INTRODUCTION
Trichomonads (Parabasalia) are flagellated protists classified into the eukaryotic supergroup Excavata (Cavalier-Smith, Reference Cavalier-Smith2002). Most of the approximately 450 described trichomonad species (Adl et al. Reference Adl, Leander, Simpson, Archibald, Anderson, Bass, Bowser, Brugerolle, Farmer, Karpov, Kolisko, Lane, Lodge, Mann, Meisterfeld, Mendoza, Moestrup, Mozley-Standridge, Smirnov and Spiegel2007) are beneficial mutualists or commensals of termites and other insects, while only a minority has been described from vertebrate intestines. Some trichomonad species, e.g. Trichomonas vaginalis, T. gallinae, Tritrichomonas foetus, and Histomonas meleagridis, are important pathogens of various internal organs of humans and domestic animals (Honigberg, Reference Honigberg and Kreier1978a, Reference Honigberg and Kreierb; McDougald and Reid, Reference McDougald, Reid and Kreier1978). Besides the endobiotic trichomonads, a few free-living representatives exist as well (Cepicka et al. Reference Cepicka, Hampl and Kulda2010; Yubuki et al. Reference Yubuki., Céza, Cepicka, Yabuki, Inagaki, Nakayama, Ionuye and leander2010). Despite their relatively low species richness outside the insect gut, trichomonads have been commonly recorded from many vertebrate species. However, true diversity of trichomonads in vertebrates is only poorly understood. Several trichomonad species have been recently newly reported even from humans and domestic animals, whose endobiotic protists have been intensively studied for a long time (Levy et al. Reference Levy, Gookin, Poore, Birkenheuer, Dykstra and Litaker2003; Cepicka et al. Reference Cepicka, Kutišová, Tachezy, Kulda and Flegr2005, Reference Cepicka, Hampl, Kulda and Flegr2006; Kutisova et al. Reference Kutisova, Kulda, Cepicka, Flegr, Koudela, Teras and Tachezy2005; Dufernez et al. Reference Dufernez, Walker, Noël, Caby, Mantini, Delgado-Viscogliosi, Ohkuma, Kudo, Capron, Pierce, Villanueva and Viscogliosi2007; Mantini et al. Reference Mantini, Souppart, Noël, Duong, Mornet, Carroger, Dupont, Masseret, Goustille, Capron, Duboucher, Dei-Cas and Viscogliosi2009). Besides humans, domestic animals and a few other vertebrates, the diversity of intestinal trichomonads has been largely understudied, a situation that surprisingly also applies for non-human primates.
The non-human primates are our closest relatives and represent a species-rich mammalian group, several of whose members are seriously endangered. Despite several recent studies (e.g., Levecke et al. Reference Levecke, Geldhof, Claerebout, Dorny, Vercammen, Cacciò, Vercuysse and Geurden2009; Stensvold et al. Reference Stensvold, Alfellani, Nørskov-Lauritsen, Prip, Victory, Maddox, Nielsen and Clark2009; Johnston et al. Reference Johnston, Gillespie, Rwego, McLachlan, Kent and Goldberg2010; Pomajbíková et al. Reference Pomajbíková, Petrželková, Profousová, Petrašová, Kišidayová, Varadyová and Modrý2010), the diversity of parasite communities in primates remains neglected in several aspects including trichomonads. So far, 9 species of intestinal trichomonads have been identified in non-human primates (e.g. Deschiens, Reference Deschiens1927; Hegner and Ratcliffe, Reference Hegner and Ratcliffe1927; Wenrich, Reference Wenrich1944a; Flick, Reference Flick1954; Abraham, Reference Abraham1961, Reference Abraham1962; Culbertson et al. Reference Culbertson, Pindak, Gardner and Honigberg1986; Pindak and de Pindak, Reference Pindak and de Pindak1998; Stark et al. Reference Stark, Phillips, Peckett, Munro, Marriott, Harkness and Ellis2008). This number is comparable to that of intestinal trichomonad species known from the other mammal groups, such as rodents, ruminants or suids. Most of the species were, however, reported only once in a single study and never have been observed again. The morphological descriptions were usually inadequate and they cannot be considered in taxonomical studies. In addition, almost no sequence data of any trichomonad from non-human primates are currently available. The lack of sequence data is especially unsatisfactory as it was recently shown that SSU rDNA and the ITS region could be used as a suitable barcode for trichomonads (e.g. Cepicka et al. Reference Cepicka, Hampl, Kulda and Flegr2006; Dufernez et al. Reference Dufernez, Walker, Noël, Caby, Mantini, Delgado-Viscogliosi, Ohkuma, Kudo, Capron, Pierce, Villanueva and Viscogliosi2007).
