirmed the existence of 5 HSP70-I genes per haploid genome in this species. Fig. 1G summarizes the genomic organization of the HSP70 locus in L. infantum, L. major, and L. tropica.

Fig. 1. Southern blot analyses of the HSP70 gene loci in Leishmania infantum (A), L. major (B), L. tropica (C), L. mexicana (D), L. amazonensis (E), and L. braziliensis (F). Genomic DNA was cleaved with the restriction enzymes: B, BamHI; H, HindIII; S, SalI. The Southern blots were sequentially hybridized with the probes: 3′-UTR I, 3′-UTR of the L. infantum HSP70-I gene; 3′-UTR II, 3′-UTR of the L. infantum HSP70-II gene; and CDS, coding region of L. infantum HSP70 genes (see Materials and Methods section for a detailed description of the sequences used as probes). (G) Deduced physical maps of the HSP70 loci in L. infantum, L. major, L. tropica, L. mexicana and L. amazonensis.
The HSP70 gene organization in L. mexicana and L. amazonensis was found to be identical (Fig. 1D,E) but different to that found in L. infantum, L. major, and L. tropica (Fig. 1A–C). The main difference was observed with the BamHI cut site: this restriction enzyme did not cut within the HSP70 locus in L. amazonensis and L. mexicana genomic DNA. However, the Southern blot analyses indicated the existence of a tandem array of HSP70 genes in these Leishmania species also. The repetitive unit was of 3·4-kb, as defined by SalI digestion. The 3·4-kb SalI band was revealed after hybridizing with either CDS or 3′-UTR I probes, indicating that in L. amazonensis and L. mexicana also most of the genes are HSP70-I type. The 3′-UTR II probe hybridized with a 4·4-kb BamHI band, indicating that the BamHI enzyme cuts in the 3′-UTR of the HSP70-II gene, but not in the rest of the locus. This result agreed with the existence of a sole HSP70-II gene also in these Leishmania species. In contrast, the densitometric analyses, comparing the intensities of the 7·8-kb and 3·4-kb SalI bands hybridizing with the CDS probe, revealed a copy number of 6–7 for the HSP70-I gene per haploid genome in L. amazonensis and L. mexicana. For these calculations, it was assumed that the 7·8-kb SalI band contains a single gene. Our data are coincident with those of Bock and Langer (Bock and Langer, 1993), who described the existence in L. amazonensis of an HSP70 tandem array composed of 7 genes. Fig. 1G summarizes the genomic organization of the HSP70 locus in L. amazonensis and L. mexicana.
The most different HSP70 genomic organization among the Leishmania species analysed in this work was found in L. braziliensis (Fig. 1F). Remarkably, the 3′-UTR II probe did not hybridized with any DNA band suggesting the absence of an HSP70-II gene in this species. By contrast, the 3′-UTR I and CDS probes hybridized with the same restriction bands suggesting that most of the HSP70 genes in L. braziliensis are of the HSP70-I type. At present is not possible to deduce the number of HSP70-I genes and additional studies using genomic clones are needed to define both the HSP70 copy number in the L. braziliensis genome and the probable existence of HSP70 gene(s) with a divergent 3′-UTR (see below).
