Introduction
The conditions that moderate the evolution of parasitic species are not as well understood as those acting on the evolution of free-living species (Criscione et al., Reference Criscione, Poulin and Blouin2005; Huyse et al., Reference Huyse, Poulin and Théron2005). This disparity is increasingly being recognized as appreciation continues to grow for how parasites shape host evolution (Woolhouse et al., Reference Woolhouse, Webster, Domingo, Charlesworth and Levin2002; Papkou et al., Reference Papkou, Gokhale, Traulsen and Schulenburg2016; Penczykowski et al., Reference Penczykowski, Laine and Koskella2016), communities (Poulin and Morand, Reference Poulin and Morand2000) and ecosystems (Marcogliese, Reference Marcogliese2004; Hatcher et al., Reference Hatcher, Dick and Dunn2012). Better understanding of attributes that set the tempo and pace of parasite evolution, such as effective population size (N e), could cast new light on foundational principles including adaptive evolution and coevolution (Criscione and Blouin, Reference Criscione and Blouin2005; Huyse et al., Reference Huyse, Poulin and Théron2005). This in turn could spark translational advances, such as new measures to control the adaptive potential of parasites and improve wildlife and human health (Huyse et al., Reference Huyse, Poulin and Théron2005; Criscione, Reference Criscione and Holland2013; Nkhoma et al., Reference Nkhoma, Nair, Al-Saai, Ashley, McGready, Phyo, Nosten and Anderson2013).
Though N e is formally defined as ‘the number of breeding individuals in an idealized population that would exhibit the same amount of dispersion of allele frequencies under random genetic drift as the population under consideration’ (Wright, Reference Wright1931, Reference Wright1938), it is generally considered to be a proxy for the number of individuals in a population that are contributing offspring to the next generation (Criscione and Blouin, Reference Criscione and Blouin2005; Criscione et al., Reference Criscione, Poulin and Blouin2005; Hedrick, Reference Hedrick2011; Harmon and Braude, Reference Harmon, Braude, Braude and Low2010; Jensen and Bachtrog, Reference Jensen and Bachtrog2011; Criscione, Reference Criscione and Holland2013). Because parasite populations are subdivided among hosts, the number of breeding individuals in a population may strongly differ from estimates derived from census counts (Criscione and Blouin, Reference Criscione and Blouin2005). Accordingly, by offering more informative perspectives on conditions such as reproductive potential and population stability (Criscione and Blouin, Reference Criscione and Blouin2005), estimates of N e can be of equal or greater value than census population size estimates in the study of parasite demography and host–parasite interactions (Papkou et al., Reference Papkou, Gokhale, Traulsen and Schulenburg2016). Similarly, N e can potentially serve as a more useful metric of evolutionary potential (Huyse et al., Reference Huyse, Poulin and Théron2005) by offering perspectives on processes that can foster adaptive responses (e.g. connectivity, gene flow) as well as processes that can impede responses by reducing the amount of variation upon which selection might act (e.g. genetic drift, inbreeding).
Remarkably few studies have examined contemporary (i.e. short-term) N e of parasites (hereafter ‘parasite N e’), particularly of metazoan (multi-cellular) parasites (Criscione, Reference Criscione and Holland2013). The majority of prior work on parasite N e has focused on coalescent-based estimation of long-term N e (e.g. Blouin et al., Reference Blouin, Dame, Tarrant and Courtney1992; Anderson et al., Reference Anderson, Hauhold, Williams, Estrada-Franco, Richardson, Mollinedo, Brockarie, Mokili, Mharakurwa, French, Whitworth, Velez, Brockman, Nosten, Ferreira and Day2000; Hughes and Verra, Reference Hughes and Verra2001; Joy et al., Reference Joy, Feng, Mu, Furuya, Chotivanich, Krettli, Ho, Wang, White, Suh, Beerli and Su2003; Crellen et al., Reference Crellen, Allan, David, Durrant, Huckvale, Holroyd, Emery, Rollinson, Aanensen, Berriman, Webster and Cotton2016). Contemporary N e has been estimated for only a handful of protozoan (unicellular) parasites, such as Plasmodium falciparum (the causative agent of malaria; Nkhoma et al., Reference Nkhoma, Nair, Al-Saai, Ashley, McGready, Phyo, Nosten and Anderson2013; Sisya et al., Reference Sisya, Kamn'gona, Vareta, Fulakeza, Mukaka, Seydel, Laufer, Taylor and Nkhoma2015) and Trypanosoma brucei (the causative agent of human African trypanosomiasis or sleeping sickness; Koffi et al., Reference Koffi, de Meeûs, Bucheton, Solano, Camara, Kaba, Cuny, Ayala and Jamonneau2009). To our knowledge, contemporary N e has only been estimated for two metazoan parasites: the nematode Ascaris lumbricoides (Criscione, Reference Criscione and Holland2013) and the phytoparasitic beet cyst nematode Heterodera schachtii (Jan et al., Reference Jan, Cracianne, Fournet, Olivier, Arnaud, Porte, Bardou-Valette, Denis and Petit2016). The effective number of breeders (N b), a parameter related to contemporary N e that is derived from single age cohorts (Waples et al., Reference Waples, Antao and Luikart2014), has been estimated in several species of schistosome trematodes (the causative agent of Schistosomiasis in humans; Gower et al., Reference Gower, Gouvras, Lamberton, Deol, Shrivastava, Mutombo, Mbuh, Norton, Webster, Stothard, Garba, Lamine, Kariuki, Lange, Mkoji, Kabatereine, Gabrielli, Rudge, Fenwick, Sacko, Dembelé, Lwambo, Tchuem Tchuenté, Rollinson and Webster2013; Steinauer et al., Reference Steinauer, Christie, Blouin, Agola, Mwangi, Maina, Mutuku, Kinuthia, Mkoji and Loker2013).
Though limited, prior work indicates that organismal and ecological factors might influence parasite N e. Work on monoecious trematodes (Prugnolle et al., Reference Prugnolle, Liu, de Meeûs and Balloux2005), for instance, suggests that self-fertilization, reproductive skew and migration between hosts act to reduce parasite N e. Higher infection intensities also correspond to higher population genetic diversity in a variety of parasitic species (Criscione et al., Reference Criscione, Poulin and Blouin2005; Doña et al., Reference Doña, Moreno-Garcia, Criscione, Serrano and Jovani2015), and because N e is expected to increase with genetic diversity, infection intensity should also be positively related to parasite N e (Doña et al., Reference Doña, Moreno-Garcia, Criscione, Serrano and Jovani2015). This expectation also underlies the prediction (Criscione et al., Reference Criscione, Poulin and Blouin2005) that a positive relationship should occur between parasite N e and the standing number of breeding adults found in definitive hosts. Further study, particularly of species that exhibit variation in life history (Criscione, Reference Criscione and Holland2013, Reference Criscione, Janovy and Esch2016), is warranted to determine whether these or other conditions consistently shape parasite N e (Criscione and Blouin, Reference Criscione and Blouin2005; Criscione et al., Reference Criscione, Poulin and Blouin2005).
Interactions between the diphyllobothridean cestode parasite Schistocephalus solidus and its second intermediate host, the three-spined stickleback fish (Gasterosteus aculeatus) – widely considered to be a ‘supermodel’ system (Heins and Baker, Reference Heins and Baker2008; Barber, Reference Barber2013) for studying evolutionary parasitology – present outstanding opportunities for testing hypotheses about factors affecting parasite N e. Schistocephalus solidus has a complex life cycle involving several species of cyclopoid copepods as initial intermediate hosts of procercoids, three-spined sticklebacks as secondary intermediate hosts of plerocercoids, and 40+ species of piscivorous birds as definitive hosts of reproducing parasites (Smyth, Reference Smyth1962). Multiple infections of S. solidus parasites are common in three-spined sticklebacks, with individuals often harbouring between one and 10 worms, though up to 140 worms have been reported in a single host (Smyth, Reference Smyth1946; Heins et al., Reference Heins, Baker and Martin2002). Schistocephalus solidus is known to impose fitness costs on three-spined stickleback hosts by altering behaviour (Barber and Huntingford, Reference Barber and Huntingford1995; Ness and Foster, Reference Ness and Foster1999), reproduction (McPhail and Peacock, Reference McPhail and Peacock1983; Bagamian et al., Reference Bagamian, Heins and Baker2004) and survival (Pascoe and Mattey, Reference Pascoe and Mattey1977; Heins et al., Reference Heins, Birden and Baker2010). There is also evidence suggesting that S. solidus are subject to pressures arising from stickleback hosts. Phylogenetic analyses have recovered signatures of evolutionary divergence between parasites infecting closely related three-spined and nine-spined stickleback hosts, suggesting that Schistocephalus are host specific (Nishimura et al., Reference Nishimura, Heins, Andersen, Barber and Cresco2011). Consistent with this, experimental cross infection trials between sympatric and allopatric pairs of three-spined stickleback and S. solidus have yielded evidence of reciprocal evolution of host resistance and parasite virulence (Kalbe et al., Reference Kalbe, Eizaguirre, Scharsack and Jakobsen2016). Evidence of discordant patterns of geographic differentiation suggests, however, that genetic variation – and thus N e – in S. solidus may instead be largely governed by factors external to stickleback hosts (Strobel et al., Reference Strobel, Alda, Sprehn, Blum and Heins2016).
