Introduction
Cestodes of the family Taeniidae (Eucestoda: Cyclophyllidea) are parasites of mammals. It contains two important genera, Taenia and Echinococcus, both of them are medically and economically important (Gori et al., Reference Gori, Armua-Fernandez, Milanesi, Serafini, Magi, Deplazes and Macchioni2015). The larval stage of the canine tapeworm, Taenia ovis, is Cysticercus ovis that is commonly known as sheep measles. The adult tapeworm lives in the gut of domestic dogs and wild canids (i.e. coyotes, foxes, wolves) (Jenkins et al., Reference Jenkins, Urwin, Williams, Mitchell, Lievaart and Armua-Fernandez2014). In the muscles of intermediate hosts, C. ovis appears as a cyst-like lesion approximately 1 cm in diameter (Ransom, Reference Ransom1913). Taenia ovis does not present a human health risk; however, T. ovis imposes economic losses due to condemnation of infected carcass. In England, annual financial losses due to sheep cysticercosis is estimated at £7 million (Eichenberger et al., Reference Eichenberger, Karvountzis, Ziadinov and Deplazes2011). Taenia ovis infection potentially presents a serious threat to sheep industry in China (Zheng, Reference Zheng2016; Shi et al., Reference Shi, He, Guo, Liu, Gao, Zhan and Zheng2016). The life cycle of T. ovis involves a canid as a definitive host and sheep or goat as an intermediate host. In the small intestine of the definitive host, the parasite matures into an adult tapeworm and, via the host's defecation, releases large numbers of eggs into the environment that may later be consumed by intermediate host species (DeWolf et al., Reference DeWolf, Peregrine, Jones-Bitton, Jansen, MacTavish and Menzies2012).
Previous studies on helminth parasites of dogs showed 7.2% in the West of Iran, 3–8% in Mashhad and 24% in Esfahan were infected with T. ovis (Dalimi et al., Reference Dalimi, Sattari and Motamedi2006; Razmi et al., Reference Razmi, Sardari and Kamrani2006; Pastechian et al., Reference Pastechian, Rasoli and Yosefi2012; Emamapour et al., Reference Emamapour, Borji and Nagibi2015). Cysticercosis caused by T. ovis has been a serious economic problem for endemic countries and many studies have been done on the development of a vaccine against this parasite (Lightowlers, Reference Lightowlers2003). No genetic information is available for T. ovis in Iran and little is known about genomic content of this parasite in the world (Shamsaddini et al., Reference Shamsaddini, Mohammadi, Mirbadie, Rostami, Dehghani, Sadeghi and Harandi2017). More comprehensive molecular studies in this area are required to improve our understanding on the genetic diversity of this species and to provide a more effective vaccine against the parasite. Mitochondrial genes are maternally inherited and are haploid (clonal). Mitochondrial DNA is exposed to an increased rate of mutations and genetic variability within individual organisms. There can be no question that mtDNA-based procedures are invaluable tools for investigating inter- and intra-specific variation in parasitic organisms (Wallace, Reference Wallace1999; Gasser, Reference Gasser2006).
In the present investigation, we tried to determine morphometric and genetic diversity of this species using two mitochondrial cytochrome c oxidase subunit 1 (CO1) and 12S ribosomal RNA (12S rRNA) genes as well as larval rostellar hook morphometry in sheep isolates of T. ovis from Iran.
Materials and methods
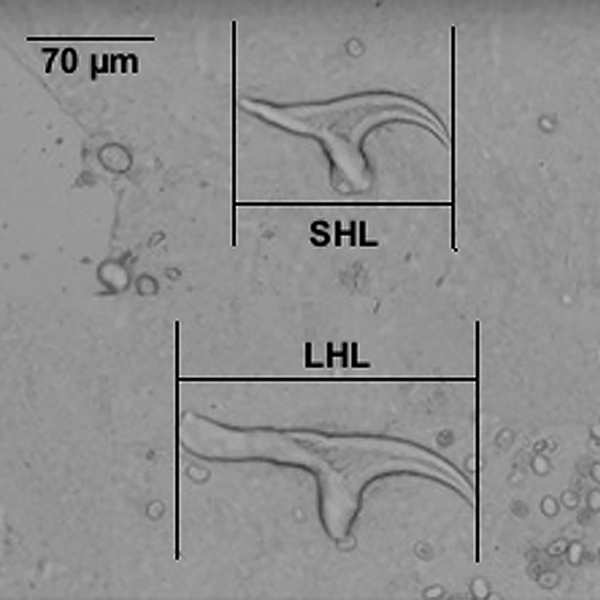
Ninety specimen of larval stage of T. ovis were collected from sheep in 18 slaughterhouses from Tehran and Alborz provinces of Iran from August 2010 to March 2011. The samples were washed three times with normal saline and stored at −20 °C until used. Morphometric study was carried out based on larval rostellar hook lengths. Each individual cysticercus was dissected under a stereomicroscope. Total lengths of each of seven large and seven small hooks per scolex were measured using a calibrated eyepiece micrometre under high-power magnification. All the measurements were made by one person (S.R.). Cluster analysis was applied to classify the subjects into homogeneous subgroups. Random-effects model was applied to estimate how much of variation of hook length was attributed to genetic differences between the subjects.