Expression of HSP70 genes in different Leishmania species
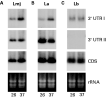
In L. infantum, the abundance of transcripts derived from HSP70-I genes increases following heat shock, whereas the levels of HSP70-II mRNAs remain unaffected (Quijada et al. 1997; Folgueira et al. 2005). We considered it of interest to analyse whether the expression pattern of the 2 types of HSP70 genes is conserved in other Leishmania species. According to the Southern blot analyses (Fig. 1), 3 different genomic organizations of the HSP70 locus were found among the 6 Leishmania species analysed. So, we selected a representative species for each type of genomic organization in order to analyse the expression of the HSP70 genes. Thus, for this study we chose L. major, L. amazonensis, and L. braziliensis. Total RNA was isolated from logarithmic phase promastigotes after incubation for 2 h at 26 °C or 37 °C, and analysed by Northern blotting. Fig. 2 shows the results after hybridizing with probes derived from the 3′-UTR I, 3′-UTR II and coding region (CDS) of L. infantum HSP70 genes. For L. major and L. amazonensis promastigotes, the Northern blots revealed that HSP70-I transcripts accumulate when the parasites are incubated at elevated temperatures (Fig. 2A,B, panel 3′UTR I). Densitometric analysis of the blots indicated that the abundance of HSP70-I transcripts is 2 to 3-fold higher in promastigotes incubated at 37 °C than in promastigotes growing at normal temperature (26 °C). In contrast, the HSP70-I transcript levels in L. braziliensis promastigotes resulted unaffected upon heat shock treatment (Fig. 2C, panel 3′UTR I). On the other hand, the steady-state levels of HSP70-II transcripts in L. major and L. amazonensis did not change after heat treatment of the promastigotes (Fig. 2A,B, panel 3′UTR II). As expected, 3′-UTR II transcripts were not detected in the blots containing RNA from L. braziliensis promastigotes (Fig. 2C, panel 3′UTR II). When the Northern blots with L. major and L. amazonensis RNAs were hybridized with the CDS probe, the accumulation of HSP70 transcripts was not evident, indicating that the HSP70-II mRNAs must be more abundant that the HSP70-I ones (Fig. 2A,B, panel CDS). These results agreed with previous studies in L. major (Lee et al. 1988). Noticeably, after probing the L. braziliensis blot with the HSP70 CDS, 2 hybridization bands very close in size were seen (Fig. 2C, panel CDS). The lower band corresponds to that hybridizing with the 3′-UTR I probe. The other band may suggest the existence in L. braziliensis of a different type of HSP70 gene(s) containing a 3′-UTR with low, or none, sequence homology to the 3′-UTR in HSP70-I or HSP70-II genes. However, the abundance of both types of HSP70 transcripts in L. braziliensis was unaffected after temperature treatment of the promastigotes.

Fig. 2. Analysis of the expression of the HSP70 genes in Leishmania major (A), L. amazonensis (B), and L. braziliensis (C). RNA samples were prepared from promastigotes grown at 26 °C (lanes 26) or incubated for 2 h at 37 °C (lanes 37), and analyzed by Northern blotting. The blots were hybridized with the probes 3′-UTR I, 3′-UTR II and CDS (see legend to Fig. 1). Bottom panel shows the ethidium bromide staining of the RNA samples loaded on agarose gel prior to transfer.
Analysis of HSP70 synthesis during heat shock
It has been estimated that HSP70 accounts for around 2·1% of the total protein in unstressed Leishmania promastigotes (Brandau et al. 1995), making it one of the most abundant proteins. Due to its abundance, no substantial increase is observed after a heat shock treatment. However, by a combination of metabolic labelling and immunoprecipitation, it has been demonstrated that the rate of HSP70 synthesis is increased during heat shock. Thus, a heat stress at 37 °C of L. donovani promastigotes results in a 6-fold increase in the synthesis rate of HSP70 (Brandau et al. 1995). Similarly, in L. infantum promastigotes the rate of HSP70 synthesis was estimated to increase 4 to 5-fold at 37 °C relative to the rate at 26 °C (Folgueira et al. 2005). For this comparative study, we wanted to analyse the de novo synthesis of the HSP70 at both 26 °C and 37 °C in promastigotes of L. major, L. amazonensis and L. braziliensis, species in which this analysis has not been previously reported and that show a different genomic organization of the HSP70 locus (Fig. 1). Fig. 3A shows an autoradiograph of the metabolically labelled proteins at 26 °C and at 37 °C in L. amazonensis, L. braziliensis and L. major promastigotes. Remarkably, total protein synthesis was drastically affected in L. amazonensis by incubation at 37 °C, with a global decrease of 41% relative to the protein synthesis at 26 °C. This finding indicates that 37 °C induces a severe heat shock in this species. Leishmania species differ in the temperature range which they resist, a feature that may be related to their ability to establish infection at different sites in the mammalian host (Zilberstein and Shapira, 1994). In L. braziliensis and L. major the overall protein synthesis was not substantially modified when promastigotes were incubated at 37 °C indicating that in these species, although they cause cutaneous infections as L. amazonensis, the protein translation is more resistant to temperature increase than in L. amazonensis promastigotes.