Recently, primates have re-attracted attention of parasitologists and thorough studies of some of their intestinal protists have been carried out (Levecke et al. Reference Levecke, Geldhof, Claerebout, Dorny, Vercammen, Cacciò, Vercuysse and Geurden2009; Stensvold et al. Reference Stensvold, Alfellani, Nørskov-Lauritsen, Prip, Victory, Maddox, Nielsen and Clark2009; Johnston et al. Reference Johnston, Gillespie, Rwego, McLachlan, Kent and Goldberg2010; Pomajbíková et al. Reference Pomajbíková, Petrželková, Profousová, Petrašová, Kišidayová, Varadyová and Modrý2010). This report describes the first molecular-phylogenetic study to investigate the diversity of intestinal trichomonads of non-human primates.
MATERIALS AND METHODS
Organisms
Thirty-five fresh fecal samples of 12 primate species were collected in 3 zoos, namely those in Brno, Pilsen and Ostrava (Czech Republic) from March to July 2005. The samples were immediately inoculated into Dobell and Leidlaw's biphasic medium (Dobell and Leidlaw, Reference Dobell and Leidlaw1926) and were cultivated at 37°C after transport to the laboratory. The isolates were maintained in xenic cultures in this medium by serial transfer every 2nd or 3rd day. In total, 30 stable trichomonad strains obtained from 11 primate species have been established (see Table 1). The remaining 5 samples were negative for trichomonads: 2 from olive baboons (Papio anubis) from the Brno Zoo, 1 from a chimpanzee (Pan troglodytes) from the Ostrava Zoo, 1 from a chimpanzee from the Brno Zoo and 1 from a Hanuman langur (Semnopithecus entellus) from the Ostrava Zoo. Strains GANG1, GSIA1, KATA1, KATASAMEC, KOMBG1, KOSBE1, MA3, MANI1, MANI2, MASP1, PAN3, SENT1, SENT2, and VARI1 have been deposited in the culture collection of the Department of Parasitology of Charles University in Prague, Czech Republic.
Table 1. List of trichomonad strains included in the study

DNA extraction, amplification, cloning and sequencing
Genomic DNA was isolated from roughly the fourth passage of cultures using the DNeasy Blood & Tissue Kit (Qiagen). Primers 16Sl (TACTTGGTTGATCCTGCC; Tachezy et al. Reference Tachezy, Tachezy, Hampl, Šedinová, Vaňáčová, Vrlík, Van Ranst, Flegr and Kulda2002) and 16SRR (TCACCTACCGTTACCTTG; Cepicka et al. Reference Cepicka, Kutišová, Tachezy, Kulda and Flegr2005) were used to amplify SSU rDNA. Primers ITSF (TTCAGTTCAGCGGGTCTTCC) and ITSR (GTAGGTGAACCTGCCGTTGG) (Cepicka et al. Reference Cepicka, Kutišová, Tachezy, Kulda and Flegr2005) were used to amplify ITS1-5.8S rDNA-ITS2. The PCR products were purified using the QIAquick PCR Purification Kit (Qiagen) and were directly sequenced on the ABI Prism 3100-Avant Genetic Analyzer (Applied Biosystems). The ITS region of the isolate GSIA1 was cloned into the pGEM®-T EASY vector (Promega). Eight clones were sequenced. The newly obtained sequences have been deposited in GenBank under Accession numbers HQ149966 – HQ150005.
Phylogenetic analyses
Pre-emptive phylogenetic analysis of obtained SSU rDNA sequences placed all but 11 of the newly obtained sequences to the Tetratrichomonas group A, as defined by Cepicka et al. (Reference Cepicka, Hampl, Kulda and Flegr2006). All these isolates also contained the 2 SSU rDNA insertions typical for group A (Cepicka et al. Reference Cepicka, Hampl, Kulda and Flegr2006). Because much of the sequence variability of tetratrichomonad group A was lost during the removal of hypervariable positions from the alignment representing broad parabasalid diversity, SSU rDNA sequences were used in 2 separate analyses. The first data set consisted of 47 SSU rDNA sequences representing all main parabasalid lineages and 11 of the new sequences which did not belong to the tetratrichomonad group A. The second data set consisted of 25 published SSU rDNA sequences of the tetratrichomonad group A, including the LP isolate, 19 newly obtained sequences placed in group A, and 3 non-group-A Tetratrichomonas sequences used as outgroups. In addition to the 2 data sets, a third data set containing ITS1-5.8S rDNA-ITS2 sequences of tetratrichomonad group A (40 obtained from GenBank and 10 new ones) was created. Sequences from each data set were aligned using the MAFFT method (Katoh et al. Reference Katoh, Misawa, Kuma and Miyata2002) with the help of the MAFFT 6 server (http://align.bmr.kyushu-u.ac.jp/mafft/online/server/) using the G-INS-i algorithm at default settings. The resulting alignments were manually edited in BioEdit 7.0.9.0 (Hall, Reference Hall1999). The lengths of the final alignments were 1323 (first data set), 1538 (second data set), and 345 (third data set) characters. The alignments are available from the corresponding author upon request.