In this study, we leveraged and extended prior assessments of genetic variation (Sprehn et al., Reference Sprehn, Blum, Quinn and Heins2015; Strobel et al., Reference Strobel, Alda, Sprehn, Blum and Heins2016) to characterize contemporary N e in S. solidus. Building on prior findings that S. solidus parasites exhibit much weaker population genetic structure than three-spined stickleback hosts across a suite of Alaskan lakes (Sprehn et al., Reference Sprehn, Blum, Quinn and Heins2015; Strobel et al., Reference Strobel, Alda, Sprehn, Blum and Heins2016), our aim was to determine the range and magnitude of contemporary N e exhibited by S. solidus in comparison to stickleback hosts. We also set out to test the hypothesis that factors external to stickleback hosts exert greater influence on parasite N e than do host attributes (e.g. host body size, host genetic variation, etc.). Specifically, we tested the hypothesis that parasite N e is positively associated with lake size, as larger lakes can potentially support larger census population sizes of intermediate fish hosts, and thus a greater number of reproductive adult parasites in definitive hosts. In addition, we tested the hypotheses that parasite N e is positively associated with host infection (i.e. the number of parasites) in stickleback hosts as well as stickleback host body size, which might allow for greater parasite loads and more efficient transfer to definitive hosts. We also assessed whether parasite N e is greater with lower host genetic diversity, as genetically depauperate hosts might be more susceptible to infection (Kaunisto et al., Reference Kaunisto, Viitaniemi, Leder and Suhonen2013). We met these objectives by deriving single-sample and temporal estimates of contemporary N e for S. solidus and three-spined stickleback hosts sampled from south-central and south-west Alaska lakes across a 9-year period (Supplementary Fig. 1). We then considered whether assumptions about population structure influence N e estimation and determined the proportion of variation in parasite N e attributable to host attributes (length, genetic diversity and N e), host infection reflecting parasite abundance (sum count of parasites across hosts), and physiographic features (lake surface area and volume).
Materials and methods
Specimens and data availability
We combined existing microsatellite data for infected three-spined stickleback fish and corresponding S. solidus parasites collected from 2006 to 2012 (Sprehn et al., Reference Sprehn, Blum, Quinn and Heins2015; Strobel et al., Reference Strobel, Alda, Sprehn, Blum and Heins2016) with new data on parasites obtained from stickleback hosts collected from 2013 to 2015. Following specimen collection protocols in Sprehn et al. (Reference Sprehn, Blum, Quinn and Heins2015), we sampled infected three-spined sticklebacks from lakes in the same three regions of south-central and south-west Alaska that were sampled in 2006–2012: Matanuska-Susitna Valley (MatSu), Kenai Peninsula (Kenai) and Bristol Bay drainage (BB) (Supplementary Fig. 1). We performed necropsies on all freshly collected fish with parasites and preserved S. solidus plerocercoids in 95% ethanol for DNA analysis.
Host infection, host size and physiographic attributes
We measured the length of each infected fish and counted the number of parasites per infected fish. Work done by Strobel et al. (Reference Strobel, Alda, Sprehn, Blum and Heins2016) involved clipping tissue from caudal fins for DNA extraction, thus an estimate of standard length (SL) was made for these individuals based on comparable measurements of whole specimens. To assess the extent of potential discrepancies between measurements and estimates, we also estimated SL for individuals with intact caudal fins following the same method. A very strong correlation was recovered between estimated and measured SL at the individual level (r = 0.997, P < 0.001) and between estimates averaged across lakes (r = 0.995, P < 0.001) as well as for each lake-by-year combination (hereafter ‘lake-year’; r = 0.998, P < 0.001), illustrating that measured and estimated SL were effectively equivalent characterizations of host size. Physiographic metrics of lake surface area and lake volume were obtained from the Alaska Department of Fish and Game (http://www.adfg.alaska.gov/index.cfm?adfg=fishingSportLakeData.main and http://www.sf.adfg.state.ak.us/fedaidpdfs/FREDF-5-R-2(2)1-C.pdf; accessed on June 2017), and the United States Fish and Wildlife Service (http://www.fwspubs.org/doi/suppl/10.3996/032014-JFWM-022/suppl_file/032014-jfwm-022.s3.pdf?code=ufws-site, https://www.fws.gov/wetlands/Documents/Status-of-Alaska-Wetlands.pdf and https://www.fws.gov/uploadedFiles/lake_Engineer.pdf, https://www.fws.gov/uploadedFiles/lake_LowerOhmer.pdf; accessed on June 2017).
Microsatellite genotyping and population genetic analyses
Gasterosteus aculeatus
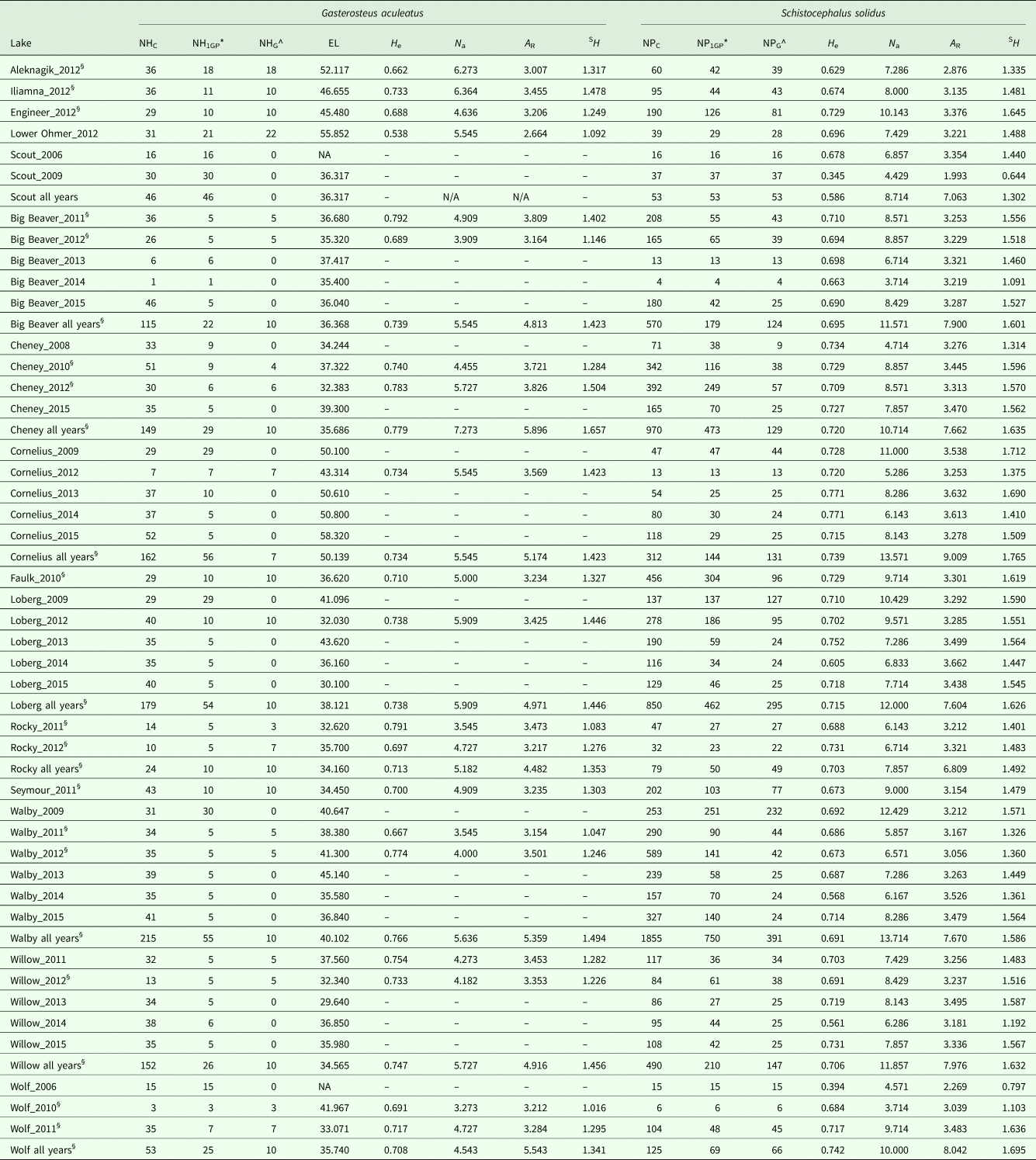
We used data from 11 microsatellite loci (Strobel et al., Reference Strobel, Alda, Sprehn, Blum and Heins2016) from 157 fish collected between 2010 and 2012 to characterize host genetic diversity and contemporary N e. To ensure consistency with work done on S. solidus parasites, we recalculated expected heterozygosity (H e) in Arlequin v. 3.5.2.2 (Excoffier and Lischer, Reference Excoffier and Lischer2010) and average number of alleles (N a), rarefied allelic richness (A R) and Shannon diversity (SH) in MSAnalyzer v. 4.05 (Dieringer and Schlštterer, Reference Dieringer2003). Hierarchical genetic differentiation was tested among lakes and regions with analyses of molecular variance (AMOVAs) with 1000 permutations in Arlequin v. 3.5.2.2.
