Published online by Cambridge University Press: 06 August 2004
Allozyme variation within and among populations of 3 species of the genus Lecithochirium (Trematoda: Hemiuridae) was studied by starch gel electrophoresis. In total, 19 loci were analysed in 7 populations. The level of genetic variability was relatively high in all populations. The percentage of polymorphic loci (0·95 criterion) observed per population varied from 21·0% to 55·5%, and expected heterozygosity levels varied from 0·082 to 0·197. All populations showed significant heterozygote deficiencies. In Lecithochirium fusiforme most of the deviations from Hardy–Weinberg proportions were within the populations and this species showed moderate population structuring (FIS=0·486, FST=0·142, Nm=1·51) and accordingly low intraspecific genetic distances (D=0·003 to 0·027). A significant lack of heterozygotes for several polymorphic loci was revealed in Lecithochirium rufoviride and Lecithochirium musculus. The most probable cause of the population genetic subdivision in L. rufoviride is the presence of at least 1 cryptic species in the populations studied. Although the lowest percentage of fixed genetic differences was that between L. fusiforme and L. musculus, two different algorithms for the construction of evolutionary trees on a matrix of genetic distances confirmed that L. fusiforme and L. rufoviride are phenetically the most closely related species.
It is traditionally considered that parasitic helminth populations are characterized by having low levels of genetic variability, as well as a strongly structured genetic diversity (Price, 1980). However, factors such as host mobility and the complexity of the life-cycle may have decisive effects on the effective population size and on the gene flow between populations, thereby determining the genetic structure of the parasite populations (Nadler, 1995). A variety of types of organization of genetic variability within populations has been confirmed in recent years by different studies, mainly carried out with nematodes. In fact, it has been found that there are as many species characterized by low genetic variability within populations and high genetic divergence between populations, as there are species characterized by high indices of genetic diversity and high gene flow between populations (Anderson, Blouin & Beech, 1998; Blouin, Liu & Berry, 1999; Hu et al. 2002). On the other hand, insofar as the mode of reproduction is determined by environmental factors, different populations of the same species of nematodes such as Strongyloides may show large differences in genetic structure (Viney, 2001). This degree of variation may be particularly likely to occur in other species, such as, for example, in helminths in which the population genetic structure may be greatly influenced by human intervention (Blouin et al. 1995; Peng et al. 1998; Folkertsma et al. 2001; Plantard & Porte, 2004).
Fewer studies of genetic structure have been carried out on trematode than on nematode populations, therefore very little is known about the genetic structure of the former (Jarne & Théron, 2001). However, there are some indications that the model of Price (1980) also considerably simplifies the actual situation of trematodes. For example, there appear to be important differences in the genetic structure of populations of trematodes with an autogenic life-cycle and those with an allogenic cycle (Criscione & Blouin, 2004), and genetic structure may also vary in the different sexes of certain dioecious digeneans (Prugnolle et al. 2003). In contrast to Price's (1980) model, some trematodes demonstrate relatively high levels of intrapopulation genetic variability (Lydeard et al. 1989; Dybdahl & Lively, 1996; Vilas, Paniagua & Sanmartín, 2003a) and, despite the fact that some schistosome populations show very little allozyme variation, recent studies using DNA markers have revealed substantial levels of genetic variability within populations of these parasites (Curtis, Sorensen & Minchella, 2002). As has been observed with nematodes, the genetic structure of populations of digenetic trematodes appears to depend greatly on factors such as host mobility, non-random distribution of the parasite, geographical and ecological characteristics of the area of transmission and the reproductive structure of the population (Mulvey et al. 1991; Sire et al. 2001; Criscione & Blouin, 2004). However, most of these studies have been carried out with parasites of terrestrial animals, and practically nothing is known about the levels of genetic variability and its spatial distribution in digenetic trematodes that are parasites of marine fish. Marine trematodes generally have a higher capacity for dispersal and more complicated life-cycles than parasites with terrestrial hosts, and therefore they may show higher levels of genetic variability. The aims of the present study were (1) to provide an estimate of the genetic variability of 7 populations of 3 species of the genus Lecithochirium – parasites of marine fish – by the use of allozyme markers, and (2) to investigate the degree of spatial population structuring in 1 of these species.