Part of each individual scolex was cut into small pieces and homogenized with an equal volume of distilled water. Each sample was frozen and thawed for six times in liquid nitrogen and 95 °C, respectively. DNA extraction was carried out using High Pure PCR Template Preparation Kit (Roche, Mannheim, Germany) according to the manufacturer's instructions. Each sample was incubated overnight in 200 µL tissue lysis buffer and 20 µg mL−1 proteinase K at 56 °C. Extracted DNA from the parasite was stored at −20 °C until used.
Two target sequences of mitochondrial DNA coding CO1 and 12S rRNA genes were amplified by PCR in a final reaction volume of 50 µL containing 250 mm each of deoxynucleotide triphosphate, 3.5 mm MgCl2, 2 units TaqDNA polymerase, 25 pmol of each primer and 4 µL (50–100 ng mL−1) of DNA template. Two primers, JB3 (forward): 5′-TTTTTTGGGCATCCTGAGGTTTAT-3′ and JB4.5 (reverse): 5′-TAAAGAAAGAACATAATGAAAATG-3′ were used to amplify an approximately 400 bp fragment of the part of CO1 gene (Bowles et al., Reference Bowles, Blair and McManus1992) under the following conditions: 5 min at 94 °C as an initial hot start step, followed by 35 cycles of 30 s at 94 °C, 45 s at 50 °C, 35 s at 72 °C, and a final extension step 10 min at 72 °C.
A taeniid-specific primer pair was designed for the amplification of 12S rRNA gene using Primer-BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). PCR was carried out using 12SRF (5′-AGGGGATAGGACACAGTGCCAGC-3′) as a forward and 12SRR (5′-CGGTGTGTACAT GAGCTAAAC-3′) as a reverse primer under the following conditions: 5 min at 94 °C as an initial hot start step, followed by 35 cycles of 30 s at 94 °C, 45 s at 57 °C, 35 s at 72 °C, and a final extension step of 10 min at 72 °C. The primers amplified an approximately 500 bp fragment of the part of 12S rRNA gene. Negative control (no DNA) was included in each experiment. The amplification reactions were done in a PCR machine (FlexCycler, Analytik Jena AG, Germany) and the amplicons were electrophoresed on 1% (W/V) agarose gel containing ethidium bromide.
All the amplicons were subjected to sequencing using ABI 3700 DNA Analyzer (Applied Biosystem, Foster, CA, USA). Sequence data were adjusted manually and complete alignments were carried out using the software BioEdit and ClustalW (Thompson et al., Reference Thompson, Gibson, Plewniak, Jeanmougin and Higgins1997; Hall, Reference Hall1999). The nucleotide and corresponding amino acid sequences of CO1 and 12S rRNA genes were submitted to NCBI GenBank (http://www.ncbi.nlm.nih.gov) (Table 1).
Table 1. Frequency distribution of Taenia ovis haplotypes from Iranian sheep for two mitochondrial CO1 and 12S rRNA genes with the corresponding GenBank accession numbers

Bayesian inference was conducted using the software MrBayes v.3.1.2 (http://mrbayes.csit. fsu.edu/index.php). Posterior probabilities (pp) were designed for 2 000 000 generations (ngen: 2 000 000). TreeviewX v.0.5.0 program (Page, Reference Page2002) was used to depict the resulting trees. The trees were run using sequences obtained in this study as well as the reference sequences available for representative Taenia species in GenBank. Echinococcus granulosus sensu stricto G1 genotype (Accession No. NC008075) was applied in the model as outgroup. MEGA7 program was used for constructing pairwise distance matrices.