Schistocephalus solidus
A combined microsatellite dataset was generated by aggregating existing data with new data from 362 parasites isolated from 83 host fish collected between 2013 and 2015. DNA extraction and locus-specific microsatellite amplification protocols followed those of Sprehn et al. (Reference Sprehn, Blum, Quinn and Heins2015). To reduce potential biases in binning and scoring alleles, we pooled the raw data for all 1749 parasites and re-scored the entire dataset according to a common panel. Due to poor amplification in the newly collected samples, we eliminated locus Scso22 in the pooled dataset, resulting in a final dataset consisting of seven loci. We used GeneMarker v. 1.90 (Softgenics, State College, PA, USA) to bin alleles and Micro-checker v. 2.2.3 (Van Oosterhout et al., Reference Van Oosterhout, Hutchinson, Wills and Shipley2004) to screen the resulting dataset for scoring errors, null alleles, large allele drop-out and heterozygote deficiency.
We calculated observed and expected heterozygosity in Arlequin v. 3.5.2.2 and calculated the average number of alleles, rarefied allelic richness and Shannon diversity by lake (i.e. all parasites from each lake) and by host (i.e. all parasites from each host) in MSAnalyzer v. 4.05. Hosts with fewer than three parasites were excluded when assessing diversity within hosts. To test for differences in parasite allelic richness and Shannon diversity among hosts within lakes and regions, we ran a two-way nested analysis of variance (ANOVA) followed by a post-hoc Tukey test for multiple comparisons in R v. 3.3.2 (R Core Team, 2016). We tested for hierarchical genetic differentiation among lakes within regions and among hosts within lakes using AMOVAs with 1000 permutations in Arlequin v. 3.5.2.2.
Estimation of Ne
Gasterosteus aculeatus
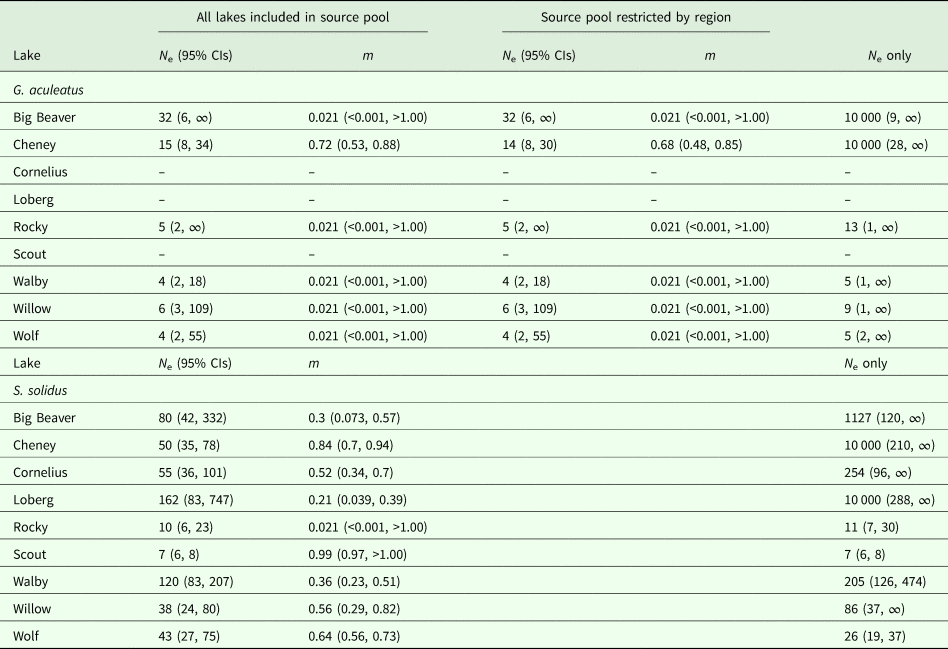
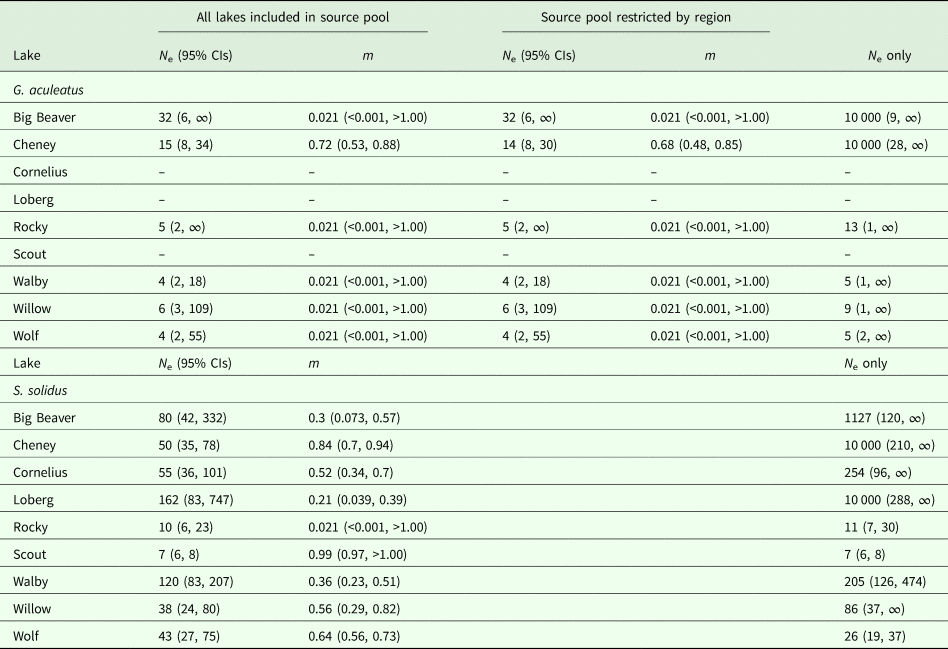
We estimated joint N e and immigration rates (m) for six lakes (Big Beaver, Cheney, Rocky, Walby, Willow and Wolf Lakes) with genotyped fish across multiple years. For each lake, generation intervals were estimated by dividing the elapsed time between sampling years by the generation time of Alaskan three-spined sticklebacks. Female three-spined sticklebacks tend to breed at age two or older, though some breed starting as early as age one (Heins et al., Reference Heins, Singer and Baker1999; Baker et al., Reference Baker, Heins, Foster and King2008). We therefore conservatively set generation time as two years, such that our samples were separated by 0.5 generations for five lakes (Beaver, Rocky, Walby, Willow and Wolf) and one generation for one lake (Cheney). We then estimated mean N e across all generations using the maximum likelihood option implemented in MLNe v. 1.0 (Wang and Whitlock, Reference Wang and Whitlock2003). For estimates of migration rates, pooled source populations were composed of fish from all lakes sampled during the same years as the respective focal population, with one exception. The source population for Cheney Lake, which was sampled in 2010 and 2012, was composed of fish from all lakes sampled in 2010, 2011 and 2012. Given the more limited dispersal capability of three-spined sticklebacks and strong signatures of population genetic structure detected by Strobel et al. (Reference Strobel, Alda, Sprehn, Blum and Heins2016), we ran a separate analysis including only lakes in the source pool that were in the same region as the focal lake. Following recommendations from Wang and Whitlock (Reference Wang and Whitlock2003), we set the maximum allowed N e at 50 000 and assumed populations were not in drift-migration equilibrium.
For comparison to temporal estimates, we also computed single-sample estimates of host N e for each lake-by-year combination. We used NeEstimator v. 2.0.1 (Do et al., Reference Do, Waples, Peel, Macbeth, Tillett and Ovenden2014) to estimate N e following the linkage disequilibrium (LD) method (Hill, Reference Hill1981). We assumed random mating and set the lowest allowable allele frequency at 0.02 to screen out rare alleles. Notably, infinite estimates of N e are sometimes recovered from the single sample LD method, which can recover less accurate estimates at larger values of N e, especially if the sample size was not adequate to resolve the difference between a large population and an infinite population (Waples and Do, Reference Waples and Do2010). Infinite estimates of N e were excluded from correlation and regression analyses.