Natural populations of adult Lecithochirium belonging to 3 different species were collected from 7 sites along the north coasts of the Iberian Peninsula and of Bretagne, France (Fig. 1). The total numbers of specimens analysed by protein electrophoresis were as follows: 613 L. fusiforme from Viana do Castelo in Portugal (LFVC), 856 from Ribeira (LFRB), 605 from Viveiro (LFVV) and 455 from Lorient (LFLR). In addition, a total of 922 L. rufoviride from Ribeira (LRRB) and 538 from Llanes (LRLN) were analysed, as were 642 specimens of the species L. musculus, collected from Ribeira (LMRB). All of the parasites were collected from a total of 175 conger eels (Conger conger), with the exception of L. musculus, specimens of which were obtained from 92 European eels (Anguilla anguilla). We considered a population as a group of individuals of one species collected from different individual hosts captured at the same location and, in accordance with Margolis et al. (1982), we use the term infrapopulation to refer to all individuals of a species in an individual host at a particular time. The small size of the parasites and the fact that we were only able to analyse a maximum of 2 enzymatic systems in any individual made necessary the collection of a large number of helminths. Therefore, despite the high prevalence and intensity of infection characteristic of these parasites, the samples analysed from each geographical location consisted of specimens collected from several individual hosts (Table 1). Furthermore, electrophoretic analysis was only made of proteins from sexually mature specimens. As an exception to the general sampling regime, only 2 eels were captured at Viana do Castelo, and most of the helminths collected were taken from 1 of these (408 and 205). However, we have previously shown that most of the genetic variation in L. fusiforme from the same geographical location is found within the infrapopulations and there are no important genetic differences between different infrapopulations obtained from the same location at a particular time (Vilas, Paniagua & Sanmartin, 2003a).

Fig. 1. Localities where Lecithochirium species were sampled in the present study. VC, Viana do Castelo (Portugal); RB, Riberia (Galicia, Spain); VV, Viveiro (Galicia, Spain); LN, Llanes (Asturias, Spain); LR, Lorient (Bretagne, France).
Table 1. Number of individual hosts, range and average number of parasites per eel of locations from which Lecithochirium populations were sampled (Some conger eels from Ribeira were the same individual host for both L. fusiforme and L. rufoviride species.)

The European eels were transported live to the laboratory where they were killed by an overdose of 2-phenoxy ethanol, and then eviscerated. The conger eels were caught by hook and immediately eviscerated in situ. Abdominal viscera were placed in individual plastic bags within Styrofoam boxes conveniently refrigerated and transported to the laboratory. The stomachs were processed and examined with a stereomicroscope. Adult Lecithochirium were collected and placed in a dish containing physiological saline, and were then identified to species level, following the description of Gibson & Bray (1986). The initial identification was confirmed by examination of permanent preparations. Thus, some specimens were previously rinsed in physiological saline, fixed in Berland's fluid and preserved in 70% ethanol. For morphological identification by optical microscope the specimens were stained with iron acetocarmine and mounted in Canada balsam. The remaining worms were maintained alive in physiological saline for 1–2 h and then frozen at −80 °C until analysis by electrophoresis.
