Introduction
Echinococcus granulosus sensu lato (s.l.) comprises a number of species which are responsible for the zoonotic disease called cystic echinococcosis (CE) (Eckert and Deplazes, Reference Eckert and Deplazes2004). From these species, E. granulosus sensu stricto (s.s.) (including the genotypes G1, G3 and their micro variants) is the most commonly found species infecting humans (Alvarez Rojas et al., Reference Alvarez Rojas, Romig and Lightowlers2014) and occurs in a wide spectrum of intermediate hosts worldwide (Cardona and Carmena, Reference Cardona and Carmena2013). Echinococcus ortleppi (G5 genotype) circulates between cattle and dogs (Romig et al., Reference Romig, Deplazes, Jenkins, Giraudoux, Massolo, Craig, Wassermann, Takahashi, de la Rue, Thompson, Deplazes and Lymbery2017), although is also able to infect humans is far less relevant than other species for human infection. The genotypes G6/7/8/10 have been conventionally clustered as E. canadensis (Nakao et al., Reference Nakao, McManus, Schantz, Craig and Ito2006). However, this nomenclature remains controversial (Nakao et al., Reference Nakao, Lavikainen and Hoberg2015) and some authors subdivide this species grouping the genotypes G6/G7 as E. intermedius, the genotype G8 as E. borealis and G10 as E. canadensis (Lymbery et al., Reference Lymbery, Jenkins, Schurer and Thompson2015). In this paper, we use the term E. intermedius to refer to the G6/7 group. The cluster (G6/7) is the second most common cause for CE in humans worldwide. Echinococcus intermedius (G6) infects mostly camels and goats with human CE, caused by this species, occurring mainly in Argentina and Africa. Echinococcus intermedius (G7) uses primarily pigs as intermediate host with high incidences in humans especially in the Baltic countries (Romig et al., Reference Romig, Deplazes, Jenkins, Giraudoux, Massolo, Craig, Wassermann, Takahashi, de la Rue, Thompson, Deplazes and Lymbery2017).
It is well documented that CE is highly endemic in South American countries including Argentina, Brazil (south), Chile, Peru and Uruguay causing serious public health issues (Moro and Schantz, Reference Moro and Schantz2006; Pavletic et al., Reference Pavletic, Larrieu, Guarnera, Casas, Irabedra, Ferreira, Sayes, Gavidia, Caldas, Lise, Maxwell, Arezo, Navarro, Vigilato, Cosivi, Espinal and Del Rio Vilas2017). In contrast, the evidence regarding the situation of CE in Bolivia is scarce and limited to the understanding that the parasite is present and causing an unknown number of cases (Schantz, Reference Schantz1972; Moro and Schantz, Reference Moro and Schantz2006). The distribution of species comprising the E. granulosus s.l. complex in the endemic countries in South America has been investigated mostly with samples from the intermediate hosts. It is known that E. granulosus s.s. is present in all the endemic South American countries; E. ortleppi has been described in Argentina, Brazil, Chile and Uruguay; E. intermedius (G6) has been found in Argentina, Chile and Peru. And E. intermedius (G7) has been isolated in Argentina, Brazil and Peru (Rosenzvit et al., Reference Rosenzvit, Zhang, Kamenetzky, Canova, Guarnera and McManus1999; Kamenetzky et al., Reference Kamenetzky, Canova, Guarnera and Rosenzvit2000; Kamenetzky et al., Reference Kamenetzky, Gutierrez, Canova, Haag, Guarnera, Parra, García and Rosenzvit2002; Guarnera et al., Reference Guarnera, Parra, Kamenetzky, García and Gutiérrez2004; Badaraco et al., Reference Badaraco, Ayala, Bart, Gottstein and Haag2008; Manterola et al., Reference Manterola, Benavente, Melo, Vial and Roa2008; Santivañez et al., Reference Santivañez, Gutierrez, Rosenzvit, Muzulin, Rodriguez, Vasquez, Rodriguez, Gonzalez, Gilman and Garcia2008; Moro et al., Reference Moro, Nakao, Ito, Schantz, Cavero and Cabrera2009; Sánchez et al., Reference Sánchez, Cáceres, Náquira, Garcia, Patiño, Silvia, Volotão and Fernandes2010; Schneider et al., Reference Schneider, Gollackner, Schindl, Tucek and Auer2010; Soriano et al., Reference Soriano, Pierangeli, Pianciola, Mazzeo, Lazzarini, Saiz, Kossman, Bergagna, Chartier and Basualdo2010; de la Rue et al., Reference de la Rue, Takano, Brochado, Costa, Soares, Yamano, Yagi, Katoh and