Schistocephalus solidus
We estimated temporal parasite N e for each focal lake using the combined dataset of genotyped parasites collected between 2006 and 2015, with source lakes from proximate years binned (e.g. Waits et al., Reference Waits, Bagley, Blum, McCormick and Lazorchak2008). The nine lakes with genotyped parasites collected across multiple years (Big Beaver, Cheney, Cornelius, Loberg, Rocky, Scout, Walby, Willow and Wolf) served as focal populations for maximum likelihood estimates of N e and migration rate (m) using the temporal method implemented in MLNe v. 1.0 (Wang and Whitlock, Reference Wang and Whitlock2003). Schistocephalus solidus parasites are thought to have similar generation times as sticklebacks (approximately 2 years) because most fish become infected during the first year of life (Heins et al., Reference Heins, Baker and Martin2002, Reference Heins, Eidam and Baker2016). Based on a 2 year generation time, temporal samples were separated by at least two generations for six lakes (Beaver, Cheney, Cornelius, Loberg, Walby and Wolf), by at least 1.5 generations for two lakes (Scout and Willow), and by only 0.5 generations for one lake (Rocky) (e.g. for Walby Lake, parasite samples from 2009, 2011, 2012, 2013, 2014 and 2015 were binned into 2009–2011, 2012–2013 and 2014–2015, corresponding to generation assignments of 0, 1.5 and 2.5, respectively). When pooling source populations, we only included data from lakes sampled during the same years as the corresponding focal population, except for Cheney, Scout and Wolf, which were sampled several years earlier than all other lakes. For these lakes, source pools contained data from lakes sampled within 2 years of when the focal lake was sampled. Settings for combined parasite N e/m estimates were the same as those used for host estimates. For comparison to temporal estimates, we also estimated N e for lake-year combinations using the single-sample LD method (Hill, Reference Hill1981) implemented in NeEstimator v. 2.0.1 with the same settings used for host estimates. Infinite estimates of N e were excluded from correlation and regression analyses.
Effects of population structure on Ne estimation
Estimating N e for each lake carries the assumption that fish and parasites are genetically structured by lake. To assess the effect this assumption has on N e estimation for fish hosts and parasites, we respectively pooled samples by year, regardless of lake, and estimated N e using the single-sample LD method implemented in NeEstimator v. 2.0.1. Migration between lakes can also cause fluctuating allele frequencies and thus influence N e estimates, yet most estimators assume minimal migration (Wang and Whitlock, Reference Wang and Whitlock2003). Accordingly, we used MLNe v. 1.0 to calculate N e under the special case of an isolated population assuming negligible influence of immigration and compared the results to estimates of joint N e and migration rate. We present all sets of estimates to demonstrate the effect that assumptions about immigration and population structure have on N e estimation. Subsequent correlation and regression analyses, however, only utilized single-sample lake-year N e and temporal lake N e estimates that accounted for immigration.
Predictors of parasite Ne
Predictor variables that might influence parasite N e were categorized as host attributes (host length, host N e and host genetic diversity), conditions internal to the host (sum count of parasites across hosts, average number of parasites per host) or lake attributes (lake volume, lake surface area). We used Pearson's correlations to test for multicollinearity among predictor variables and to determine bivariate relationships between predictors and parasite N e (Gagne et al., Reference Gagne, Hogan, Pracheil, McIntyre, Hain, Gilliam and Blum2015) in R v. 3.3.2.
Redundancy analysis (RDA), a multiple linear regression ordination method (Rao, Reference Rao1964), was used to determine the relative influence of host attributes, conditions internal to the host and lake attributes on lake-year and lake estimate of parasite N e. RDA modelling was carried out with the vegan package for R (Oksanen et al., Reference Oksanen, Blanchet, Friendly, Kindt, Legendre, McGlinn, Minchin, O'Hara, Simpson, Solymos, Stevens, Szoecs and Wagner2016). We estimated the adjusted coefficient of determination (R adj2) for each model and statistical significance was determined using permutation tests to compare observed and randomized model R adj2. We conducted variance partitioning with partial RDAs to estimate the variance in lake-year and lake N e that was independently explained by each predictor variable (Legendre, Reference Legendre2008; Peres-Neto and Legendre, Reference Peres-Neto and Legendre2010). The RDAs were conducted on the subset of populations with finite estimates of S. solidus N e and corresponding estimates of host genetic diversity (Table 1). Further, an atypical outlier estimate of parasite N e recovered for Big Beaver Lake in 2015 was excluded from the RDA of lake-year N e estimates.
Table 1. Sampling and microsatellite diversity

NHC = number of hosts collected, NH1GP = number of hosts with ⩾1 genotyped parasite, NHG = number of hosts genotyped, EL = estimated average fish length, H e = expected heterozygosity, N a = number of alleles, A R = rarefied allelic richness, SH = Shannon diversity, NPC = number of parasites collected, NP1GP = all parasites from fish with ⩾1 genotyped parasite, NPG = number of parasites genotyped. * counts used for Pearson's correlations and RDA analysis; ^ sample used for genetic analysis; § populations used in RDAs.
Results
Genetic diversity and population structure
Gasterosteus aculeatus
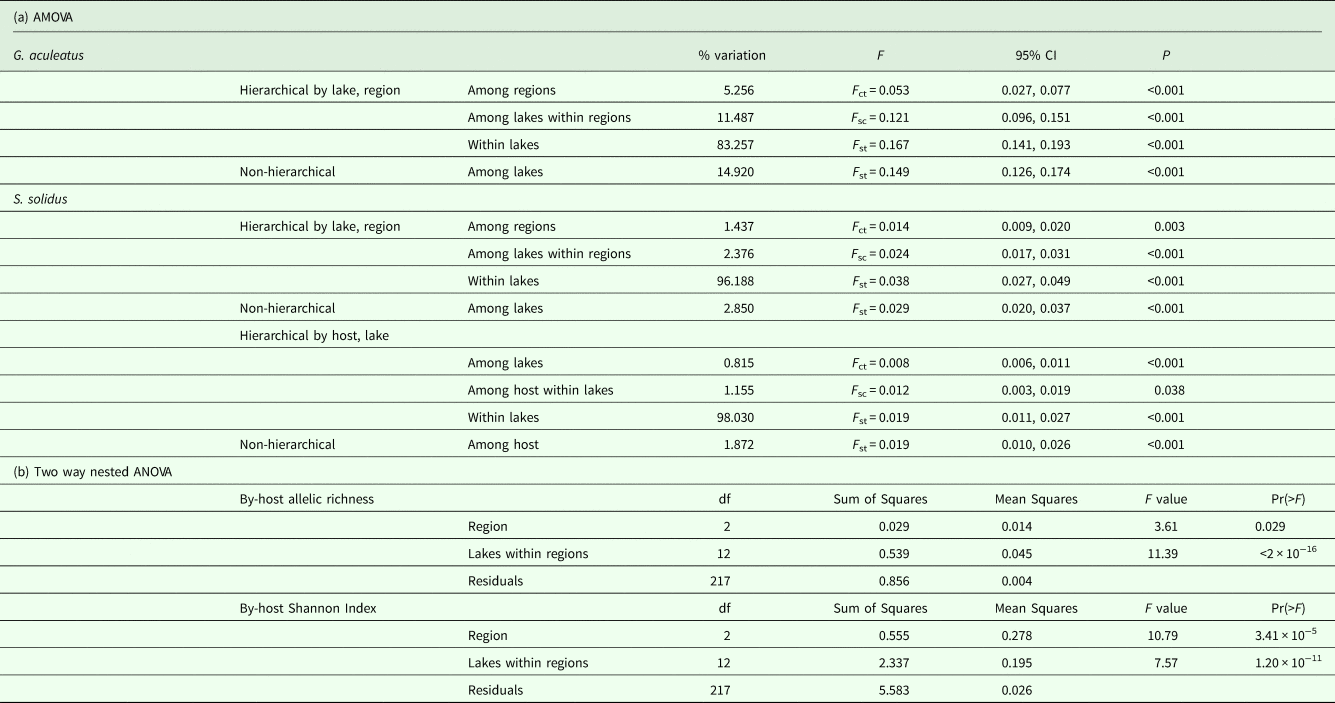
Consistent with prior work (Strobel et al., Reference Strobel, Alda, Sprehn, Blum and Heins2016), measures of host genetic diversity varied across lakes and regions. The highest estimates of H e, N a, A R and SH were observed in fish from Cheney Lake in the MatSu region, whereas the lowest H e, A R and SH were recovered from Lower Ohmer lake, and the lowest N a was recovered from Engineer Lake, which are both located in the Kenai region (Table 1). A higher proportion of genetic variation occurred among lakes within regions (F SC = 0.121, P < 0.001) than among regions (F CT = 0.053, P < 0.001) (Table 2).