Results
Linear measurement of larval hooks was done in 90 sheep isolates. The mean ± s.d. for total length of large and small hooks were 174.1 ± 6.4 µm (range 158.0–195.0) and 116.7 ± 5.4 µm (range 100.8–132.3), respectively (Fig. 1). Random-effect model analysis indicated that 75 and 69% of variations in large and small hook lengths are attributable to differences in individual isolates, respectively (P < 0.001). Interclass correlation coefficients (ICCs) showed that there were no significant association between small and large hook length variation and the variability within either of CO1 and 12S rRNA genes. Results revealed that 35% of variation in large hook length is associated with CO1 gene variation (P = 0.07, Table 2).

Fig. 1. Measurement of the large and small rostellar hooks of T. ovis as used in the present study.
Table 2. Interclass correlation coefficients (ICCs) obtained using random-effect model analysis for large and small hook lengths and the variability in CO1 and 12S rRNA genes of sheep isolates of T. ovis

PCR amplification was successfully performed on all of the isolates for both CO1 (400 bp) and 12S rRNA (450 bp) genes. Total number of mutations was 10 and nine in CO1 and 12S rRNA genes that occurred in 10 and nine polymorphic sites, respectively. CO1 gene showed 11 representative profiles (haplotypes) designated as IRTOCO1–IRTOCO11 and 12S rRNA gene showed nine representative profiles designated as IRTOSR1–IRTOSR9. All different sequences from CO1 and 12S rRNA of T. ovis were identified and submitted to GenBank under the accession numbers JX134111–JX134122 and JX134123–JX134131, respectively (Table 1). The level of pairwise nucleotide variation between individual haplotypes of CO1 gene is determined to be 0.3–1.1%, while the overall nucleotide variation among all 11 haplotypes was 2.6% (Fig. 2). For 12S rRNA sequence data, the level of pairwise nucleotide variation was found to be 0.2–1.0% and the overall nucleotide variation was determined as 2.1% among nine haplotypes.

Fig. 2. Pairwise comparison of nucleotide sequence differences (%) in CO1 and 12S rRNA genes among 90 T. ovis isolates and other related taeniids.
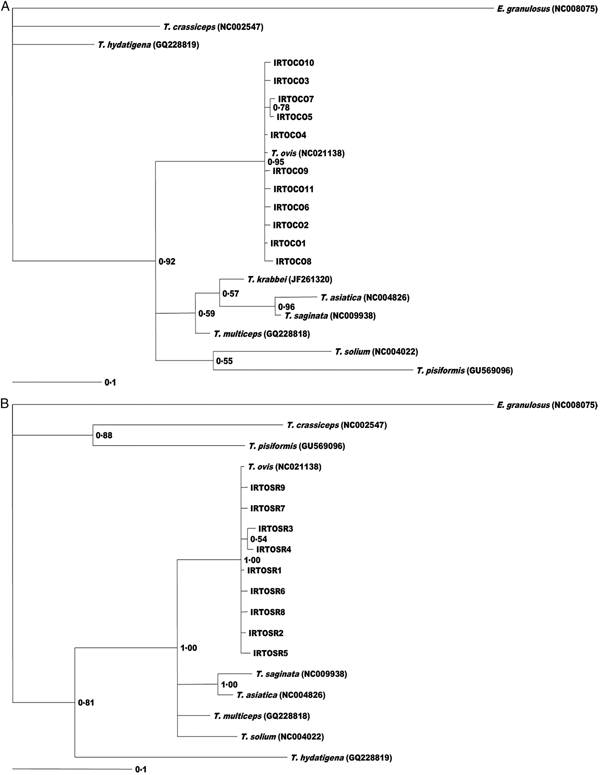
The consensus trees based on phylogenetic analysis of CO1 and 12S rDNA sequence data showed that all T. ovis isolates are in a single clade with high statistical support (pp: 0.95; 0.1) comprised of all haplotypes (Fig. 3).

Fig. 3. Genetic relationships of sheep isolates of T. ovis from the present study and other published sequences for taeniids and E. granulosus as outgroup. The relationships were inferred based on phylogenetic analysis of cox1 (A) and 12SrRNA (B) data using Bayesian inference. The accession numbers and sources of sequences are shown in Table 1. Nodal support is given as a P value.