Takahashi2011; Balbinotti et al., Reference Balbinotti, Santos, Badaraco, Arend, Graichen, Haag and Zaha2012; Sánchez et al., Reference Sánchez, Cáceres, Náquira, Miranda, Samudio and Fernandes2012; Armua-Fernandez et al., Reference Armua-Fernandez, Castro, Crampet, Bartzabal, Hofmann-Lehmann, Grimm and Deplazes2014; Espinoza et al., Reference Espinoza, Salas, Vargas, Freire, Diaz, Sánchez and Venegas2014; Monteiro et al., Reference Monteiro, Botton, Tonin, Azevedo, Graichen, Noal and de la Rue2014; Cucher et al., Reference Cucher, Macchiaroli, Baldi, Camicia, Prada, Maldonado, Avila, Fox, Gutiérrez, Negro, López, Jensen, Rosenzvit and Kamenetzky2016; Alvarez Rojas et al., Reference Alvarez Rojas, Ebi, Paredes, Acosta-Jamett, Urriola, Roa, Manterola, Cortes, Romig, Scheerlinck and Lightowlers2017; Deplazes et al., Reference Deplazes, Rinaldi, Alvarez Rojas, Torgerson, Harandi, Romig, Antolova, Schurer, Lahmar, Cringoli, Magambo, Thompson, Jenkins, Thompson, Deplazes and Lymbery2017; Corrêa et al., Reference Corrêa, Stoore, Horlacher, Jiménez, Hidalgo, Alvarez Rojas, Figueiredo Barros, Bunselmeyer Ferreira, Hernández, Cabrera and Paredes2018) and more recently, for the first time in Ecuador (Ramos-Sarmiento and Chiluisa-Utreras, Reference Ramos-Sarmiento and Chiluisa-Utreras2020). In the case of Bolivia, this data is limited to two human cases reported as E. granulosus s.s. (Kamenetzky et al., Reference Kamenetzky, Gutierrez, Canova, Haag, Guarnera, Parra, García and Rosenzvit2002; Jarovsky et al., Reference Jarovsky, Brito, Monteiro, Azevedo, Botton, Mimica, Arnoni, Sáfadi, Berezin, Salgado Filho, Almeida and Rue2020).
Bolivia shares borders with all endemic countries in South America except for Uruguay. From all Bolivian borders, the ones with Argentina (773 km) and Peru (1000 km) are the most relevant for the proximity with areas where CE is endemic, like Salta and Jujuy in Argentina (Casas et al., Reference Casas, Otero, Céspedes, Sosa and Santillán2013; Salud, Reference Salud2019) and Puno in Peru (Cabrera, Reference Cabrera2007). For this study, we have collected cyst parasite material isolated from cattle, sheep, pigs and humans. We also analysed dog faecal samples infected with E. granulosus s.l. from Bolivia. Most of the samples were originated in La Paz Department. We present here the genetic characterization of such samples as a first step to produce data which can contribute to a better understanding of the distribution of the species comprising E. granulosus s.l. present in Bolivia.
Material and methods
Parasite material
A total of 35 cysts, identified as the larval stage of E. granulosus s.l., were collected from livestock including 7 cysts from sheep, 3 cysts from pigs and 25 cysts from cattle. Parasite material was collected from established abattoirs with veterinary inspection, except for sheep samples which were collected directly in an ‘open-air’ indigenous abattoir with no veterinary inspection. Additionally, three cysts from CE human patients were also included in the study. Microscopic analysis of cyst fluid was performed for discrimination of fertile and infertile cysts based on the presence or absence of protoscoleces. Furthermore, six dog faecal samples positive for E. granulosus s.l. were also included in this study. These dog faecal samples were identified from a parallel study from which 14 out of 131 samples were found to contain Taenia spp. eggs at microscopic examination (samples were fixed with formalin 10%). The information of the species, organ and fertility (for cysts) and collection place of all the samples used in this study can be seen in Table 1. A more detailed geographic origin of the samples isolated within La Paz Department including cysts isolated from 2 sheep, 18 cattle and all dog faecal samples can be seen in the map in Supplementary Fig. 1. Samples from two other cattle were originated in Punata, in the Cochabamba Department and 3 cysts from 2 pigs were originated in Santa Cruz Department (Table 1). Human cysts were collected after surgery from hospitals in Oruro, La Paz and Potosi and sent to the laboratory in La Paz. Ethics clearance for the use of human samples was provided by the Universidad Mayor de San Andres, La Paz, Bolivia.