Fig. 3. Analysis of the de novo synthesis of HSP70 in Leishmania species. (A) Promastigotes of L. amazonensis (La), L. braziliensis (Lb), and L. major (Lmj) were metabolically labelled for 1 h at either 26 °C (lanes 26) or 37 °C (lanes 37), and the proteins were analysed by SDS-PAGE and autoradiography. Also, the proteins samples were analysed by Western blotting using an anti-HSP70 antibody (bottom panel). (B) The HSP70 of new synthesis was determined by immunoprecipitation of the HSP70 in the proteins extracts of these Leishmania species. The immunoprecipitates were analysed by SDS-PAGE and either autoradiography (de novo panel) or Western blotting with the anti-HSP70 antibody (TP panel). (C) Plot of the ratios of de novo synthesized HSP70 compared to total amount of HSP70; the de novo/TP ratio at 26 °C was set as 1.
Finally, we determined the rate of de novo HSP70 synthesis at both temperatures in these three Leishmania species. An anticipated but not yet demonstrated observation of this study was that the steady state level of HSP70 does not substantially change after heat stress in L. amazonensis, L. braziliensis and L. major promastigotes (Fig. 3A, panel HSP70). Therefore, it was necessary to perform a combination of metabolic labelling and immunoprecipitation to determine the rate of HSP70 synthesis in parasites incubated at 37 °C (Fig. 3B,C). It was found that there was a preferential translation of HSP70 at 37 °C in L. braziliensis and L. major, accounting for a 2-fold increase in the de novo HSP70 synthesis at 37 °C relative to the synthesis detected at 26 °C. In contrast, in L. amazonensis the rate of HSP70 synthesis was similar at both 26 °C and 37 °C. Nevertheless, taking into account the decrease in the global protein synthesis observed at 37 °C, it is concluded that the HSP70 mRNA translation in L. amazonensis is resistant to the partial blockage of the global translational process observed at 37 °C.
DISCUSSION
Given that the genome of L. major has been sequenced (Ivens et al. 2005) and this information is available at the GeneDB website (http://www.genedb.org/; Aslett et al. 2005), we performed a bioinformatics study of the HSP70 gene locus in the L. major genome database. According to the L. major database, the HSP70 locus is located in chromosome 28 and consists of 2 HSP70 genes. The genomic organization is similar to that determined by Southern blotting in this work, i.e. the locus is composed of 2 types of genes (HSP70-I and HSP70-II) and the relative position of the genes is identical. The HSP70-I and HSP70-II genes correspond to the L. major database entries with systematic names LmjF28.2780 and LmjF28.2770, respectively. However, a clear difference exists between the L. major genome database and our results: there is no coincidence in the number of HSP70-I genes. Our analysis supports the existence of five HSP70-I genes in L. major, in close agreement with a previous report that described the existence in the L. major genome of a tandem array consisting of 4 HSP70 genes (Lee et al. 1988). Sequence analysis of these genes indicated that they possess a 3′-UTR homologous to the L. infantum HSP70-I gene (Quijada et al. 1997). Furthermore, the discrepancy between our results and those of the L. major genome Project cannot be due to the use of a different L. major strain, since in both studies the strain Friedlin has been used. A plausible explanation is that the copy number of HSP70-I genes in the L. major genome has been underestimated because of the collapse of multiple tandem repeats into fewer copies during assembly of sequence data, a frequent problem associated with reiterated genes (El-Sayed et al. 2005). It is likely that similar discrepancies will emerge when other tandemly linked, repeated genes are analysed in the L. major database.