Horizontal starch gel electrophoresis was used to assess genetic polymorphism in populations of the Lecithochirium species. Electrophoresis and staining procedures were according to Vilas et al. (2002). In total, 22 enzyme systems were screened and 17 had sufficient activity for scoring. These represent 19 presumptive loci: aconitase (ACO, EC. 4.2.1.3, encoded by the locus Aco), adenosine deaminase (ADA, EC. 3.5.4.4, Ada), adenylate kinase (AK, EC. 2.7.4.3, Ak), aldolase (ALD, EC. 4.1.2.13, Ald), diaphorase (DIA, EC. 1.8.1.4, Dia), fumarase (FH, EC. 4.2.1.2, Fh-1 and Fh-2), glutamate dehydrogenase (GDH, EC. 1.4.1.3, Gdh), glutamate oxalate transaminase (GOT, EC. 2.6.1.1, Got-2), glucose phosphate dehydrogenase (GPD, EC. 1.1.1.49, Gpd), glucose phosphate isomerase (GPI, EC. 5.3.1.9, Gpi), isocitrate dehydrogenase (IDH, EC. 1.1.1.42, Idh), malate dehydrogenase (MDH, EC. 1.1.1.37, Mdh-1), nucleoside phosphorylase (NP, EC. 2.4.2.1, Np), malic enzyme (ME, EC. 1.1.1.40, Me), manose phosphate isomerase (MPI, EC. 5.3.1.8, Mpi), phosphogluconate dehydrogenase (PGD, EC. 1.1.1.44, Pgd), and phosphoglucomutase (PGM, EC. 2.7.5.1, Pgm-1 and Pgm-2). Two enzymes were analysed per specimen, except for ADA, ALD and AK, which were analysed individually. Only 1 locus for MDH (Mdh-1) was able to be interpreted under the electrophoretic conditions used. No nucleoside phosphorylase activity was detected in any of the populations of L. rufoviride studied, and a portion of each of these populations did not show any enzymatic activity at the Pgm-2 locus. Thus, for comparisons between L. rufoviride and other species, the NP enzyme was not taken into account and for the allozyme analysis, only those individuals that expressed the Pgm-2 locus in each of the populations under study, were considered. To control for possible enzymatic contamination from the host tissue, samples of stomach from Conger conger and Anguilla anguilla were subject to eletrophoresis for each of the 17 enzymatic systems and the results were compared with those obtained for the parasites. Only the allozyme corresponding to the presumed Got-1 locus of L. rufoviride coincided in relative mobility with the allozyme with highest activity in the host; furthermore this locus was not able to be interpreted in most of the samples of the other two species and was therefore excluded from the study. Genotypes were inferred from the banding patterns on the basis of conformity with known protein structures (Vilas, Paniagua & Sanmartín, 2002a). Genotypes and allele frequencies were determined by direct counts on the gels. Alleles were designated different numbers representing their relative mobility from the origin. At each locus the allele corresponding to the most common allozyme from L. rufoviride from Ribeira was designated 100 (−100 in the case of electrophoretic mobility to the cathode).
Genetic variation was expressed in terms of the proportion of loci whose most common allele occurred no more frequently than 0·99 (P99) and 0·95 (P95), and also as the observed mean heterozygosity per locus (Ho), mean heterozygosity per locus expected under Hardy–Weinberg equilibrium (He) and the mean number of alleles per locus (A). The percentage of fixed genetic differences between species was calculated as the percentage of loci in the total number of loci sampled in which there were no alleles common to the taxa under study.
The GENEPOP population genetics software (version 3.2; Raymond & Rousset, 1995) was used to calculate Wright's F-statistics – using Weir & Cockerham's (1984) procedure – and also to test compliance with Hardy–Weinberg expectations in the sample populations, using Fisher's exact test. Estimation without bias of exact P-values was made using the Markov-chain method, following the algorithm of Guo & Thompson (1992). The FIT statistic quantifies the global deviation from the Hardy–Weinberg equilibrium in a subdivided population and it can be partitioned into FIS, which quantifies the deviation within populations, and FST, which reflects heterogeneity among populations because it is the correlation between random gametes within populations relative to the array of gametes in all populations. Thus, the coefficient FST is an estimate of the deviation from the Hardy–Weinberg proportions caused by population subdivision. Values of FIS range between −1 and 1; negative values indicate an excess of heterozygotes, positive values indicate heterozygote deficiency, and FIS=0 when the genotypic frequencies of the population fit to the values predicted under the Hardy–Weinberg equilibrium. Values of FST vary between 0 and 1, so that if all the populations have the same allele frequencies, then FST=0, whereas FST=1 when all populations are fixed for different alleles. Two dendrograms of the populations studied were constructed by bootstrapping among the loci and calculating a consensus of 1000 trees obtained using two different algorithms on Nei's genetic distances: the UPGMA (unweighted pair-group method) and the neighbour-joining methods (Michener & Sokal, 1957; Saitou & Nei, 1987). The dendrograms were constructed using the DISPAN program (Ota, 1993). An indirect estimate of gene flow between the populations of Lecithochirium fusiforme was calculated using Weir & Cockerham's FST-estimator θ, using the expression: M=(1−FST)/4FST; M is an estimate of the product Nem (amount of gene flow), where Ne is the effective population size and m is the proportion of migrant individuals in the population (Slatkin, 1993).