Table 1. Origin, host species and fertility of Echinococcus cysts and faecal dog samples from Bolivia used for the genetic analysis in this study with the corresponding genotype and cox1 haplotype
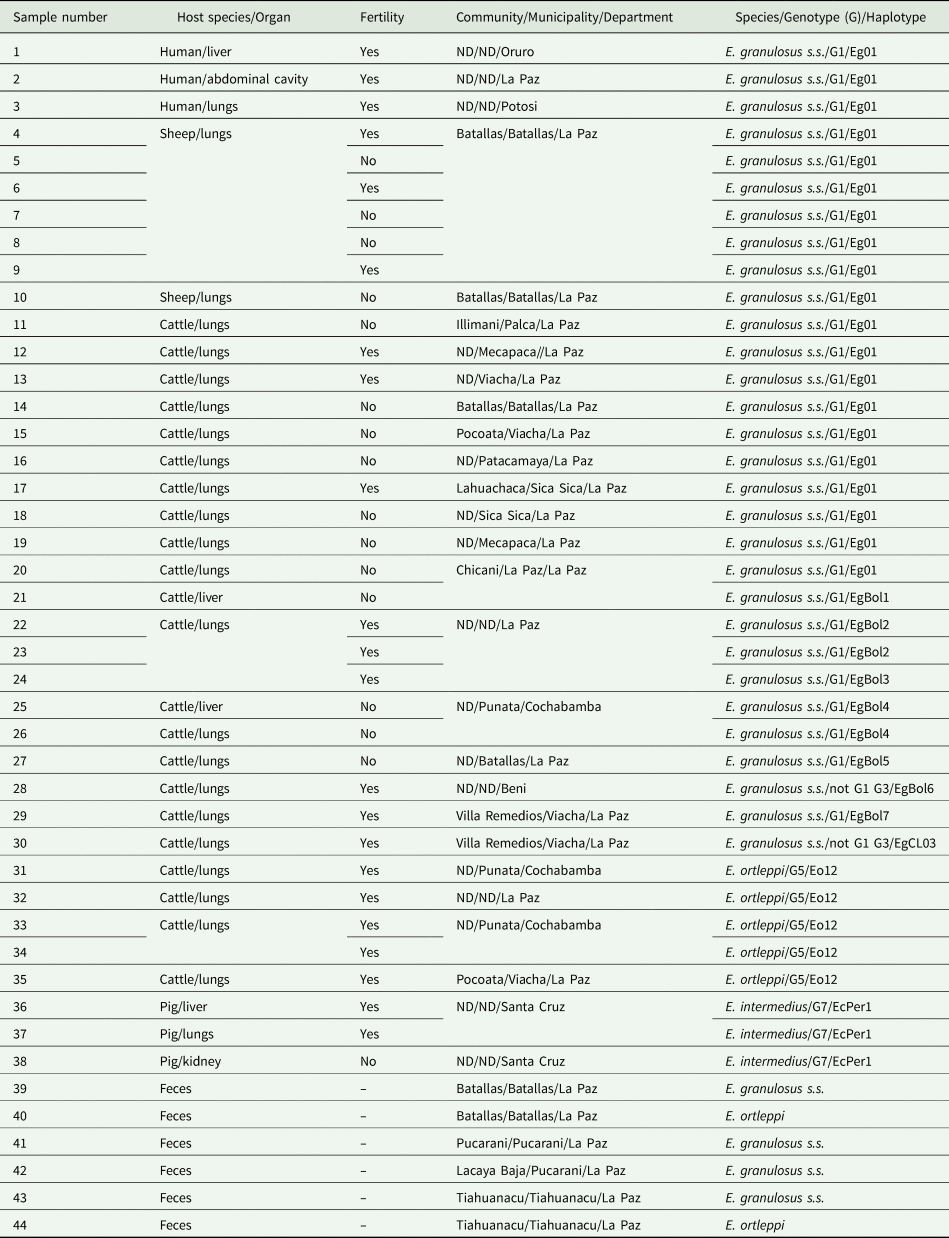
ND, no data.
Molecular analysis
Genomic DNA was isolated from the germinal layer of each cyst using the DNA mini kit (Qiagen). DNA was used as a template for polymerase chain reaction (PCR) to amplify and sequence 1609 bp of the cox1 gene of E. granulosus s.l. using the primers previously described (Hüttner et al., Reference Hüttner, Nakao, Wassermann, Siefert, Boomker, Dinkel, Sako, Mackenstedt, Romig and Ito2008). If no PCR product was amplified, then two overlapping fragments were produced by PCR (5′ and 3′ fragments) and sequenced (Alvarez Rojas et al., Reference Alvarez Rojas, Ebi, Gauci, Scheerlinck, Wassermann, Jenkins, Lightowlers and Romig2016). PCR products were visualized in a 2% agarose gel stained with GelRed and purified using the MinElute PCR purification kit (Qiagen) for sequencing in both directions using Sanger technology (Microsynth, Switzerland). The final sequence of the cox1 gene (1609 bp) from each cyst was built with the software Geneious (Kearse et al., Reference Kearse, Moir, Wilson, Stones-Havas, Cheung, Sturrock, Buxton, Cooper, Markowitz, Duran, Thierer, Ashton, Meintjes and Drummond2012) using the cox1 haplotype EG01 (accession number: JQ250806) as reference. Sequencing chromatograms were thoroughly checked, only sequences with single peaks were used for subsequent analysis. The identification of genotypes (G-system) was performed comparing the cox1 gene from each isolate with the original reference sequences (366 bp) for E. granulosus s.s. G3 (M84663), E. equinus G4 (M84664), E. ortleppi G5 (M84665), E. intermedius G6 (M84666) and E. intermedius G7 (M84667) from Bowles et al. (Reference Bowles, Blair and McManus1992). The original reference sequence uploaded to GenBank for E. granulosus s.s. G1 (M84661) is incorrect as this is identical to G3 (M84663). The sequence M84661 was manually corrected following Fig. 1 from the original publication and used as a reference for E. granulosus s.s. G1 (Bowles et al., Reference Bowles, Blair and McManus1992). The identification of new cox1 haplotypes (1609 bp) and haplotype network design was accomplished comparing data produced in this study with all same length cox1 sequences of E. granulosus s.l. deposited in GenBank using TCS and PopArt (Clement et al., Reference Clement, Posada and Crandall2000; Leigh and Bryant, Reference Leigh and Bryant2015). For the 14 dog faecal samples microscopically positive for taeniid eggs, DNA was isolated using the alkaline lysis method as previously described (Štefanić et al., Reference Štefanić, Shaikenov, Deplazes, Dinkel, Torgerson and Mathis2004) and further used as a template for multiplex PCR and sequencing to discriminate between E. granulosus s.l. (band of 117 bp) and other cestodes comprising Taenia spp. (267 bp) (Trachsel et al., Reference Trachsel, Deplazes and Mathis2007).