Phylogenetic trees were constructed by maximum parsimony (MP), Fitch-Margoliash distance method with maximum likelihood distances (MLdist), maximum likelihood (ML), and Bayesian methods. The models of nucleotide substitution for ML and MLdist methods were chosen by a hierarchical nested likelihood ratio test implemented in Modeltest 3.7 (Posada and Crandall, Reference Posada and Crandall1998). The models were selected as follows: TrN+I+Γ for the first data set, GTR+I+Γ for the second data set and F81+I+Γ for the third data set. MP and MLdist trees were constructed in PAUP* 4.0b10 (Swofford, Reference Swofford2002) by 10 replicates of heuristic search in which the starting tree was obtained by the stepwise addition procedure with a random order of taxon addition and swapped using the tree bisection and reconnection (TBR) algorithm. The trees were bootstrapped with 1000 replicates. ML trees were constructed in Phyml (Guindon and Gascuel, Reference Guindon and Gascuel2003) and were bootstrapped with 1000 replicates. Bayesian analyses were performed using MrBayes 3.1 (Huelsenbeck and Ronquist, Reference Huelsenbeck and Ronquist2001). Base frequencies, rates for the 6 different types of substitution, the proportion of invariable sites and the shape parameter of the gamma correction for the rate heterogeneity (approximated by 4 discrete categories) were allowed to vary. A co-varion model was used to allow rate heterogeneity along the tree. The number of generations of Markov chain Monte Carlo was 2×106 for the first and third data set, and 3×106 for the second data set (until the average standard deviation of split frequencies was lower than 0·01); the trees were sampled every 100th generation. The first 5000 (first and third data set) or 7500 (second data set) trees were discarded as burn-in.
RESULTS
Trichomonads were present in approximately 85% in primocultures on the second day after the inoculation.
Figure 1 shows the phylogenetic tree of Parabasalia constructed from SSU rDNA sequences on the basis of the first data set. The tree topology was similar to the topology obtained by Cepicka et al. (2010). Classes Trichonymphea, Trichomitea, Cristamonadea, Spirotrichonymphea and Trichomitea formed robust clades with all the used methods. The class Tritrichomonadea formed an unsupported clade only in ML analysis (bootstrap value 39), whereas it was paraphyletic, having Cristamonadea as an inner branch, in the other analyses. The internal topology of Tritrichomonadea was not well resolved. The close relationship between Tritrichomonadea and Cristamonadea was recovered by all methods, although without strong support. The class Trichomonadea was always paraphyletic, having Trichonymphea as an inner branch. However, the position of Trichonymphea was not well supported by any method. The strain KOMBG1 formed a well-supported clade with Hypotrichomonas acosta. The strain KATA1 robustly clustered with Trichomitus batrachorum. Sequences of strains GANG1, KOSBE1 and VARI1 were identical with that of Pentatrichomonas hominis. The strain KATASAMEC was placed to the Tetratrichomonas gallinarum species complex and occupied a sister position of the GPO strain. Sequences of the MA3, MA4, MANI1, MANI2, and MASP1 strains were identical and formed a novel tetratrichomonad lineage (here called novel Tetratrichomonas lineage 1) that clustered, albeit weakly, with the ZUBR strain (Tetratrichomonas lineage 11). Tetratrichomonad group A was recovered by all methods with low support.

Fig. 1. Phylogenetic tree of Parabasalia based on SSU rDNA. The tree was constructed from the first data set by ML method under the TrN+I+Γ substitution model and is unrooted. Values at the nodes represent statistical support (MP bootstrap value/MLdist bootstrap value/ML bootstrap value/Bayesian posterior probability). Asterisks indicate nodes with bootstrap support under 50%. The newly obtained sequences are in bold.