Discussion
Ovine cysticercosis infection has been a common problem of sheep and goat in the world, however our knowledge on the intraspecific genetic variation of the causative agent, T. ovis is limited to a couple of studies on a few number of isolates (Gasser et al., Reference Gasser, Zhu and McManus1999; Zhang et al., Reference Zhang, Hu, Jones, Allsopp, Beveridge, Schindler and Gasser2007). Gasser et al. (Reference Gasser, Zhu and McManus1999) demonstrated the genetic relationship of mitochondrial ND1 gene of T. ovis isolated from New Zealand sheep compared with eight other Taenia species (Gasser et al., Reference Gasser, Zhu and McManus1999). Recently complete mitochondrial genome of T. ovis has been released in NCBI GenBank (Nakao et al., Reference Nakao, Lavikainen, Iwaki, Haukisalmi, Konyaev, Oku and Ito2013). The present study was conducted to determine morphological and genetic variation of T. ovis on relatively large number of sheep isolates. Sequence heterogeneity of two mitochondrial CO1 and 12S rRNA genes within T. ovis isolates of sheep were investigated in the present study.
In her classical work on the taxonomic revision of different Taenia species, Verster (Reference Verster1969) found the range of large and small hook lengths as 170–191 and 111–127 µm, respectively. The existing hook length data for T. ovis in the literature is reviewed in Table 3 (Verster, Reference Verster1969; Loos-Frank, Reference Loos-Frank2000). Total large and small rostellar hook length of C. ovis isolates obtained from the present study were in agreement with the existing hook data for this parasite. ICCs obtained from differences in rostellar hook length indicated that a large part of the differences in rostellar hook length is significantly attributable to differences among individual isolates of T. ovis (P < 0.001). This means that observed hook length differences among T. ovis isolates had not occurred by chance and rostellar hook length was varied based on statistically significant variations among individual isolates.
Table 3. Summary of present data and comparison of large and small hook lengths of T. ovis from different studies

In tapeworms, the genetic basis of rostellar hook development is poorly understood. There were no association between the length of rostellar hooks and the genetic diversity in CO1 and 12S rRNA genes. However, marginally significant results showed that 35% of variation in the large hook length is associated with CO1 gene variation (P = 0.07, Table 2).
CO1 sequence analysis of 11 T. ovis profiles of the present study revealed 10 nucleotide substitutions of which five substitutions were nucleotide transversions. As shown in Table 4, six amino acid changes were seen due to 10 nucleotide substitutions within the haplotypes. There is no evidence that any change in the amino acid composition of cytochrome c oxidase might affect the function of this enzyme or parasite adaptation. However, it has been shown in other parasitic species that even a single amino acid change could affect the biological fitness of the organism (Tachibana et al., Reference Tachibana, Matsumoto, Cheng, Tsukamoto and Yoshihara2004; Otsuki et al., Reference Otsuki, Kaneko, Thongkukiatkul, Tachibana, Iriko, Takeo and Torii2009). Further studies are required to evaluate biological consequences of mitochondrial gene variation in taeniid species.
Table 4. Nucleotide substitutions and the corresponding amino acid changes in 11 CO1 sequence profiles of T. ovis of sheep in Iran compared with the reference CO1 sequence from New Zealand (accession number NC021138)

*No substitution.
The study showed a lower degree of variation in CO1 and 12S rRNA genes of T. ovis in comparison with other Taenia species. Figure 2 shows pairwise comparison of CO1 and 12S rRNA nucleotide differences between taeniid species and T. ovis isolates of the present study. Pairwise comparison of T. ovis isolates from the present study and available mitochondrial sequences from New Zealand sheep isolate showed 0.0–0.8 and 0.0–0.5% nucleotide difference in CO1 and 12S rRNA genes, respectively (Nakao et al., Reference Nakao, Lavikainen, Iwaki, Haukisalmi, Konyaev, Oku and Ito2013). Intraspecific variation within different isolates of T. multiceps, T. hydatigena and T. taeniaeformis showed 0.2–5.6, 0.3–3.4 and 0.3–4.1% CO1 nucleotide differences, respectively (Okamoto et al., Reference Okamoto, Bessho, Kamiya, Kurosawa and Horii1995; Avcioglu et al., Reference Avcioglu, Yildirim, Duzlu, Inci, Terim and Balkaya2011; Varcasia et al., Reference Varcasia, Jia, Yan, Manunta, Pipia, Garippa and Schuster2012, Reference Varcasia, Pipia, Dessì, Zidda, Tamponi, Pau and Boufana2016; Boufana et al., Reference Boufana, Scala, Lahmar, Pointing, Craig, Dessì and Varcasia2015). This presents relatively higher level of intraspecific variations than that of T. ovis isolates from different localities. Additional studies from other parts of the world are needed to provide a comprehensive picture of intraspecific variability of T. ovis.