Fig. 1. Distribution of E. granulosus s.s., E. ortleppi and E. intermedius (G7) and host species infected in different departments in Bolivia according to the data presented in this study.
Results
Echinococcus granulosus s.s. was identified in all 7 cysts from sheep (from La Paz), also in 20 out of 25 cysts from cattle (17 from La Paz, 2 from Cochabamba and 1 from Beni) and in all 3 cysts from humans (from Oruro, La Paz and Potosi) (Table 1). When assigning genotypes (G-system) to these sequences, based on 366 bp of the cox1 gene, 28 out of the 30 samples identified as E. granulosus s.s. show 100% identity with the 366 bp of the original reference sequence for G1 and two other differ by a single nucleotide with either the genotypes G1 or G3 (Table 1). Echinococcus ortleppi (G5) was identified in the remainder 5 cysts from cattle (3 cysts were collected in La Paz and 2 others in Cochabamba) (Table 1). Echinococcus intermedius (G7) was identified in the three cysts available from pigs collected in Santa Cruz (Table 1). The presence of protoscoleces in samples identified as E. granulosus s.s. was reported in 3 out of 3 human samples, in 3 out 7 cysts (43%) from sheep and in 8 out of 20 cysts (40%) from cattle. Protoscoleces were observed in all five samples identified as E. ortleppi and in all the three samples identified as E. intermedius (G7). From the 6 dog faecal samples used in this study, E. granulosus s.s. was identified in 4 samples (1 from Batallas, 2 from Pucarani and 1 from Tiahuanacu) and E. ortleppi in 2 dogs (1 from Batallas and 1 from Tiahuanacu) (Supplementary Fig. 1). It was not possible to amplify the full length of the cox1 gene of E. granulosus s.l. in the dog faecal samples due to the use of formalin as a fixative. Taenia hydatigena was found in five dog faecal samples and Taenia pisiformis in two cases (not shown). One dog faecal sample showed a mixed infection with E. granulosus s.s. and T. hydatigena while another faecal sample showed a co-infection with E. ortleppi and T. hydatigena (not shown). A schematic representation of the species of E. granulosus s.l. found in different departments of Bolivia in this study is shown in Fig. 1.
From the 30 isolates identified as E. granulosus s.s. in the cyst samples, 20 of them were recognized as the cox1 haplotype Eg01 (Accession number JQ250806) (Table 1). The cox1 haplotype EgCL03 (KX227118) (Alvarez Rojas et al., Reference Alvarez Rojas, Ebi, Paredes, Acosta-Jamett, Urriola, Roa, Manterola, Cortes, Romig, Scheerlinck and Lightowlers2017) was identified in a single sequence in this study (Table 1). The remainder 9 sequences identified as E. granulosus s.s. do not share 100% homology with any sequence deposited in GenBank and they are named here as EgBol1-EgBol7 (Accession numbers: MT072973-MT072979) (Table 1). Two cattle presented mixed infection with two different cox1 haplotypes of E. granulosus s.s (Table 1). The isolates identified as E. ortleppi in 5 cysts from cattle had an identical sequence, this sequence is 100% homologous with the cox1 haplotype Eo12 (KU743926) originally identified in Namibia (Addy et al., Reference Addy, Wassermann, Banda, Mbaya, Aschenborn, Aschenborn, Koskei, Umhang, De La Rue, Elmahdi, Mackenstedt, Kern and Romig2017). The isolates identified as E. intermedius (G7) have the same sequence sharing 100% homology with the cox1 haplotype EcPer1 (AB777924) identified in Peru and also in the sequence of the complete mitochondrial genomes of E. intermedius (G7) from Poland, France, Spain, Romania and Mexico (Laurimäe et al., Reference Laurimäe, Kinkar, Romig, Omer, Casulli, Umhang, Gasser, Jabbar, Sharbatkhori, Mirhendi, Ponce-Gordo, Lazzarini, Soriano, Varcasia, Rostami Nejad, Andresiuk, Maravilla, González, Dybicz, Gawor, Šarkūnas, Šnábel, Kuzmina and Saarma2018).