Table 2. Summary of AMOVA and ANOVA results

(a) Summary of hierarchical AMOVA results for G. aculeatus fish and S. solidus parasites. (b) Summary of two way nested ANOVA results for allelic richness and Shannon diversity of S. solidus parasites pooled by host.
Schistocephalus solidus
Observed variation in parasite genetic diversity differed from that found in hosts. The highest estimate of parasite H e was recovered for Wolf Lake, whereas the highest N a estimate was recovered for Walby Lake, which are both located in the MatSu region (Table 1). The highest A R and SH estimates were recovered for Cornelius Lake in the MatSu region (Table 1). The lowest H e and SH estimates were recovered for Scout Lake in the Kenai region, whereas the lowest N a and A R estimates were recovered for Aleknagik Lake in the Bristol Bay region (Table 1). Like the pattern detected in fish hosts, a greater proportion of genetic variation in S. solidus occurred among lakes within regions (F SC = 0.024, P < 0.001) than among regions (F CT = 0.014, P = 0.003) (Table 2). When parasites were delineated by host, however, more variation occurred among host fish within lakes (F SC = 0.012, P = 0.038) than among lakes (F CT = 0.008, P < 0.001) (Table 2). A two-way nested ANOVA revealed significant differences in host-level rarefied allelic richness among regions (F 2,217 = 3.61, P = 0.029) (Table 2) and among lakes within regions (F 12,217 = 11.39, P < 0.001) (Table 2), although post-hoc pairwise Tukey tests between regions were not significant (Kenai-BB: P = 0.938, MatSu-BB: P = 0.178, MatSu-Kenai: P = 0.0880). There were also significant differences in host-level SH among regions (F 2,217 = 10.8, P < 0.001) and among lakes within regions (F 12,217 = 7.57, P < 0.001). Post hoc tests were significant for Kenai-BB (P = 0.004) and MatSu-BB (P < 0.001), whereas the MatSu-Kenai comparison was not significant (P = 1.000).
N e estimation
Gasterosteus aculeatus
Host N e varied substantially across lake-years, with infinite N e estimates recovered for 15 lake-year sets of samples and finite estimates ranging from 2 (Engineer_2012) to 315 (Aleknagik_2012) recovered for five lake-year sample sets according to the LD single-sample method (Table 3). When samples from all lakes were pooled by year, reflecting a more panmictic population structure, all estimates were finite and bounded by finite confidence intervals (Table 3). Temporal estimates were finite for all five lakes, ranging from 4 (Walby and Wolf) to 32 (Big Beaver) (Table 4), with all but two estimates containing finite upper bounds. Estimates of migration ranged from 0.021 to 0.72, although a migration rate with bounded confidence intervals was only recovered for one lake (Cheney) (Table 4). Restricting the source pool to only lakes within the same region as the focal population yielded identical estimates, with the exception of N e estimates for Cheney Lake, which were <7% apart from one another (Table 4). When migration was disregarded, all N e estimates remained finite but the upper confidence intervals for all estimates became infinite (Table 4).
Table 3. Single year estimates of N e

Estimates of effective population size for G. aculeatus and S. solidus based on the single sample LD method. CI = confidence interval. CIs were produced using the jackknife method.
Table 4. Temporal estimates of N e
Temporal estimates of effective population size for G. aculeatus and S. solidus for lakes across all years.
Schistocephalus solidus
Single-sample LD estimates of parasite N e were infinite for 11 lake-year sets of samples and finite for 32 lake-year sets, with values ranging from 1 (Scout_2009, Wolf_2006 and Wolf_2010) to 2937 (Big Beaver_2015) (Table 3). Comparable to hosts, a greater fraction of parasite N e estimates were bounded by finite confidence intervals when samples for all lakes were pooled by year instead of by lake-year (44% vs 23%, respectively) (Table 3). Temporal estimates were finite for all nine lakes, ranging from 7 (Scout) to 162 (Loberg), with all estimates bounded by finite confidence intervals (Table 4). Migration rates ranged from 0.021 (Rocky) to 0.99 (Scout), with bounded confidence intervals recovered for seven of nine lakes (Table 4). When migration was disregarded for parasites, most estimates became unbounded (66%) (Table 4).
Predictors of parasite N e
We recovered evidence of covariation and collinearity among predictor variables within each category. As expected, different predictors of host genetic diversity were correlated (Supplementary Table 1), therefore host heterozygosity, which is less susceptible to the vagaries of sample size, was retained for use in RDAs. Lake volume and surface area also were correlated (Supplementary Table 1). Information on surface area was more readily available for the study lakes, therefore it was retained for use in RDAs. Average numbers of parasites per host was found to be collinear with the sum of parasites totalled across hosts (Supplementary Table 1), so only the sum metric was retained for use in RDAs. No predictor variables were correlated with lake estimates of host N e (Supplementary Table 1), and correlations between predictor variables and host N e were unreliable at the lake-year level due to low sample size (Supplementary Table 1). Host N e was not retained for use in the RDAs to avoid concerns about small sample sizes (i.e. only a few bounded, finite estimates of host N e were recovered; Tables 3 and 4), and because we did not detect a pairwise relationship between host and parasite N e estimates (Supplementary Table 1).
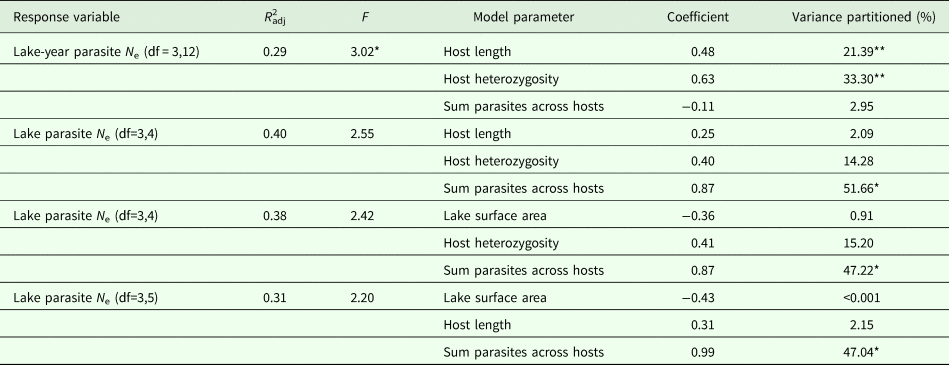
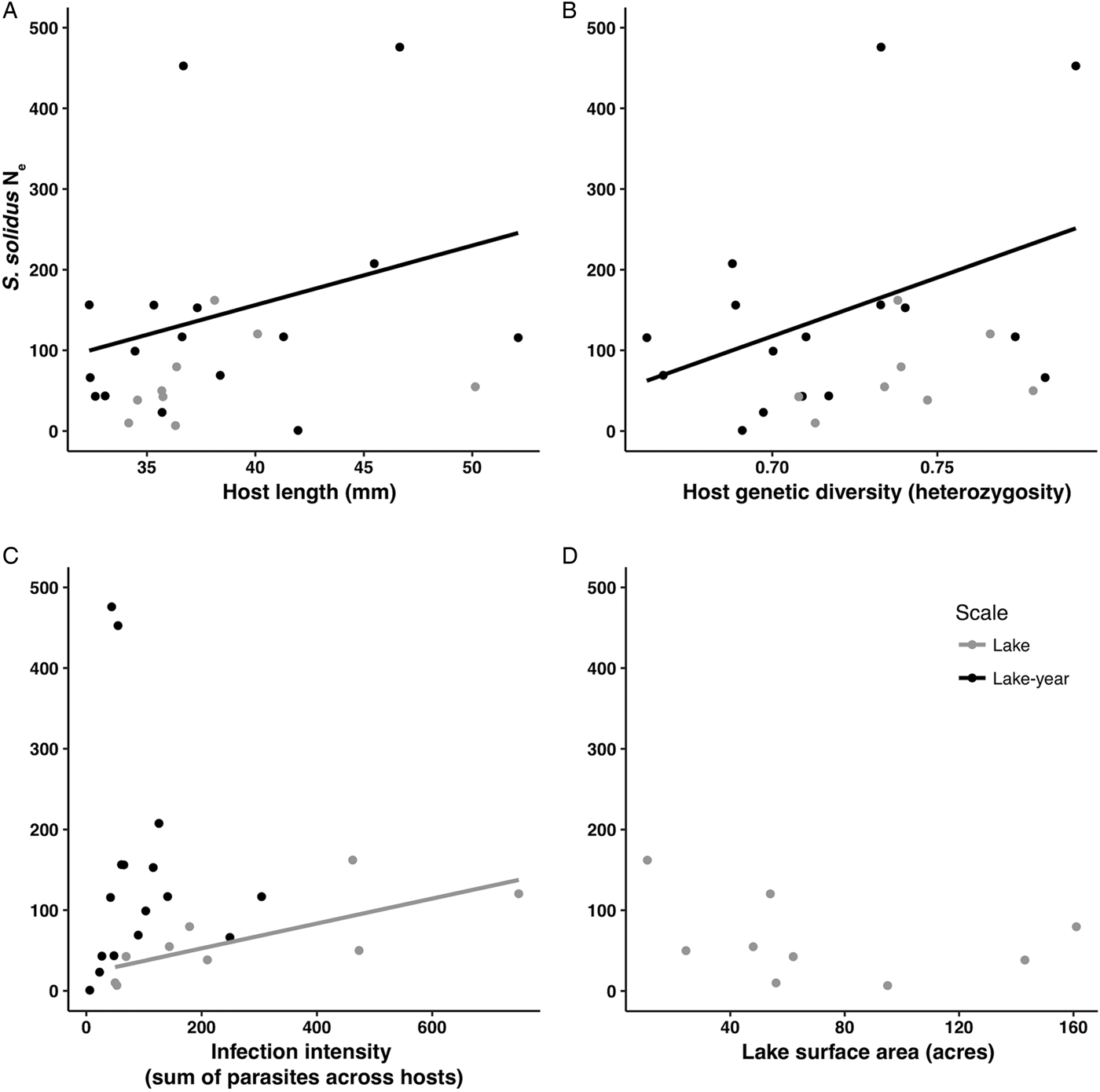
The RDA models explained 29–40% of observed variation in estimates of parasite N e depending on the estimation method (Table 5). Host length and heterozygosity best explained variation in lake-year estimates of parasite N e, contributing 21.39 and 33.30% to the observed variance, respectively. The sum of parasites across hosts best explained variation in lake estimates of parasite N e. This variable consistently contributed >47% to the observed variance, regardless of the other predictor variables included in the lake level model (Table 5). Though not statistically significant, host heterozygosity explained another 14.28–15.20% of the observed variance in lake estimates of parasite N e. Host length and lake surface area consistently explained relatively little of the observed variance in lake estimates of parasite N e (2.09–2.15% and <0.001–0.91%, respectively).