The Southern blot analyses reported here indicate that the HSP70 gene locus is highly conserved in the different species of Leishmania, excepting L. braziliensis. For most species, the locus is composed of 2 types of HSP70 genes: 5-6 HSP70-I genes and an HSP70-II gene, the latter always located at the 3′-end of the gene cluster. Nevertheless, restriction polymorphisms differentiate the HSP70 locus from Old and New World Leishmania species. Thus, the HSP70 loci in L. infantum, L. major and L. tropica have essentially the same structure and the repetition units of the tandem are defined by either BamHI or SalI restriction enzymes. In contrast, the HSP70 locus in the New World species L. mexicana and L. amazonensis lacks the BamHI restriction site but conserves the SalI site. For L. braziliensis, our present data and those previously published (Zurita et al. 2003) are insufficient to determine the genomic organization of the HSP70 genes. However, our Southern blot analysis demonstrates that this species lacks HSP70-II genes and, therefore, that the L. braziliensis HSP70 locus is structurally different to the HSP70 loci in the other Leishmania species. According to the genomic structure of the HSP70 locus, the Leishmania species analyzed in this study may be clustered in 3 groups. The first cluster includes the Old World species L. infantum, L. major and L. tropica. The second cluster includes the New World species of the subgenus Leishmania (L. mexicana and L. amazonensis). Finally, L. braziliensis, New World species of the subgenus Viannia, possesses the most divergent HSP70 locus. It should be necessary to analyse other species of the subgenus Viannia to see whether the L. braziliensis HSP70 gene organization is conserved among species of this subgenus. Nevertheless, the clustering denoted by the organization of the HSP70 locus is similar to that obtained after analysing in different isolates of Leishmania the mini-exon gene loci (Fernandes et al. 1994), the sequence of DNA and RNA polymerase genes (Croan et al. 1997), and the chromosomal karyotype (Britto et al. 1998). In those studies, the L. (V.) braziliensis complex represents the earliest divergent species of the genus Leishmania, a fact that agrees with the current beliefs on the origin and evolution of the genus Leishmania (Momen and Cupolillo, 2000). As a conclusion of our study, we think that the HSP70 locus may be a useful genetic marker aiding for Leishmania species identification.
At the level of gene expression, differences were also found when comparing the expression pattern in species of the subgenus Leishmania with the expression in L. braziliensis, pointing to differences in the regulatory mechanisms. It is well established that transcripts derived from HSP70-I genes accumulated in response to heat treatment in both L. major (Lee et al. 1988; this work) and L. infantum (Quijada et al. 1997). Here, we found that this is also the case for HSP70-I mRNAs in L. amazonensis but not for those of L. braziliensis. As occurs for most genes in kinetoplastids (Clayton, 2002), the regulation of HSP70 genes in Leishmania occurs at the post-transcriptional level (Brandau et al. 1995; Quijada et al. 1997). Taking into account this feature, it must be concluded that L. braziliensis lacks the regulatory mechanisms responsible of the temperature-dependent stabilization of the HSP70-I transcripts observed in the species of the subgenus Leishmania. However, when the rate of HSP70 synthesis was analysed, it became evident that L. braziliensis promastigotes have the capacity to translate preferentially the HSP70 transcripts during heat stress, as do other Leishmania species. Further studies are needed to establish the genomic structure of the HSP70 locus in L. braziliensis, the genome sequencing project is ongoing (Laurentino et al. 2004) and, more importantly, to determine the mechanisms of gene expression.
We gratefully acknowledge financial support from the Ministerio de Ciencia y Tecnología (BFU2005-08432-C02-01) and grants from the Fondo de Investigaciones Sanitarias (C03/04 and PI050973). Also, an institutional grant from Fundación Ramón Areces is acknowledged.