The haplotype network for E. granulosus s.s. built with data from Bolivia from the present study and also cox1 sequences from Chile, Peru, Argentina and Brazil is shown in Fig. 2. The network has the typical star-like shape with the cox1 haplotype Eg01 in the centre. This haplotype is present in 20 sequences from Bolivia (this study), 33 from Chile, 16 from Peru, 10 from Argentina and 6 from Brazil. Other cox1 haplotypes found in more than one country in South America included EgCL03 (in Chile, Argentina and Bolivia), EgCL04 (in Chile and Argentina) and the haplotype ARG12 (in Argentina and Brazil). A total of 27 cox1 haplotypes in Fig. 2, including the ‘new’ haplotypes from Bolivia, differ only in a single nucleotide with the sequence of Eg01.
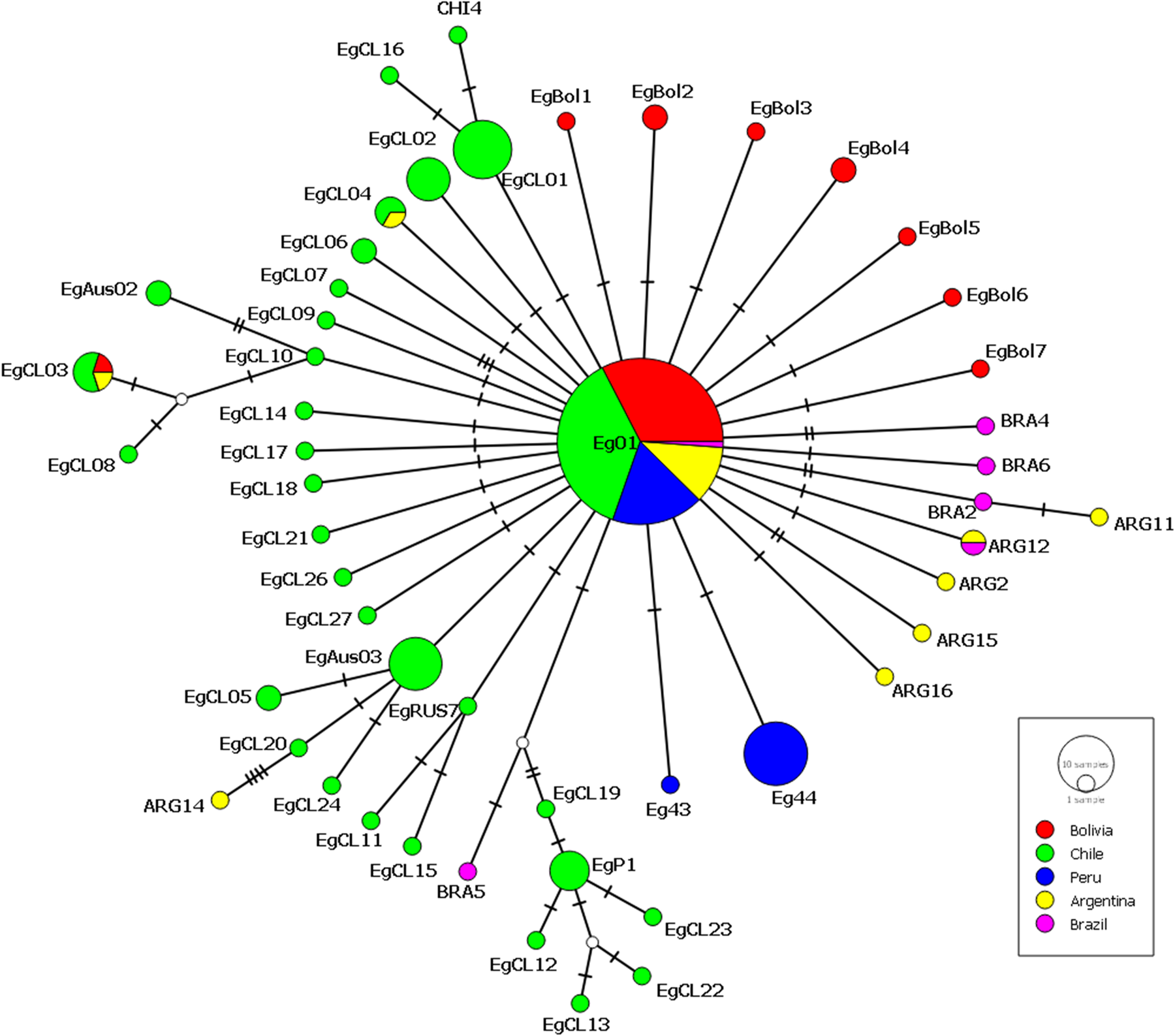
Fig. 2. Haplotype network built with 1609 bp of the cox1 gene of E. granulosus s.s. isolated in Bolivia in this study (red circles) (MT072973-MT072979) together with similar data from Chile: haplotypes EgCL1-21 (Alvarez Rojas et al., Reference Alvarez Rojas, Ebi, Paredes, Acosta-Jamett, Urriola, Roa, Manterola, Cortes, Romig, Scheerlinck and Lightowlers2017), EgCL22-27 (Hidalgo et al., Reference Hidalgo, Stoore, Pereira, Paredes and Alvarez Rojas2019). Haplotypes found in Chile (Alvarez Rojas et al., Reference Alvarez Rojas, Ebi, Paredes, Acosta-Jamett, Urriola, Roa, Manterola, Cortes, Romig, Scheerlinck and Lightowlers2017) but first described in Australia: EgAus02 (KT968703) and EgAus03 (KT968704) (Alvarez Rojas et al., Reference Alvarez Rojas, Ebi, Gauci, Scheerlinck, Wassermann, Jenkins, Lightowlers and Romig2016); Russia: EgRUS7 (AB777904) (Konyaev et al., Reference Konyaev, Yanagida, Nakao, Ingovatova, Shoykhet, Bondarev, Odnokurtsev, Loskutova, Lukmanova, Dokuchaev, Spiridonov, Alshinecky, Sivkova, Andreyanov, Abramov, Krivopalov, Karpenko, Lopatina, Dupal, Sako and Ito2013) and Nepal EgP1 (AB522646). From Peru: haplotypes Eg43 (AB688620) and Eg44 (AB688621) (Yanagida et al., Reference Yanagida, Mohammadzadeh, Kamhawi, Nakao, Sadjjadi, Hijjawi, Abdel-Hafez, Sako, Okamoto and Ito2012). Also included in the network is the sequence of the cox1 gene from isolates from Argentina: ARG1-ARG17 (KX039937- KX039953), Chile: CHI1-CHI4 (KX039961-KX039964), Brazil: BRA1-BRA6 (KX039955- KX039960) and haplotype AB1 (KX039954) (Laurimäe et al., Reference Laurimäe, Kinkar, Romig, Omer, Casulli, Umhang, Gasser, Jabbar, Sharbatkhori, Mirhendi, Ponce-Gordo, Lazzarini, Soriano, Varcasia, Rostami Nejad, Andresiuk, Maravilla, González, Dybicz, Gawor, Šarkūnas, Šnábel, Kuzmina and Saarma2016). Eg01 includes the cox1 sequence from ARG1, ARG3-10, ARG17, BRA1 and CHI2 (Laurimäe et al., Reference Laurimäe, Kinkar, Andresiuk, Haag, Ponce-Gordo, Acosta-Jamett, Garate, Gonzàlez and Saarma2016). EGCL04 is 100% homologous to ARG13, ARG12 = BRA3, EgCL01 = CHI1, EgCL02 = CHI3. The size of the circles is proportional to the number of sequences in which each haplotype has been identified, the Eg01 haplotype includes 30 sequences from Bolivia (this study), 33 from Chile (Alvarez Rojas et al., Reference Alvarez Rojas, Ebi, Paredes, Acosta-Jamett, Urriola, Roa, Manterola, Cortes, Romig, Scheerlinck and Lightowlers2017), 16 from Peru (Yanagida et al., Reference Yanagida, Mohammadzadeh, Kamhawi, Nakao, Sadjjadi, Hijjawi, Abdel-Hafez, Sako, Okamoto and Ito2012), 10 from Argentina and 1 from Brazil (Laurimäe et al., 2016).