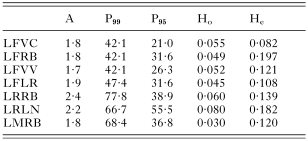
The 7 populations of the genus Lecithochirium under study showed relatively high levels of variability. In each of them, at least 2 alleles were revealed for most of the loci; the variability shown by L. fusiforme for the Pgm-1 locus was particularly high, with up to 6 different allozymes being revealed in 2 of the 4 locations studied for this species (Table 2). According to the most restrictive criteria, the percentage of polymorphic loci in the 3 species studied was never lower than 20% and reached up to 55·5% in L. rufoviride from Llanes. The latter species was, in fact the most variable, although the highest level of heterozygosity expected in the 7 geographical populations studied – according to the Hardy–Weinberg equilibrium – corresponded to L. fusiforme from Ribeira (Table 3). For the latter species, of which 4 populations were studied, the mean percentage of polymorphic loci (P95) was 27·6 and the mean heterozygosity expected was 0·127. These values are high compared with the levels of genetic variability shown by other trematodes, and similar to those shown by populations of non-parasitic invertebrates. The species L. rufoviride and L. musculus were characterized by high genetic polymorphism, but also showed the greatest difference between the numbers of heterozygotes observed and those expected under the Hardy–Weinberg equilibrium, as they revealed highly significant deficits of heterozygotes.
Table 2. Allele frequencies at 19 enzyme loci in populations of Lecithochirium species (LFVC: Lecithochirium fusiforme from Viana do Castelo; LFRB: L. fusiforme from Ribeira; LFVV: L. fusiforme from Viveiro; LFLR: L. fusiforme from Lorient; LRRB: Lecithochirium rufoviride from Ribeira; LRLN: L. rufoviride from Llanes; LMRB: Lecithochirium musculus from Ribeira. Average number of individuals analysed per locus are shown in parentheses.)

Table 3. Indices of genetic variability for populations of Lecithochirium species (A: mean number of alleles per locus; P99 and P95: proportion polymorphic loci at 0·99 and 0·95 criteria, respectively; Ho and He: observed and expected mean heterozygosity per locus, respectively. Population abbreviations as in Table 2.)
Although the global levels of genetic variability were high, all populations studied showed large deviations from the Hardy–Weinberg equilibrium for various loci, always towards a heterozygote deficiency. In the case of L. fusiforme, the Aco, Ada, Fh-2, Gpd, Me, Pgd and Pgm-1 loci showed statistically significant deficits of heterozygotes, compared with those predicted under the Hardy–Weinberg equilibrium, in all populations in which these loci were polymorphic (except for the Pgd locus in the Viana do Castelo population) (Table 4). By contrast, the Gpi, Got, Idh, Mdh, and Pgm-2 loci differed very little from the values predicted under the equilibrium, although of these loci, only Gpi showed an important degree of polymorphism (Table 2). The heterozygote deficiencies were even higher in the other two species. Thus, statistically very significant deviations from the Hardy–Weinberg equilibrium were revealed for most of the poylmorphic loci in L. rufoviride in the 2 populations studied. The Ak and Pgd loci showed a statistically significant, total lack of heterozygotes in both the Ribeira and Llanes populations. Furthermore, no heterozygotes were found for the Dia and Idh loci in at least 1 of these locations, and in both cases the heterozygote deficiency was statistically significant. Thus, for example, both populations showed 2 alleles for the Idh locus, at moderate frequencies (34 and 100), but only 1 individual heterozygotic for this locus was detected in 1 of the populations, and it constituted the most common allele and a different allozyme of very low frequency (39). The single population of L. musculus studied was also characterized by showing large deviations from the values predicted under Hardy–Weinberg equilibrium, towards a heterozygote deficiency for most of the polymorphic loci, and as with L. rufoviride, the population of L. musculus showed a highly significant lack of heterozygotes for the polymorphic Ak and Gdh loci (Table 4).
Table 4. Estimates of deviations from Hardy–Weinberg proportions (FIS) and Fisher's test values within populations of species of the genus Lecithochirium (Population abbreviations as in Table 2.)