All remaining strains of trichomonads from primates belonged to Tetratrichomonas group A. The LP strain from human (Mantini et al. Reference Mantini, Souppart, Noël, Duong, Mornet, Carroger, Dupont, Masseret, Goustille, Capron, Duboucher, Dei-Cas and Viscogliosi2009) was regarded as a member of group A on the basis of 2 insertions in the SSU rDNA sequence that are typical for the group (Cepicka et al. Reference Cepicka, Hampl, Kulda and Flegr2006). Figure 2 shows the phylogenetic tree of Tetratrichomonas group A constructed from SSU rDNA sequences on the basis of the second data set. The tetratrichomonad lineages defined by Cepicka et al. (Reference Cepicka, Hampl, Kulda and Flegr2006) were robustly recovered. However, relationships between particular tetratrichomonad lineages were generally unsupported, correspondingly to results of previous studies (Cepicka et al. Reference Cepicka, Hampl, Kulda and Flegr2006; Dufernez et al. Reference Dufernez, Walker, Noël, Caby, Mantini, Delgado-Viscogliosi, Ohkuma, Kudo, Capron, Pierce, Villanueva and Viscogliosi2007; Mantini et al. Reference Mantini, Souppart, Noël, Duong, Mornet, Carroger, Dupont, Masseret, Goustille, Capron, Duboucher, Dei-Cas and Viscogliosi2009). The trichomonad LP formed a basal branch of tetratrichomonad group A. The sequence of the PAN11 strain was identical with that of the Tetratrichomonas sp. PB strain (lineage 8). SENT1 and SENT4 strains had identical SSU rDNA sequences and belonged to Tetratrichomonas lineage 7. Strains GSIA1, GSIA2, and GSIA3 had identical sequences and clustered within Tetratrichomonas group 10. The sequence of the trichomonad MMV-2003 obtained from GenBank (Accession number AY247747) from a black crested gibbon (Nomascus concolor) was placed within lineage 10 as well. The remaining 13 strains from non-human primates formed a novel tetratrichomonad lineage (here called the novel Tetratrichomonas lineage 2), which was closely related to lineage 10. The mean genetic distance (p-distance) between lineage 10 and the new lineage was 1·0%.
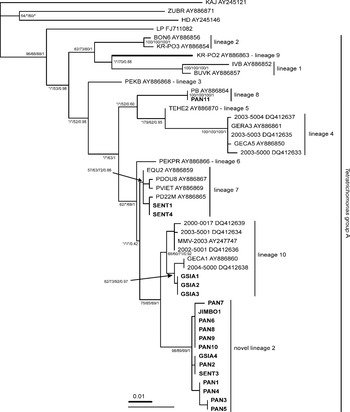
Fig. 2. Phylogenetic tree of Tetratrichomonas group A based on SSU rDNA. The tree was constructed from the second data set by ML method under the GTR+I+Γ substitution model and was rooted with Tetratrichomonas KAJ, ZUBR and HD. Values at the nodes indicate statistical support (MP bootstrap value/MLdist bootstrap value/ML bootstrap value/Bayesian posterior probability). Asterisks indicate nodes with bootstrap support under 50%. Tetratrichomonas lineages as defined by Cepicka et al. (Reference Cepicka, Hampl, Kulda and Flegr2006) are indicated. The branch length of Tetratrichomonas sp. KR-PO2 was reduced by one half. The newly obtained sequences are in bold.
Figure 3 shows the phylogenetic tree of Tetratrichomonas group A constructed from ITS1-5.8S rRNA-ITS2 on the basis of the third data set. All Tetratrichomonas lineages appeared monophyletic and well supported. However, relationships between the particular lineages remained mostly unresolved, as in a previous study (Cepicka et al. Reference Cepicka, Hampl, Kulda and Flegr2006). Contrary to the SSU rDNA analysis, the trichomonad LP formed a robust clade with lineages 4 and 5. The sister branch of this clade was formed by the KR-PO2 strain (lineage 9), although with low support. The phylogenetic position of the newly obtained strains was in agreement with the SSU rDNA analysis. SENT1 and SENT4 strains with identical sequences were placed within lineage 7. The PAN11 strain was in a sister position to the PB strain. The ITS region of the GSIA1 strain was subcloned because it was impossible to directly sequence the PCR product. Sequences of 7 clones were almost identical, differing in at most 2 positions. However, their consensual sequence differed from the sequence of the clone 1_10 in 6 substitutions and 1 indel, which indicated that either intragenomic polymorphism or 2 different, though closely related, organisms were present within the GSIA1 strain. Both of the sequences branched within lineage 10. Strains PAN2, PAN3, PAN6, PAN9, and JIMBO1 formed a robust clade closely related to lineage 10. The mean genetic distance between lineage 10 and the new clade was 3·8%.

Fig. 3. Phylogenetic tree of tetratrichomonad group A based on ITS1-5.8S rDNA-ITS2. The tree was constructed from the third data set by ML method under the F81+I+Γ substitution model and is unrooted. Values at the nodes indicate statistical support (MP bootstrap value/MLdist bootstrap value/ML bootstrap value/Bayesian posterior probability). Asterisks indicate nodes with bootstrap support under 50%. Tetratrichomonas lineages as defined by Cepicka et al. (Reference Cepicka, Hampl, Kulda and Flegr2006) are indicated. The newly obtained sequences are in bold.