Discussion
Data from the present study show that at least three species of the E. granulosus s.l. complex are circulating in Bolivia. The cosmopolitan species E. granulosus s.s. was found in the majority of the isolates examined, including the human samples. Interestingly, a similar fertility rate was observed in E. granulosus s.s. cysts isolated from sheep and cattle (around 40%). It is usually accepted that fertility of E. granulosus s.s. cysts is much higher in sheep than in cattle (Thompson, Reference Thompson, Thompson, Deplazes and Lymbery2017). The relatively high fertility of cysts from cattle in this study suggests that this species could play an important role in the transmission of the parasite in Bolivia. In this study cysts from only 2 sheep were included. The sheep population in Bolivia reaches 7 416 185 animals (2017) from which a high proportion is slaughtered at home compared to cattle. In the last decades, the sheep population in the Altiplano has decreased due to high infection with Fasciola hepatica (Plata, Reference Plata2005). The finding of E. ortleppi and E. intermedius (G7) in Bolivia extends the knowledge of the distribution of these parasites in South America (Fig. 2). It is known that both species can also cause infection in humans (Alvarez Rojas et al., Reference Alvarez Rojas, Romig and Lightowlers2014). Echinococcus ortleppi was found in cattle from two different areas of Bolivia (La Paz and Cochabamba), which are located approximately 380 km from each other. This suggests that E. ortleppi could, potentially, be widely spread in Bolivia. It is well known that E. ortleppi produce fertile cysts in cattle unlike the low fertility observed when E. granulosus s.s infects cattle (Thompson et al., Reference Thompson, Kumaratilake and Eckert1984; Balbinotti et al., Reference Balbinotti, Santos, Badaraco, Arend, Graichen, Haag and Zaha2012), all the cysts identified as E. ortleppi in this study were fertile (Table 1). The total number of cattle in Bolivia is estimated to be 9 304 572 animals, from which 4 010 258 are located in Santa Cruz. Cattle population in La Paz reaches 548 841 animals, most of them in the Altiplano, north of La Paz (MDR&T, 2017). Interestingly, E. intermedius (G7) was found in all three fertile cysts available from pigs in this study. The isolates were collected in Santa Cruz and all three cysts produced the same cox1 sequence which shares 100% homology with G7 isolates previously found in Peru and also in different European countries. The total number of pigs in Bolivia (in 2014) has been estimated to be 2 941 827 from which 892 232 are in Santa Cruz (MDR&T, 2014). An important number of pigs in Bolivia are raised outdoors with no veterinary inspection (Callisaya, 2018). Most likely these pigs are slaughtered at home continue facilitating the transmission of the parasite. Further investigations of population-based prevalences have to be established to estimate the relative contribution of the different intermediate hosts to the transmission.