Statistical analysis (using the F-statistics) of the L. fusiforme populations revealed that most of the deviation from the Hardy–Weinberg equilibrium took place within each population and that of all the variation, only 14% could be attributed to differences among populations (Table 5). With exception of the Ada and Pgm-1 loci, there were few differences among populations in the gene frequencies. The estimated number of immigrants per generation in the 4 populations was 1·51. In accordance with the moderate values of estimated FST, Nei's genetic distances between populations of L. fusiforme were small, and varied from 0·004±0·003 for the Viano do Castelo and Lorient populations and 0·027±0·013 for the populations from Viana do Castelo and Viveiro (Table 6).

Table 6. Matrix of Nei's genetic distances below the diagonal, and the associated standard errors above diagonal, among populations of Lecithochirium species (Population abbreviations as in Table 2.)

The percentage of fixed genetic differences between L. rufoviride and L. fusiforme was 27·8, between L. rufoviride and L. musculus, 55·5; L. fusiforme and L. musculus did not share alleles for 15·8% of their loci. According to the latter finding, it may appear that L. fusiforme and L. musculus are genetically the most similar of the species studied. However, the results were different when the allele frequencies were used to estimate interspecific genetic distances, suggesting that estimation of fixed genetic differences is an inaccurate method of establishing molecular similarities between these species. In fact, the maximum distance between L. fusiforme and L. rufoviride was 1·148±0·343, between L. fusiforme and L. musculus 1·955±0·529, and between L. rufoviride and L. musculus 1·925±0·572 (Table 6). When the distance matrix was used to study evolutionary relationships between species, it was found that L. rufoviride and L. fusiforme were phenetically the closest species, as deduced from the dendrograms obtained by 2 different algorithms, the UPGMA and neighbour-joining methods (Fig. 2).

Fig. 2. UPGMA (A) and neighbour-joining (B) dendrograms showing evolutionary relationships among Lecithochirium populations on Nei's genetic distances (D). Population abbreviations as in Table 2.
The geographical populations under study, belonging to the genus Lecithochirium, showed relatively high levels of genetic variability, similar to those shown by free-living invertebrates (Ward, Skibinski & Woodwark, 1992). Furthermore, samples of L. fusiforme collected from fairly widely separated locations did not reveal large differences in their allele frequencies. Thus the greatest variation observed was between individuals belonging to the same location and only a small fraction of the genetic variation was due to differences between specimens from different geographical locations. The degree of genetic variability detected in the species studied contrasts with the prediction that hermaphroditic trematodes integrate populations with high levels of inbreeding because of a breeding structure that is determined by the existence of hermaphroditism and clonal amplification. Furthermore, populations of L. fusiforme are subject to large demographic fluctuations over time (Vilas & Paniagua, 2004), which should reinforce the fundamental effects of genetic drift, i.e. the decrease in the genetic variability within the population and the increase in genetic differences among populations. The results obtained therefore suggest the existence of mechanisms that counteract the loss of genetic variability produced by inbreeding and caused by both the mode of reproduction and genetic drift.
Assuming a low mutation rate and that the effect of natural selection is small, the degree of spatial genetic differentiation will basically depend on the equilibrium established between the diversifying effect of genetic drift and the mixing produced by gene flow (Slatkin, 1987). One of the factors which explains the absence of large genetic differences between geographically separated populations and which, furthermore, partly explains the high estimated genetic variability, is the existence of higher levels of gene flow than those considered in Price's model. It has been suggested that the main factor determining the gene flow among parasite populations is the mobility of the host (Blouin et al. 1995; McCoy et al. 2003; Criscione & Blouin, 2004), which is certainly favoured in the marine environment. The conger eel (Conger conger), the main definitive host of L. fusiforme, and also the snails and copepods which constitute the intermediate hosts of this parasite, probably have a low capacity for dispersal in the study area; however, small, more highly mobile fish, which are paratenic hosts of the parasite, may play an important role in the life-cycle of this species (Vilas et al. 2003a). In fact, non-parasitic platyhelminths, which lack the possibility of dispersion facilitated by a host, appear to comprise genetically highly structured populations, even on a microspatial scale (Herbert & Payne, 1985; Dynes, Flemming & Murchie, 2001; Pongratz, Gerace & Michiels, 2002).