The level of nucleotide variation in CO1 and 12S rRNA between T. ovis haplotypes from the present study and eight other Taenia species was found to be 11.3–17.8 and 5.3–16.3%, respectively. This is in agreement with the expected variations in CO1 in the genus Taenia that has been estimated at 6.3–15.8% by McManus and Bowles, Reference McManus and Bowles1994 (McManus and Bowles, Reference McManus and Bowles1994). Currently T. ovis sensu lato is believed to include two subspecies namely T. ovis ovis and T. ovis krabbei. In the present study, 14.0–15.2% pairwise nucleotide difference in CO1 was documented between T. ovis ovis isolates of the present study and T. ovis krabbei. Pairwise comparison of T. ovis with each of the four zoonotic Taenia species (T. saginata, T. solium, T. multiceps and T. asiatica) showed 11.3–14.3% nucleotide difference. Sweatman and Henshall (Reference Sweatman and Henshall1962) found that sheep and goat were refractory to infection with T. ovis krabbei and deers were refractory to T. ovis ovis infection (Sweatman and Henshall, Reference Sweatman and Henshall1962). Verster (Reference Verster1969) described morphological differences in the male reproductive system (cirrus pouch extension and layers of testes) between the two subspecies. Our mitochondrial genetic results support Sweatman and Henshall (Reference Sweatman and Henshall1962) experimental evidence of biological distinctness of T. ovis ovis and T. ovis krabbei. CO1 nucleotide difference between the two subspecies is equal or greater than that of observed differences between T. ovis ovis and either of the more distinct species like T. saginata and T. multiceps (Varcasia et al., Reference Varcasia, Jia, Yan, Manunta, Pipia, Garippa and Schuster2012; Rostami et al., Reference Rostami, Salavati, Beech, Sharbatkhori, Babaei, Saedi and Harandi2013, Reference Rostami, Salavati, Beech, Babaei, Sharbatkhori and Harandi2015). The nucleotide difference between T. ovis ovis and T. asiatica is 13.6% that is lesser than the corresponding difference between T. ovis ovis and T. ovis krabbei. Therefore, based on the present molecular, biological, morphological and intermediate host differences (Lavikainen et al., Reference Lavikainen, Haukisalmi, Lehtinen, Henttonen, Oksanen and Meri2008), the two subspecies could be considered distinct species as T. ovis (Cobbold, Reference Cobbold1869) (Ransom, Reference Ransom1913), and Taenia krabbei (Moniez, Reference Moniez1879). Likewise in other taeniid species, e.g. T. multiceps, the existence of considerable intraspecific variations among isolates from Turkey (Erzurum), Italy (Sardinia) and the UK (Wales) warrants further investigations (Varcasia et al., Reference Varcasia, Jia, Yan, Manunta, Pipia, Garippa and Schuster2012; Boufana et al., Reference Boufana, Scala, Lahmar, Pointing, Craig, Dessì and Varcasia2015).
The dendrogram generated by phylogenetic analysis uniformly clustered all CO1 and 12S rRNA haplotypes into a single clade together with T. ovis reference sequence. Taenia krabbei, T. asiatica, T. saginata and T. multiceps were clustered together as a distinct subclade.
Conclusion
Phylogenetic analysis has confirmed the distinctness of T. ovis ovis and T. ovis krabbei as separate species. Bayesian analyses using CO1 and 12S rRNA genes of nucleotide sequences represented phylogenetic trees with similar topologies. However, 12S rRNA tree was slightly different due to the lack of 12S rRNA gene sequence data for T. krabbei. To promote our knowledge on the nature of genetic variation in this taeniid tapeworm, further investigations are required to characterize mitochondrial and nuclear genes from T. ovis as well as T. krabbei isolates from different endemic countries around the world.
Author ORCIDs
Majid Fasihi Harandi http://orcid.org/0000-0003-3257-5389
Acknowledgements
The authors wish to thank all veterinary staff of different abattoirs that help collecting parasite specimens for this study.
Financial support
This work was financially supported by the Vice-Chancellor for Research, Kerman University of Medical Sciences, grant number 90-072.
Conflict of interest
None.
Ethical standards
Not applicable.