Echinococcus granulosus s.s. was also found in 4 faecal dog samples (Batallas, Pucarani and Tiahuanacu) while E. ortleppi was present in 2 samples (Batallas and Tiahuanacu) demonstrating active transmission of both species in La Paz Department. The presence of T. hydatigena in 5 dog faecal samples (together with the finding of E. granulosus s.l.) described in this study shows that some dogs are actively been fed or have access to livestock offal in La Paz Department. Dog samples included in this study were taken from a parallel study investigating intestinal parasites from 131 samples in the La Paz department. Although this is not intended to be a prevalence study, it is possible to say that at least 4.5% (6 out of 131) of the dogs sampled were infected with E. granulosus s.l. This number could have been potentially higher if a more sensitive method for isolation/concentration of taeniid eggs would have been used, for example, the flotation and sieving method widely used in previous studies (Mathis et al., Reference Mathis, Deplazes and Eckert1996; Trachsel et al., Reference Trachsel, Deplazes and Mathis2007). Previous reports of the prevalence of E. granulosus s.l. in dogs from Bolivia are rather limited. These include a vague report of 50% of infection in dogs near La Paz after arecoline purgation in the 1970's (Moro and Schantz, Reference Moro and Schantz2006); a report of 23.9% of positive dogs using copro-ELISA in Tupiza (Potosi) in 2011 (Villena, Reference Villena2011); and the most recent report of prevalence indicating values of between 3.4 and 30% using copro-ELISA and copro-Western blot, in border towns with Argentina (Casas et al., Reference Casas, Otero, Céspedes, Sosa and Santillán2013). Future studies of the prevalence of E. granulosus s.l. in dogs from Bolivia are urgently needed to have a better understanding of the epidemiology of the parasite in this country.
The analysis of 1609 bp of the cox1 gene from the cysts from this study identified a vast majority as the haplotype Eg01. Two animals showed infection with more than one haplotype as it has been previously described (Hidalgo et al., Reference Hidalgo, Stoore, Pereira, Paredes and Alvarez Rojas2019). It is important to mention that the biological significance of the different haplotypes of E. granulosus s.s. remains unknown and no data is available concerning differences in virulence or host/parasite interaction or antigenicity which could affect the response to vaccines or diagnostic methods. In the meantime the description of new cox1 haplotypes of E. granulosus s.s. worldwide contributes to the general knowledge of how the parasite spread worldwide. The haplotype EG01 was originally described by Yanagida et al. (Reference Yanagida, Mohammadzadeh, Kamhawi, Nakao, Sadjjadi, Hijjawi, Abdel-Hafez, Sako, Okamoto and Ito2012) as the likely founder population of E. granulosus s.s. which was disseminated from the Middle East with the export of livestock after their domestication. It is usually located at the centre of haplotype networks in different countries where similar studies have been performed including Iran, Jordan, Peru, Russia, Chile and Australia (Yanagida et al., Reference Yanagida, Mohammadzadeh, Kamhawi, Nakao, Sadjjadi, Hijjawi, Abdel-Hafez, Sako, Okamoto and Ito2012; Konyaev et al., Reference Konyaev, Yanagida, Nakao, Ingovatova, Shoykhet, Bondarev, Odnokurtsev, Loskutova, Lukmanova, Dokuchaev, Spiridonov, Alshinecky, Sivkova, Andreyanov, Abramov, Krivopalov, Karpenko, Lopatina, Dupal, Sako and Ito2013; Romig et al., Reference Romig, Ebi and Wassermann2015; Alvarez Rojas et al., Reference Alvarez Rojas, Ebi, Gauci, Scheerlinck, Wassermann, Jenkins, Lightowlers and Romig2016; Alvarez Rojas et al., Reference Alvarez Rojas, Ebi, Paredes, Acosta-Jamett, Urriola, Roa, Manterola, Cortes, Romig, Scheerlinck and Lightowlers2017). The haplotype network built with the cox1 sequences from E. granulosus s.s. from this study and the ones available in GenBank from South American countries shows that 27 haplotypes differ by a single nucleotide with the central haplotype Eg01 (Fig. 2). Although, there is no evidence that the population of E. granulosus s.s. differ significantly between South American countries, it is obvious that most of the cox1 haplotypes are uniquely identified with particular countries. Since the Eg01 cox1 haplotype remains as the most commonly found in South America, we speculate here that it could have been the most common variant of the parasite infecting livestock and also dogs imported into the Americas with the Spanish colonization more than 500 years ago. The parasite and especially the one belonging to the Eg01 haplotype adapted to a variety of climates in South America and random mutations accumulated over time (in the cox1 and other genes) which now we identify as different haplotypes of the parasite. Livestock (including cattle, sheep and pigs) first arrived in La Española Island (currently in the Dominican Republic) 1 year after the arrival of Columbus. Later, livestock also arrived in Brazil with the Portuguese colonization (Primo, Reference Primo2004) and also directly to Uruguay and Argentina in subsequent trips from Spain (Beteta Ortiz, Reference Beteta Ortiz1999). After the colonization, most livestock was concentrated in what is currently known as Panama, from where it was distributed south to Venezuela, Colombia, Ecuador and Peru (Primo, Reference Primo1992; Villalobos Cortés et al., Reference Villalobos Cortés, Martinez and and Delgado2009). After conquering the Inca Empire, livestock was mostly concentrated in Peru from where it was distributed to the territories known now as Bolivia, Chile and Argentina. Since the discovery of silver in Potosi (Bolivia) this city was known to be a hub for livestock some of which was brought to the Americas from Argentina and Uruguay (Escobari de Querejazu, Reference Escobari de Querejazu1985). Also, La Paz was a connecting point between the commercial routes that led from Potosí and Oruro to Lima. These events show that the Bolivian territory was an important hub for livestock distribution suggesting that the parasite has been present in Bolivia for centuries and it has been mostly neglected in this country.