DISCUSSION
Although non-human primates are our closest relatives, investigation of their intestinal parasites has been neglected for decades. Most of the studies focused on their intestinal trichomonads were published before the introduction of molecular phylogenetics into protistology, and were thus limited to light microscopy (e.g. Deschiens, Reference Deschiens1927; Wenrich, Reference Wenrich1944a; Flick, Reference Flick1954; Abraham, Reference Abraham1961, Reference Abraham1962; Reardon and Rininger, Reference Reardon and Rininger1968; Culbertson et al. Reference Culbertson, Pindak, Gardner and Honigberg1986; Pindak and de Pindak, Reference Pindak and de Pindak1998). Unfortunately, the morphological descriptions were mostly inadequate, too brief or confusing. Although intestinal parasites of non-human primates have attracted more attention during the last years, new reports pertaining to intestinal trichomonads almost ceased. Only 4 studies of intestinal trichomonads of non-human primates were published recently (Lilly et al. Reference Lilly, Mehlman and Doran2002; Carmona et al. Reference Carmona, Bermudez, Gutierrez-Espeleta, Porras and Ortiz2005; Stark et al. Reference Stark, Phillips, Peckett, Munro, Marriott, Harkness and Ellis2008; Lankester et al. Reference Lankester, Kiyang, Bailey and Unwin2010), the 2 former being almost uninformative. Such a paucity of data on this group contrasts with literature addressing the diversity of Entamoeba (Suzuki et al. Reference Suzuki, Kobayashi, Murata, Tajima, Hashizaki, Yanagawa and Takeuchi2008; Tachibana et al. Reference Tachibana, Yanagi, Akatsuka, Kobayash, Kanbara and Tsutsumi2009), Blastocystis (Stensvold et al. Reference Stensvold, Alfellani, Nørskov-Lauritsen, Prip, Victory, Maddox, Nielsen and Clark2009; Parkar et al. Reference Parkar, Traub, Vitali, Vitali, Elliot, Levecke, Robertson, Geurden, Steele, Drake and Thompson2010), Giardia (Levecke et al. Reference Levecke, Geldhof, Claerebout, Dorny, Vercammen, Cacciò, Vercuysse and Geurden2009; Johnston et al. Reference Johnston, Gillespie, Rwego, McLachlan, Kent and Goldberg2010), entodiniomorphid ciliates (Pomajbíková et al. Reference Pomajbíková, Petrželková, Profousová, Petrašová, Kišidayová, Varadyová and Modrý2010; Tokiwa et al. Reference Tokiwa, Modrý, Ito, Pomajbíková, Petrželková and Imai2010) or nematodes (e.g. Cutillas et al. Reference Cutillas, Callejon, de Rojas, Tewes, Ubeda, Ariza and Guevara2009; Hasegawa et al. Reference Hasegawa, Sato, Fujita, Nguema, Nobusue, Miyagi, Kooriyama, Takenoshita, Noda, Sato, Morimoto, Ikeda and Nishida2010; Krief et al. Reference Krief, Vermeulen, Lafosse, Kasenene, Nieguitsila, Berthelemy, L'Hostis, Bain and Guillot2010). The reason for such a discrepancy is most likely connected to a methodological bias. At present, coproscopical methods are mostly employed to study the intestinal parasites of non-human primates. They are optimized for the detection of cysts of protists and eggs of helminths (Greiner and McIntosh, Reference Greiner, McIntosh, Huffman and Chapman2009). However, the ability to form true cysts was demonstrated only for a few trichomonad species (Hampl et al. Reference Hampl, Cepicka, Flegr, Tachezy and Kulda2007). The vast majority of trichomonads produce only trophozoites that remain undetected by the coprological methods. On the other hand, a PCR-based approach, which has been used for trichomonads from various other host taxa, has been employed only once in the case of non-human primates (Stark et al. Reference Stark, Phillips, Peckett, Munro, Marriott, Harkness and Ellis2008). In the present study, we used this promising approach to study the diversity of cultivated trichomonad strains obtained from the feces of various non-human primate species.
Our results show that intestinal trichomonads are rather common in various species of non-human primates. To exclude possible cultivation bias, i.e. the loss of uncultivable trichomonad species during early passages, we created polyxenic cultures in the Dobell and Leidlaw's (Reference Dobell and Leidlaw1926) biphasic medium. Our previous experience showed that most trichomonad species are able to survive at least a few passages under such conditions. To detect trichomonads, the cultures were microscopically controlled the second day after the inoculation of feces into the medium. We are therefore convinced that the results correspond to the actual occurrence of these trichomonads among examined individuals.