There exist other reasons apart from gene flow, which may explain the levels of genetic variability and the moderate values of FST calculated, such as natural selection and also the high prevalence and intensity of parasitization shown by L. fusiforme in C. conger (Vilas & Paniagua, 2004). In general, it is considered that allozymes are neutral genetic markers, but it is possible that a relatively high proportion of protein polymorphism is maintained by natural selection (Karl & Avise, 1992; Pogson, Mesa & Boutilier, 1995). If this is true, natural selection may have a homogenizing effect on the population genetic structure of L. fusiforme, as the stomach of the conger eel is probably a relatively stable environment over geographical distance.
It is interesting that all of the populations under study, corresponding to different locations, did not conform to Hardy–Weinberg expectations, as large, statistically significant, deficits of heterozygotes were revealed. The most likely cause of these deviations from the Hardy–Weinberg expectations is the mode of reproduction of these hermaphroditic helminths. However, if this was the main cause of the low frequencies of heterozygotes detected in L. fusiforme, it would be expected that all the loci would show deviations from the Hardy–Weinberg predictions, of similar magnitude, because the inbreeding generated by selfing or crossing between genetically identical individuals is a phenomenon that affects the whole genome. However, the values of the FIS statistic for different loci were very different and various loci even showed genotypic frequencies that were in consistent with the values expected under the Hardy–Weinberg equilibrium. This suggests that there may be other, more important factors that account for the deficiencies of heterozygotes detected.
Despite there being various loci that reflect the genotypic deficits, and it therefore being unlikely that natural selection acts independently on these loci, it is theoretically possible that these loci reflect the effect of natural selection acting on an unknown gene situated close to them on the chromosome. The deviations from the Hardy–Weinberg equilibrium may be also explained by the Wahlund effect, favoured by the existence of paratenic hosts in the life-cycle. However, because an association between FIS and FST was not detected it does not appear that the heterozygote deficit detected was caused by the mixture of genetically differentiated, geographically different populations. These results suggest that other, more important factors accounting for the heterozygote deficits may also have effects at the infrapopulational level. For example, it is possible that the pooling of parasites across eels at each geographical location in our sampling regime produced the Wahlund effect. However, because there were no great differences in the allele frequencies between different infrapopulations collected from the same location and at the same time (Vilas et al. 2003a; Vilas, Sanmartin & Paniagua, 2004), the analysis of parasites obtained from different eels does not appear to be an important factor in causing the deviations from Hardy–Weinberg equilibrium within each geographical population. Furthermore, the population of L. fusiforme from Viana do Castelo, that was practically represented by a single infrapopulation, revealed heterozygote deficiencies as detected at the other three locations studied for this species. In fact, the heterozygote deficit in L. fusiforme populations already occurred at the level of individual host (Vilas et al. 2003a). Thus it appears likely that the observed heterozygote deficit within local populations of this trematode was largely due to the deviation from the Hardy–Weinberg equilibrium that existed within each infrapopulation, rather than to the presumed Wahlund effect caused by mixing of infrapopulations. Recently, Plantard & Porte (2004) also considered that the heterozygote deficit detected in a plant-parasitic nematode at the level of individual host argues against its generation through a Wahlund effect. However, in the case of L. fusiforme the heterozygote deficits detected within each eel could be caused by the occasional mixing of genetically different temporal and spatial samples (Vilas et al. 2003a).
Populations of L. rufoviride and L. musculus showed a total absence of heterozygotes, which is highly significant for various polymorphic loci. The two geographical populations of L. rufoviride studied revealed a profound genetic subdivision for the same loci. The results shown here confirm – and extend to other loci – the subdivision of populations previously shown for the Idh locus, which was not related to the presence of the sibling species Lecithochirium furcolabiatum (Vilas, Paniagua & Sanmartín, 2002b). Given that the observed genetic subdivision in both populations is strongly associated with a certain variation in the morphology of the preoral lobe, which differs from that shown by the said sibling species (unpublished observation), the most likely explanation for the lack of heterozygotes detected in the L. rufoviride populations is the existence of at least one cryptic species. In a previous study we considered it unlikely that the presence of null phenotypes, observed for the zone of electrophoretic variation corresponding to the Pgm-2 locus in a local population of L. rufoviride, was due to the existence of a sibling species that differed in the number of loci that coded for PGM (Vilas et al. 2001). However, on applying the results obtained in this study to other loci, we must evaluate this hypothesis more closely, especially if we take into account that when individual carriers of the null phenotype were excluded from the analysis, both geographical populations studied showed genotypic frequencies consistent with the values expected under the Hardy–Weinberg equilibrium for the Pgm-2 locus and did not reveal the homozygote excess expected if there was a high frequency of recessive null alleles in the sample.