To conclude, the finding of three different species of E. granulosus s.l. have important implications in the design and implementation of control measures. For example, it is known that the improved general slaughtering hygiene without any specific control measure per se has reduced the presence of E. ortleppi (G5) in parts of Central and Western Europe (Deplazes et al., Reference Deplazes, Rinaldi, Alvarez Rojas, Torgerson, Harandi, Romig, Antolova, Schurer, Lahmar, Cringoli, Magambo, Thompson, Jenkins, Thompson, Deplazes and Lymbery2017). Recently, a pilot control programme in Lithuania, based on the deworming of dogs in specific times before the slaughtering of pigs, has achieved a strong reduction of E. intermedius (G7) (Šarkūnas et al., Reference Šarkūnas, Vienažindienė, Alvarez Rojas, Radziulis and Deplazes2019). However, in areas with extensive sheep production, it is known that further measures have to be applied to control E. granulosus s.s. (e.g. registration and regular deworming of dogs, public health education, construction of elimination ditches for the correct disposal of offal, etc.) (Craig et al., Reference Craig, Hegglin, Lightowlers, Torgerson, Wang, Thompson, Deplazes and Lymbery2017). Also, it remains unclear if the EG95 vaccine which prevents the infection in livestock, based on E. granulosus s.s., is effective to prevent the infection with different species of E. granulosus s.l. For example, it has been suggested that differences in the antigenicity of the EG95 proteins from E. intermedius G6 could make the current vaccine less effective against this species (Alvarez Rojas et al., Reference Alvarez Rojas, Gauci and Lightowlers2013). Therefore, the use of the EG95 vaccine, based in E. granulosus s.s., in a control programme where multiple species occur, as is the case of Bolivia, would need to be thoroughly monitored. Data reported here represents the first step to understand the epidemiological situation of CE in Bolivia. However, a systematic sample collection of parasite material in different regions from a diverse number of intermediate hosts and humans is needed to understand the real distribution of different species of E. granulosus s.l. in Bolivia.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182020000529
Acknowledgements
The project ‘Cystic echinococcosis in Bolivia’ is a joint research between the University of Zurich and the Universidad Mayor San Andres in Bolivia. A mobility grant of the Leading House of the Latin American Region ‘Epidemiological investigation of cystic echinococcosis in humans and animals in Bolivia’ is directly linked to this project and has been financed by the Leading House of the Latin American Region/State Secretariat for Education, Research and Innovation (SERI). We acknowledge the authorities of the Faculty of Medicine, Universidad Mayor de San Andrés, for administrative collaboration for this joint initiative; to the Arq Anna Hilda Portugal Peña and the staff of the Centro Municipal de Faeno, Gobierno Autónomo Municipal de La Paz, for their collaboration with collecting of samples from cattle; to Javier Larico and Ever Quispe, from the Servicio Nacional de Sanidad Agropecuaria e Inocuidad Alimentaria (SENASAG) at La Paz, for their collaboration with this project. Our gratefulness to Dr George Garcia for providing parasite material collected from a human. Special acknowledgements to the authorities and inhabitants of the Municipalities of Batallas, Pucarani and Tiahuanacu for their collaboration during the fieldwork.
Financial support
The analyses of the samples were financed by the Institute of Parasitology, University of Zurich. The sample collection, transportation and parasitological analysis were funded by IINSAD and the Research Laboratory of Parasitology, Faculty of Medicine, Universidad Mayor de San Andrés. Cristian A. Alvarez Rojas is a recipient of the mobility grant funded by the Leading House of the Latin American Region/State Secretariat for Education, Research and Innovation (SERI).
Conflict of interest
The authors declare no conflicts of interest.
Ethical standards
Ethics clearance for the use of human samples was provided by the Universidad Mayor de San Andres, La Paz, Bolivia.