According to their distribution in phylogenetic trees, the studied trichomonad isolates from non-human primates belong to 8 or 9 distinct species. As far as we know, 9 trichomonad species have been reported from the intestine of primates, including humans (Deschiens, Reference Deschiens1927; Cleveland, Reference Cleveland1928; Wenrich, Reference Wenrich1944a; Flick, Reference Flick1954; Abraham, Reference Abraham1961, Reference Abraham1962; Culbertson et al. Reference Culbertson, Pindak, Gardner and Honigberg1986; Pindak and de Pindak, Reference Pindak and de Pindak1998; Stark et al. Reference Stark, Phillips, Peckett, Munro, Marriott, Harkness and Ellis2008). However, all but 1 of these species either are clearly distinct from our isolates or were inadequately described and thus cannot be considered as valid in taxonomic studies. The morphological evaluation of the presented material is beyond the scope of this study and will be published elsewhere.
The KOMBG1 strain from a northern greater galago (Otolemur garnettii) clustered with Hypotrichomonas acosta from a snake, and probably represents an undescribed species within this genus. The genus Hypotrichomonas belongs to a small though evolutionarily distinct parabasalid lineage called Hypotrichomonadea (Cepicka et al. Reference Cepicka, Hampl and Kulda2010). H. acosta, which is the only member of the Hypotrichomonas whose cell structure has been examined by electron microscopy, possesses an interestingly reduced costa (a striated fibre that underlies the undulating membrane in some trichomonads). As we have successfully established a stable culture of a second Hypotrichomonas species, a future TEM study of the KOMBG1 isolate is desirable.
The KATA1 strain from a ring-tailed lemur (Lemur catta) is closely related to Trichomitus batrachorum, suggesting that it belongs to the genus Trichomitus, the second genus of Hypotrichomonadea. So far, 2 Trichomitus species have been reported from primates, though each of them only once. T. wenyoni was originally described from laboratory rodents (Wenrich and Nie, Reference Wenrich and Nie1949) and has been also reported from a rhesus macaque (Flick, Reference Flick1954). However, no picture or drawing that would corroborate the observation was included in the latter study. We thus consider the presence of T. wenyoni in primates as uncertain (see also Culbertson et al. Reference Culbertson, Pindak, Gardner and Honigberg1986). The second species of the genus Trichomitus from primates, T. fecalis, was described from a single human subject (Cleveland, Reference Cleveland1928). It is morphologically very similar to T. batrachorum from amphibians, reptiles and leeches (Wenrich, Reference Wenrich1944b). It is also infectious for tadpoles and frogs (Cleveland, Reference Cleveland1928), suggesting that the two species may actually be conspecific. As the diversity of the genus Trichomitus is considerably undersampled in molecular-phylogenetic studies (SSU rDNA sequences of only 2 isolates of T. batrachorum are currently available from the whole genus), we cannot decide to which Trichomitus species the KATA1 isolate belongs solely on the basis of the sequence data.
Pentatrichomonas hominis has a rather broad host range and has been repeatedly found in many mammals including humans and non-human primates (Wenrich, Reference Wenrich1944a; Flick, Reference Flick1954; Reardon and Rininger, Reference Reardon and Rininger1968). Our strains GANG1 from an Angola colobus (Colobus angolensis), VARI1 from a ruffed lemur (Varecia variegata) and KOSBE1 from a common marmoset (Callithrix jacchus) have identical or almost identical SSU rDNA sequences with P. hominis isolated from cattle and humans. We thus confirmed the presence of this wide-host-range species in non-human primates.
The rest of the isolates belonged to the species-rich genus Tetratrichomonas. The second strain from a ring-tailed lemur, KATASAMEC, surprisingly branched within Tetratrichomonas gallinarum. This trichomonad, once thought to be a specific intestinal parasite of birds, has been recently found also in human oral cavity, bronchi and sputum (Kutisova et al. Reference Kutisova, Kulda, Cepicka, Flegr, Koudela, Teras and Tachezy2005). It was also shown that T. gallinarum represents, in fact, an intricate complex of multiple species, some of which display considerable host specificity (Cepicka et al. Reference Cepicka, Kutišová, Tachezy, Kulda and Flegr2005). The KATASAMEC strain formed an independent lineage, closely related, although without good bootstrap support, to the GPO strain isolated from chicken (Cepicka et al. Reference Cepicka, Kutišová, Tachezy, Kulda and Flegr2005). Since the diversity of the basal members of the T. gallinarum species complex has not yet been satisfactorily investigated, it is currently impossible to determine whether the KATASAMEC strain represents an independent species.
Strains MA3, MA4, MANI1, MANI2 and MASP1 formed a novel Tetratrichomonas lineage. The isolates had identical sequences, originated from 2 different zoos and were obtained from 3 cercopithecine species (lion-tailed macaque – Macaca silenus, crested macaques – Macaca nigra and mandrill – Mandrillus sphinx). Since no close relative of the novel lineage is known, it might represent a new species. However, a morphological study should be performed before its formal description.