Although L. musculus also revealed a statistically significant lack of heterozygotes for various polymorphic loci, careful consideration must be made of whether or not this constitutes the existence of a cryptic species. Unlike L. rufoviride, genetic subdivision has not been confirmed in other locations and, although there exist at least two different morphotypes of L. musculus in the population under study (Vilas, Paniagua & Sanmartín, 2003b), no association between the phenotypic and genotypic variation has been found to date. Furthermore, the analyses carried out with L. fusiforme showed that populations of these organisms are characterized by the existence of large deficits of heterozygotes, the cause of which is not necessarily related to the presence of cryptic species. Given that the intensities of infection of A. anguilla by L. musculus were lower than those corresponding to species that mainly parasitize C. conger, it is possible that the Wahlund effect is more important in this case. The species L. musculus, the main definitive host of which is A. anguilla, is considerably distanced – from an evolutionary point of view – from the other two species, which prefer C. conger. The fact that L. fusiforme and L. rufoviride are phenetically the two most closely related species supports the conclusion of Gibson & Bray (1979) that L. fusiforme should not be placed in a separate genus from L. rufoviride on the basis of the lack of a presomatic pit. However, both species, often mistaken for each other in the past because of their morphological similarities (Gibson & Bray, 1986), maintain a relatively large genetic distance as they are sympatric species that parasitize the same definitive host. This may be associated with the existence of different evolutionary rates at morphological and molecular levels.
The results shown here are not consistent with the predictions made using Price's (1980) model and suggest that factors such as the existence of paratenic hosts and of various intermediate hosts in the life-cycle of helminth parasites determine the genetic structure of their populations. There exist very few studies of the genetic variation within and between trematode populations, however, there are reasons to believe that the ecological diversity of these organisms gives rise to a greater variety of types of organization of genetic variability in space than were previously thought to exist.
We thank G. Alvarez (USC) for help with the data analysis. We gratefully acknowledge Divina Gómez and Candelas Paniagua for their assistance in obtaining samples from France and Asturias respectively. The manuscript was improved by useful comments from three anonymous reviewers. This study was financially supported by grant BOS2000-0331 (Ministerio Español de Ciencia y Tecnología; Plan Nacional de Investigación Científica, Desarrollo e Innovación Tecnológica, I+D+I) and PGIDT01PXI20307PN (Xunta de Galicia, Spain).
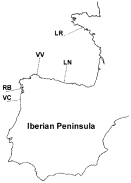
Fig. 1. Localities where Lecithochirium species were sampled in the present study. VC, Viana do Castelo (Portugal); RB, Riberia (Galicia, Spain); VV, Viveiro (Galicia, Spain); LN, Llanes (Asturias, Spain); LR, Lorient (Bretagne, France).

Table 1. Number of individual hosts, range and average number of parasites per eel of locations from which Lecithochirium populations were sampled

Table 2. Allele frequencies at 19 enzyme loci in populations of Lecithochirium species
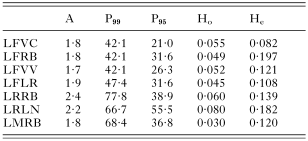
Table 3. Indices of genetic variability for populations of Lecithochirium species

Table 4. Estimates of deviations from Hardy–Weinberg proportions (FIS) and Fisher's test values within populations of species of the genus Lecithochirium

Table 5. Wright's F-statistics obtained by the method of Weir & Cockerham (1984) for Lecithochirium fusiforme populations

Table 6. Matrix of Nei's genetic distances below the diagonal, and the associated standard errors above diagonal, among populations of Lecithochirium species
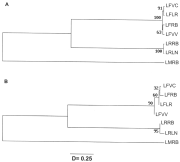
Fig. 2. UPGMA (A) and neighbour-joining (B) dendrograms showing evolutionary relationships among Lecithochirium populations on Nei's genetic distances (D). Population abbreviations as in Table 2.