Table 5. RDA models for lake-year and lake estimates of N e
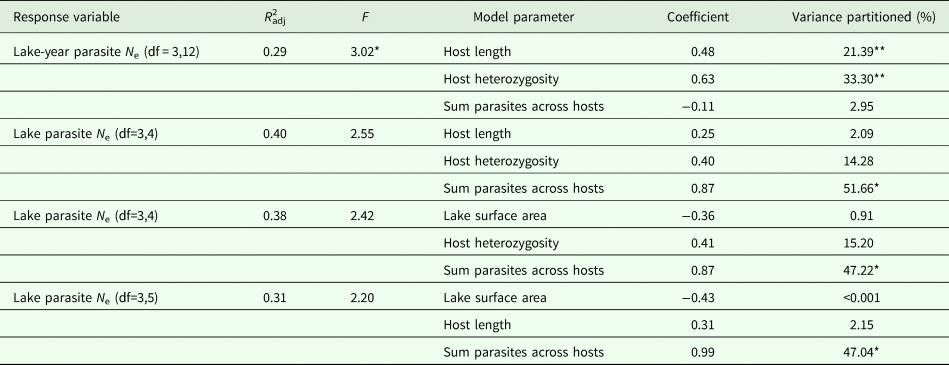
Results of the full model RDAs and variance partitioning of each model parameter. *P < 0.01, **P < 0.05.
Discussion
In this study, we characterized the range and magnitude of contemporary N e exhibited by S. solidus relative to three-spined stickleback hosts to offer further perspective on the conditions that set the tempo and pace of parasite evolution. Prior studies demonstrating reciprocal evolution of host resistance and parasite virulence suggested that N e might reflect host characteristics, whereas patterns of S. solidus genetic variation suggested that N e might instead reflect factors external to stickleback hosts. Our results indicate that N e of both S. solidus parasites and their three-spined stickleback hosts are relatively small and highly variable among lakes. We also found evidence that parasite N e is better predicted by host attributes and factors internal to hosts. Contrary to expectation, we detected a positive association between lake-year estimates of parasite N e and host genetic diversity, though we also detected a positive association between lake estimates of parasite N e and a proxy measure of host infection reflecting parasite abundance (the sum count of parasites per host). This finding, alongside evidence of relatively small N e and low genetic diversity (Sprehn et al., Reference Sprehn, Blum, Quinn and Heins2015; Strobel et al., Reference Strobel, Alda, Sprehn, Blum and Heins2016), suggests that further work on selfing and inbreeding in S. solidus would be revelatory next steps for understanding conditions that govern parasite N e. We also found, however, that assumptions about population genetic structure and connectivity affected estimates of N e. For both three-spined sticklebacks and S. solidus, temporal estimates calculated under the assumption that migration did not influence fluctuations in allele frequencies differed from joint estimates of N e and migration rate. Similarly, pooling samples from different lakes, representing the assumption that there is no population genetic structure among lakes, resulted in substantively different estimates than those calculated for each lake. This affirms the importance of characterizing conditions like connectivity to better scrutinize and interpret estimates of contemporary N e (Baalsrud et al., Reference Baalsrud, Sæther, Hagen, Myhre, Ringsby, Pärn and Jensen2014), as well as the importance of utilizing complementary approaches to characterize N e (Gagne et al., Reference Gagne, Tinker, Gustafson, Ralls, Larson, Tarjan, Miller and Ernest2018).
N e of Alaskan Gasterosteus aculeatus
Our estimates of contemporary N e in Alaskan three-spined sticklebacks were consistent with prior estimates of contemporary and long-term N e in other stickleback populations. Araguas et al. (Reference Araguas, Vidal, Pla and Sanz2012), for example, recovered comparable or lower values of contemporary N e of three-spined sticklebacks from the Iberian Peninsula. Though relatively small, our estimates nonetheless fall on the low end of the range of long-term N e estimates for other Nearctic stickleback populations (Reusch et al., Reference Reusch, Wegner and Kalbe2001; Mäkinen et al., Reference Mäkinen, Cano and Merilä2006; Caldera and Bolnick, Reference Caldera and Bolnick2008), as well as for a wider range of freshwater fish species (DeWoody and Avise, Reference DeWoody and Avise2000). Some discordance between contemporary and long-term N e is expected because estimates of long-term N e correspond more to historical conditions and processes (e.g. post-glacial range expansion) whereas estimates of contemporary N e reflect conditions acting on the parental generation (i.e. single-sample estimators) or across the generations sampled (i.e., temporal estimators) (Hare et al., Reference Hare, Nunney, Schwartz, Ruzzante, Burford, Waples, Ruegg and Palstra2011; Criscione, Reference Criscione and Holland2013).
Several evolutionary and ecological factors may be constraining N e of three-spined stickleback in Alaskan lakes. Broadly speaking, freshwater fish species tend to exhibit smaller N e estimates than do marine species, possibly due to factors like greater isolation, bottleneck effects and stronger selection pressures that reduce genetic diversity and thus N e (DeWoody and Avise, Reference DeWoody and Avise2000). Consistent with this observed phenomenon, smaller N e values have been detected for freshwater three-spined stickleback populations than marine and estuarine populations (Reusch et al., Reference Reusch, Wegner and Kalbe2001; Mäkinen et al., Reference Mäkinen, Cano and Merilä2006; Caldera and Bolnick, Reference Caldera and Bolnick2008). In addition, inbreeding rates are thought to be higher in relatively small populations of freshwater fishes (Aeschlimann et al., Reference Aeschlimann, Häberli, Reusch, Boehm and Milinski2003; Frommen et al., Reference Frommen, Luz, Mazzi and Bakker2008), like those examined in this study. Pressures from interactions with invasive fishes (i.e. competition, predation) as well as habitat degradation arising from pollution, water withdrawals and climate change (Waits et al., Reference Waits, Bagley, Blum, McCormick and Lazorchak2008; von Hippel, Reference von Hippel2008; Araguas et al., Reference Araguas, Vidal, Pla and Sanz2012) might also be imposing constraints (e.g. bottlenecks) that reduce stickleback N e in the study area.