The remaining 19 strains belonged to tetratrichomonad group A as defined by Cepicka et al. (Reference Cepicka, Hampl, Kulda and Flegr2006). Group A contains several Tetratrichomonas lineages that have been isolated from various mammals and tortoises and have been shown to represent distinct, though mostly undescribed, species (Cepicka et al. Reference Cepicka, Hampl, Kulda and Flegr2006). Our analyses also have clearly shown that the LP strain, which has been isolated from human pleural empyema (Mantini et al. Reference Mantini, Souppart, Noël, Duong, Mornet, Carroger, Dupont, Masseret, Goustille, Capron, Duboucher, Dei-Cas and Viscogliosi2009), belongs to Tetratrichomonas group A as well. The LP strain thus represents the second species from the respiratory system of humans in addition to T. gallinarum. The isolates from non-human primates occupied 4 different positions within tetratrichomonad group A. Strains SENT1 and SENT4 from Hanuman langurs (Semnopithecus entellus) were placed within lineage 7, which corresponds to Tetratrichomonas buttreyi and has been previously observed in wild and domestic pigs and a horse. The PAN11 strain from a common chimpanzee (Pan troglodytes) belongs to lineage 8, whose only previous representative, the PB strain, was isolated from a desert warthog kept in a zoo (Cepicka et al. Reference Cepicka, Hampl, Kulda and Flegr2006).
Three strains from siamangs (Symphalangus syndactylus) and the trichomonad MMV-2003 (GenBank Accession number AY247747) obtained from a black crested gibbon branched within lineage 10, which represents so far undescribed Tetratrichomonas species from cattle and tortoises (Cepicka et al. Reference Cepicka, Hampl, Kulda and Flegr2006). In addition, all but 1 isolate from chimpanzees together with the GSIA4 isolate from a siamang and the SENT3 isolate from a Hanuman langur formed the novel Tetratrichomonas lineage 2, closely related to lineage 10. It is currently impossible to decide whether lineage 10 and the new tetratrichomonad lineage are conspecific and a further morphological or infection study is necessary to satisfactorily address this question.
To assess the host specificity of any symbiotic organisms based on the data from captive animals is always complicated; trichomonads are no exception. Contacts with novel hosts, environments and ecological conditions as well as differences in food composition might distort the original trichomonad-host relationships. On the other hand, the transmission of intestinal non-cyst-forming trichomonads requires relatively intimate contact between hosts, which might preclude accidental transmissions. The distribution of our trichomonad strains in phylogenetic trees points to 3 interesting and unexpected aspects related to the host specificity of trichomonads.
(1) Some of the strains clustered within Tetratrichomonas lineages 7, 8, 10 and 15, which are dominated by trichomonads from non-primate hosts, i.e. pigs, cattle, tortoises, lizards, and birds (Cepicka et al. Reference Cepicka, Hampl, Kulda and Flegr2006). This observation means that at least some species of primates may be infected by intestinal trichomonads of these hosts and the host range of the 4 Tetratrichomonas lineages is wider than previously expected by Cepicka et al. (Reference Cepicka, Hampl, Kulda and Flegr2006). A similar situation was also described in the case of Blastocystis hominis (Stensvold et al. Reference Stensvold, Alfellani, Nørskov-Lauritsen, Prip, Victory, Maddox, Nielsen and Clark2009). Yet, at least 2 of the novel Tetratrichomonas lineages may turn out to be specific for primates. Host specificity of most intestinal trichomonads is unclear. Most species are assumed to be confined to a relatively narrow group of hosts, e.g. Dientamoeba fragilis to humans and gorillas (Stark et al. Reference Stark, Phillips, Peckett, Munro, Marriott, Harkness and Ellis2008), and Tetratrichomonas limacis to snails (Cepicka et al. Reference Cepicka, Hampl, Kulda and Flegr2006). On the other hand, at least one generalist species, Pentatrichomonas hominis from a wide range of warm-blooded hosts including humans, is known as well (see Honigberg, Reference Honigberg and Kreier1978b). Moreover, several trichomonad species parasitizing various non-primate hosts have been detected in the respiratory tract of humans (e.g. Cepicka et al. Reference Cepicka, Kutišová, Tachezy, Kulda and Flegr2005; Kutisova et al. Reference Kutisova, Kulda, Cepicka, Flegr, Koudela, Teras and Tachezy2005; Duboucher et al. Reference Duboucher, Caby, Dufernez, Chabé, Gantois, Delgado-Viscogliosi, Billy, Barré, Torabi, Capron, Pierce, Dei-Cas and Viscolgiosi2006). Our data indicate that many trichomonad species may be, in fact, less specific than currently assumed.