N e of Alaskan Schistocephalus solidus
The recovered estimates of contemporary N e in S. solidus were consistently low but nonetheless exhibited considerable variation. No prior attempts have been made to estimate contemporary N e in S. solidus or any closely related species, but the estimates that we recovered are consistent with expectations that subdivision of parasites among hosts constrains parasite N e by increasing reproductive variance (Criscione and Blouin, Reference Criscione and Blouin2005). The recovered estimates also are comparable to published accounts of N e in other metazoan parasites. Criscione (Reference Criscione and Holland2013), for example, estimated contemporary N e for the nematode parasite A. lumbricoides in human households, which exhibited values ranging from fewer than 20 to more than 300. Similarly, estimates of contemporary N e of a phytoparasitic beet cyst nematode (Heterodera schachtii) were also low with substantial variation between plants, with most finite estimates falling below 500 (Jan et al., Reference Jan, Cracianne, Fournet, Olivier, Arnaud, Porte, Bardou-Valette, Denis and Petit2016). This comparison should be interpreted with some caution, however, because of differences in scale between the studies; N e estimates for S. solidus were produced by pooling parasites across hosts from the same lake, whereas Jan et al. (Reference Jan, Cracianne, Fournet, Olivier, Arnaud, Porte, Bardou-Valette, Denis and Petit2016) estimated N e for individual hosts (i.e. individual plants).
Effects of population structure and connectivity on N e estimation
Assumptions about the underlying structure and extent of migration between populations influenced estimates of host and parasite N e. For example, we recovered infinite upper 95% confidence interval (CI) bounds for most estimates of host N e (Table 3) when samples were grouped by lake-year (corresponding to the assumption of differentiation among lakes), whereas all of the estimates were bounded by finite intervals when samples were pooled by year (corresponding to the assumption of no differentiation). Similar outcomes were recovered for S. solidus when samples were pooled by year rather than lake-year (Table 3). Greater frequency of bounded N e estimates might simply be an outcome of increased sample sizes resulting from pooling samples. Estimates derived from pooling samples might also more closely reflect underlying population structure. This may be more likely for S. solidus than G. aculeatus, given their respective potential for dispersal (i.e. high and low, respectively) and observed patterns of population genetic structure (i.e. weak and strong, respectively) (Sprehn et al., Reference Sprehn, Blum, Quinn and Heins2015; Strobel et al., Reference Strobel, Alda, Sprehn, Blum and Heins2016). This difference was captured in our joint N e/m analyses, which consistently recovered higher estimates of migration for S. solidus than G. aculeatus (Table 4). We also recovered greater frequencies of bounded temporal estimates for both S. solidus and G. aculeatus when accounting for migration (Table 4). This further indicates the importance of accounting for connectivity in estimates of N e, even when there is evidence of extensive population differentiation as has been found for G. aculeatus (Strobel et al., Reference Strobel, Alda, Sprehn, Blum and Heins2016). It is also consistent with expectations that migration shifts estimates of local (i.e. lake-level) N e towards that of the entire metapopulation (Wang and Whitlock, Reference Wang and Whitlock2003).
Predictors of parasite N e
We did not find strong support for the hypothesis that factors external to stickleback hosts exert influence on parasite N e. Factors external to hosts might be expected to exert a downward cascade of influence on parasite N e. For example, lake size might influence parasite N e by determining host abundance, but multivariate RDAs showed that lake size did not significantly contribute to observed variance in parasite N e. Lake size also was not related to other conditions that might be subject to physiographic constraints, such as a lake-level measure of host infection (i.e. sum count of parasites per lake; Pearson's r = −0.391, df = 7, P = 0.298; Supplementary Table 1).
We did find support for the hypothesis that host attributes exert influence on parasite N e. We detected a significant association between lake-level estimates of parasite N e and host size (Table 5). This is consistent with the expectation that parasite census size should increase with host size, such that the standing number of reproductive individuals is greater in larger hosts (Criscione et al., Reference Criscione, Poulin and Blouin2005). A relationship between host size and parasite N e might arise because larger individuals suffer from a greater number of infections vis-à-vis higher consumption rates of infected copepods. We did not, however, detect a pairwise relationship between host size and the sum count of parasites per host, which was strongly correlated with the average number of parasites per host (Supplementary Table 1). This is not especially surprising since we only examined infected hosts in this study. Accordingly, this relationship would be better characterized by assessing both infected and uninfected hosts. Though we did not detect a relationship between host N e and parasite N e (possibly because the high proportion of unbounded infinite N e estimates for hosts limited the scope of the comparison), we did detect a relationship between host genetic diversity and parasite N e. The positive association recovered between host heterozygosity and estimates of parasite N e (Table 5 and Fig. 1) runs contrary to the notion that greater genetic diversity might offer a host some protection against parasitism (Kurtz et al., Reference Kurtz, Kalbe, Aeschlimann, Häberli, Wegner, Reusch and Milinski2004; Kaunisto et al., Reference Kaunisto, Viitaniemi, Leder and Suhonen2013). It is possible that any benefit provided by genetic diversity is nominal if parasites are genetically homogeneous and if the most common host genotypes are already resistant to parasite infection (van Baalen and Beekman, Reference van Baalen and Beekman2006). Further exploration of the observed relationship is warranted, however, considering that there is evidence to the contrary according to other measures of host genetic diversity (e.g. Kurtz et al., Reference Kurtz, Kalbe, Aeschlimann, Häberli, Wegner, Reusch and Milinski2004). There is also evidence of genetic heterogeneity in S. solidus (Sprehn et al., Reference Sprehn, Blum, Quinn and Heins2015; Strobel et al., Reference Strobel, Alda, Sprehn, Blum and Heins2016) as well as adaptive variation in S. solidus virulence and stickleback host resistance to parasitism (Kalbe et al., Reference Kalbe, Eizaguirre, Scharsack and Jakobsen2016; Weber et al., Reference Weber, Steinel, Shim and Bolnick2017).
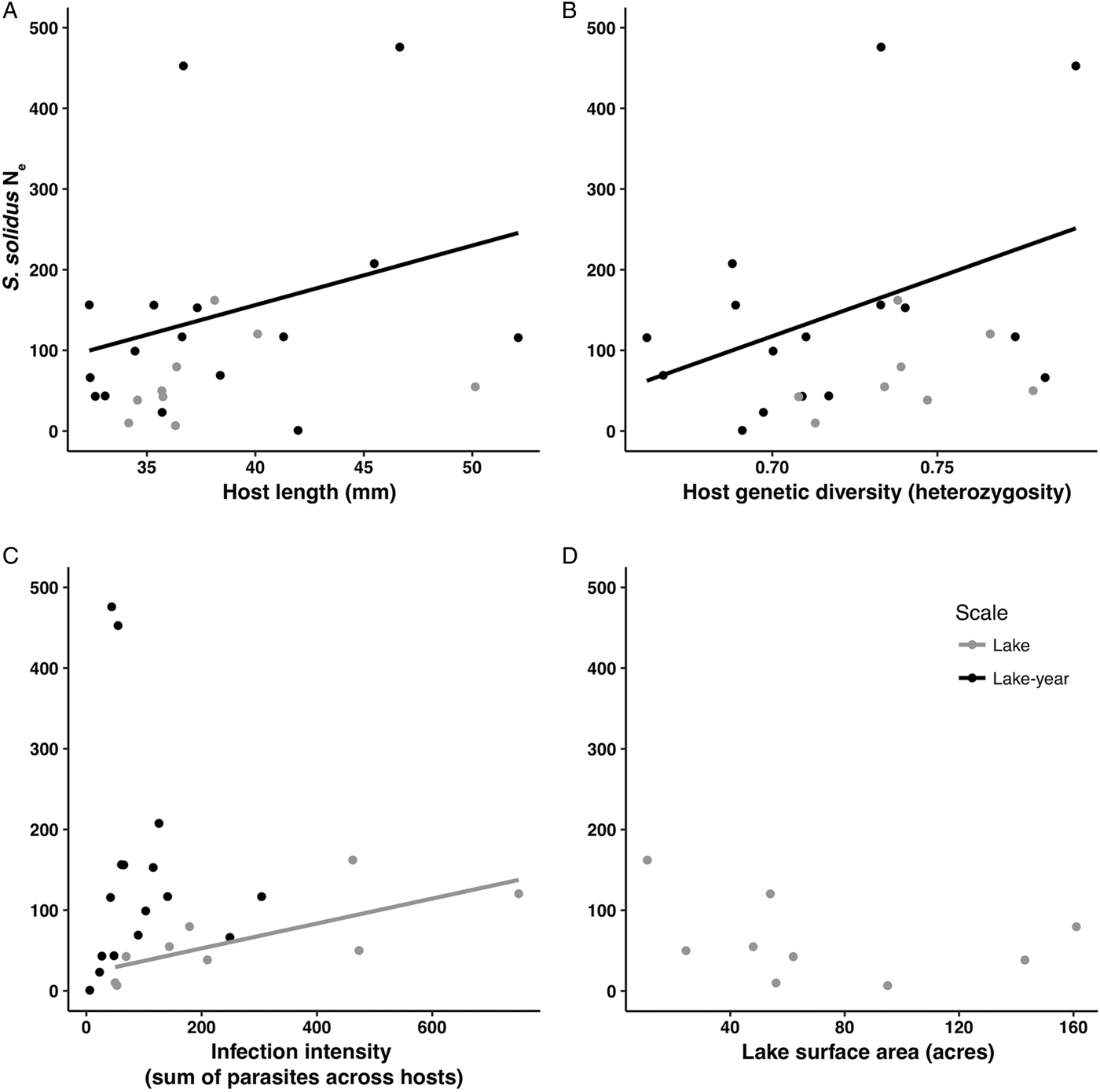
Fig. 1. Scatterplots showing relationships between parasite N e and the four predictor variables used in the RDAs, A: host length (mm), B: host genetic diversity (expected heterozygosity), C: infection intensity (sum count of parasites across hosts), D: lake surface area (acres). Both lake (grey) and lake-year (black) levels are shown. Trend lines are based on ordinary least squares linear regressions and reflect the significant relationships demonstrated in the RDAs (Table 5).
We additionally found support for the hypothesis that factors internal to the host exert influence on parasite N e. Pairwise correlation and redundancy analyses identified the sum of parasites across hosts within a lake as a significant predictor of parasite N e. Notably, post-hoc RDAs including the average number of parasites as a predictor variable recovered models similar to those from RDAs run with the sum of parasites across hosts as a predictor variable, though model support was lower (Supplementary Table 3) suggesting that average number of parasites is a weaker predictor of N e compared to the sum of parasites across hosts. As with host size, this is consistent with the expectation that higher infection intensities reflect higher parasite census sizes, which might elevate N e by ensuring a greater number of breeding individuals in hosts (Criscione et al., Reference Criscione, Poulin and Blouin2005; Doña et al., Reference Doña, Moreno-Garcia, Criscione, Serrano and Jovani2015).
Improving estimation of parasite N e
Several factors may have affected our estimates of genetic diversity and N e. First, only a subset of available fish and parasites were chosen for genotyping. In order to ensure adequate sample sizes for analyses of host characteristics, we focused on hosts that were infected with seven or more parasites (Sprehn et al., Reference Sprehn, Blum, Quinn and Heins2015; Strobel et al., Reference Strobel, Alda, Sprehn, Blum and Heins2016). As a consequence, measures of genetic diversity and parasite N e corresponded to only a subset of the total pool of samples, which also served as the basis for our proxy measure of host infection (i.e. sum parasites across hosts). This may have reduced our capacity to assess the influence of host attributes and host infection on parasite N e, as it may not have captured the full range of variation in host responses to infection by S. solidus (Kalbe et al., Reference Kalbe, Eizaguirre, Scharsack and Jakobsen2016). Thus our inferences are likely conservative, and might have been stronger (i.e. predictors might have explained a greater proportion of observed variance) had we considered all available specimens. Examining parallel subsets of the available data might have similarly improved our inferences, though reduced datasets also can increase the likelihood of recovering spurious relationships.
Additionally, greater consideration should be given to the influence of generation time on estimates of contemporary N e. Both single-sample and temporal estimators assume discrete generations (Waples and Yokota, Reference Waples and Yokota2007; Waples and Do, Reference Waples and Do2008), an assumption likely not met by most natural populations, including the host and parasite populations that were examined in this study. Single-sample estimators utilizing the LD method do, however, present a close approximation of generational N e if a random sample of mixed-age adults are considered, as opposed to a cohort of same-aged individuals, which favours estimation of N b (i.e. the effective number of breeders per reproductive cycle) (Robinson and Moyer, Reference Robinson and Moyer2013; Waples et al., Reference Waples, Antao and Luikart2014). The samples of sticklebacks examined in this study probably correspond to mixed-age adults because spawning age varies among individuals and some fish spawn across several reproductive cycles (Greenbank and Nelson, Reference Greenbank and Nelson1959; Saito and Nakano, Reference Saito and Nakano1999; Baker et al., Reference Baker, Heins, Foster and King2008). Consistent with this observation, pooling S. solidus parasites across hosts within a lake probably approximates mixed age classes, partly because fish largely consume infected copepods as juveniles during their first year of life and because plerocercoids can live for 2 years or more in the stickleback body cavity (Heins et al., Reference Heins, Singer and Baker1999, Reference Heins, Birden and Baker2010, Reference Heins, Eidam and Baker2016; Heins and Baker, Reference Heins and Baker2008). In species with overlapping generations, temporal estimates of N e are less likely to be affected by assumptions of generation time (Waples and Yokota, Reference Waples and Yokota2007) if samples are taken at least three to five generations apart. This suggests that temporal estimation of parasite and host N e might be improved by increasing the time period of data collection and comparison. This is more relevant for our temporal estimates of host N e, which are based on samples taken 2 or fewer years apart. Our estimates of parasite N e are more robust, considering that samples spanned 2 to 7 year periods (i.e. approximately one to three generations). Nonetheless, efforts to increase the period of time over which data are available (i.e. via further sampling) could improve estimates of parasite N e by mitigating possible bias due to overlapping generations.
Limitations and suggestions for future studies
Other aspects of our study design may have limited our ability to detect associations with parasite N e. For example, our sampling regime might not have been adequate for assessing landscape-scale phenomena. It is possible that the size range of the lakes included in our study might not have been large enough to capture the relationship between physiography and parasite N e. This concern could be addressed by assessing whether stickleback host demography varies with lake size. It is also possible, of course, that we might have overlooked factors that temper the influence of physiographic factors on parasite N e. Work on monoecious trematodes (Prugnolle et al., Reference Prugnolle, Liu, de Meeûs and Balloux2005) suggests that it would be worthwhile to assess ecological and genetic factors, such as migration between hosts, that can moderate the influence of physiography.
Greater consideration should also be given to factors that can influence variance in reproductive success (i.e. reproductive skew). Parasite populations tend to be highly aggregated, with most hosts harbouring a small number of parasites and a small minority of hosts harbouring a large number of parasites (Boulinier et al., Reference Boulinier, Ives and Danchin1996; Poulin, Reference Poulin2007). Aggregation and crowding (Read, Reference Read1951) might consequently reduce parasite N e by restricting reproduction to only a small fraction of parasites in heavily infected hosts (Poulin, Reference Poulin2007). Some evidence conversely suggests, however, that aggregation might have the opposite effect, where N e increases because sex ratios equalize as infection intensity increases (Morand et al., Reference Morand, Pointier, Borel and Theron1993). While our study was not designed to assess either condition (i.e. we did not examine uninfected individuals that would register counts of zero intensity), it is likely that both aggregation and crowding contribute to the observed variation in S. solidus N e. Estimates based on the ratio of variance to mean infection intensity (Supplementary Table 2) suggest that aggregation (variance/mean >1) occurs in as many as 36% of lake-year samples and 67% of lake samples (Poulin, Reference Poulin2007). Similarly, estimates based on residuals from a regression of log-transformed variance against log mean intensity (Supplementary Table 2) suggest that aggregation occurs in approximately half of our lake-year and lake sample sets (Poulin, Reference Poulin2013). We also found evidence of crowding in 18% (74 of 403) of stickleback hosts (Supplementary Table 2), with crowding defined as an infection intensity ⩾1 standard deviation above the mean. While undoubtedly too high, these estimates illustrate that further study of both conditions would be a fruitful endeavour. Not only would it shed additional light on stickleback–cestode interactions, it would also help reveal how reproductive variance influences parasite N e (Criscione and Blouin, Reference Criscione and Blouin2005). It might also be fruitful to examine metrics of parasite body mass – in addition to parasite abundance – to further characterize relationships between infection and parasite N e. It is conceivable, for example, that higher infection intensity might reduce inbreeding by providing a greater abundance of unrelated mates, but crowding might also skew reproductive output in favour of larger worms (Criscione and Blouin, Reference Criscione and Blouin2005). Assessment of other related metrics of parasitism, such as measures of host condition (e.g. standard mass indices) that serve as proxy metrics of host resilience to infection, would also likely shed further light on how interactions with hosts influence parasite N e.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182018002226
Author ORCIDs
Hannah M. Strobel, 0000-0002-3278-4087; David C. Heins, 0000-0002-6175-8777.
Acknowledgements
We would like to thank Dr Thomas P. Quinn at the University of Washington for providing stickleback specimens that were used in this study and prior studies. We would also like to thank Dr Erick Gagne for guidance on statistical analyses, and we would like to thank Megan Sekiya for assisting with laboratory-based data collection.
Financial support
Funding was provided by the Newcomb College Institute of Tulane University through the Newcomb Grant Program. Grants were awarded to H.M.S. and S.H. with faculty sponsors D.C.H. and M.J.B.
Conflict of interest
None.
Ethical standards
All collections were made under Fish Resource Permits issued by the Alaska Department of Fish and Game and in accordance with approved animal use protocols from Tulane University and the University of Washington.