(2) Trichomonads of hominoid and non-hominoid primates occupied strikingly different positions in our phylogenetic trees. All strains from hominoids, i.e. chimpanzees, siamangs and a black crested gibbon (GenBank sequence AY247747) belonged exclusively to Tetratrichomonas group A. On the contrary, the strains from non-hominoid primates occupied diverse positions and only 2 strains, SENT1 and SENT4 from Hanuman langur belonged to Tetratrichomonas group A.
(3) The closely related humans and chimpanzees possess different intestinal trichomonads. Three trichomonad species have been reported from the human intestine, namely Pentatrichomonas hominis, Trichomitus fecalis and Dientamoeba fragilis. Only P. hominis has been detected also in chimpanzees (Myers and Kuntz, Reference Myers and Kuntz1972). In contrast, all but 1 strain that we isolated from captive chimpanzees belonged to the novel Tetratrichomonas lineage 2. Although 2 different Tetratrichomonas species (unrelated to our strains from primates) have been retrieved from the human respiratory tract (Cepicka et al. Reference Cepicka, Kutišová, Tachezy, Kulda and Flegr2005; Kutisova et al. Reference Kutisova, Kulda, Cepicka, Flegr, Koudela, Teras and Tachezy2005; Mantini et al. Reference Mantini, Souppart, Noël, Duong, Mornet, Carroger, Dupont, Masseret, Goustille, Capron, Duboucher, Dei-Cas and Viscogliosi2009), none has been found in the human intestine.
Most intestinal trichomonads are considered to be harmless commensals in mammalian hosts, but only little data are available to support this assumption. As demonstrated very recently, humans and chimpanzees share not only agents of malaria (Prugnolle et al. Reference Prugnolle, Duran, Neel, Ollomo, Ayala, Arnathau, Etienne, Mpondi-Ngole, Nkoghe, Leroy, Delaporte, Peeters and Renaud2009), but also a broad spectrum of gastrointestinal parasites (Cutillas et al. Reference Cutillas, Callejon, de Rojas, Tewes, Ubeda, Ariza and Guevara2009; Hasegawa et al. Reference Hasegawa, Sato, Fujita, Nguema, Nobusue, Miyagi, Kooriyama, Takenoshita, Noda, Sato, Morimoto, Ikeda and Nishida2010; Krief et al. Reference Krief, Vermeulen, Lafosse, Kasenene, Nieguitsila, Berthelemy, L'Hostis, Bain and Guillot2010). On the other hand, the community of their intestinal commensal/mutualistic protists differs substantially, which was demonstrated particularly for ciliates (Kortland: comment in Stahl et al. Reference Stahl, Dunbar, Homewood, Ikawa-Smith, Kortlandt, McGrew, Milton, Paterson, Poirier, Sugardjito, Tanner and Wrangham1984; Pomajbíková et al. Reference Pomajbíková, Petrželková, Profousová, Petrašová, Kišidayová, Varadyová and Modrý2010; Tokiwa et al. Reference Tokiwa, Modrý, Ito, Pomajbíková, Petrželková and Imai2010). An appealing scenario is that these differences were brought about by significant changes in the human diet during evolution. The hominid diet before the introduction of fire was more similar to that of great apes, characterized by a high fibre content (Stahl et al. Reference Stahl, Dunbar, Homewood, Ikawa-Smith, Kortlandt, McGrew, Milton, Paterson, Poirier, Sugardjito, Tanner and Wrangham1984; Wrangham and Conklin-Brittain, Reference Wrangham and Conklin-Brittain2003; Carmody and Wrangham, Reference Carmody and Wrangham2009) and both the ciliates and trichomonads seems to be to some extent influenced by diet composition (Ratcliffe, Reference Ratcliffe1928; Pomajbíková et al. Reference Pomajbíková, Petrželková, Profousová, Petrašová, Kišidayová, Varadyová and Modrý2010). We might suggest that our ancestors harboured trichomonads similar to other hominoids, especially chimpanzees, as was hypothesized also for entodiniomorphid ciliates that are present in chimpanzees, but absent in humans (Kortland: comment in Stahl et al. Reference Stahl, Dunbar, Homewood, Ikawa-Smith, Kortlandt, McGrew, Milton, Paterson, Poirier, Sugardjito, Tanner and Wrangham1984). However, to reveal the extent of the discrepancy of trichomonad species occurring in humans and chimpanzees, data from wild great apes are urgently needed.
ACKNOWLEDGEMENTS
We would like to express our sincere thanks to the primate curators and Zoo-keepers from the Pilsner, Ostrava and Brno Zoos for their help with the sample collection. This work was supported by the Ministry of Education, Youth and Sport of the Czech Republic (project MSM0021620828) and Czech Science Foundation (project 206/09/0927).






