INTRODUCTION
Blood parasites (Plasmodium spp. and Haemoproteus spp.) are found throughout the world (Young et al. Reference Young, Garvin and McDonald1993; Murata, Reference Murata2002; Valkiūnas et al. Reference Valkiūnas, Iezhova, Brooks, Hanelt, Brant, Sutherlin and Causey2004; Reullier et al. Reference Reullier, Pérez-Tris, Bensch and Secondi2006). Although the haematozoa of Neotropical birds have been surveyed in many areas (Woodworth-Lynas et al. Reference Woodworth-Lynas, Caines and Bennet1989; Young et al. Reference Young, Garvin and McDonald1993; Valkiūnas, Reference Valkiūnas1997; Matta et al. Reference Matta, Basto, Gutierrez, Rodríguez and Greiner2004; Ribeiro et al. Reference Ribeiro, Sebaio, Branquinho, Marini, Vago and Braga2005; Durrant et al. Reference Durrant, Beadell, Ishtiaq, Graves, Olson, Gering, Peirce, Milensky, Schmidt, Gebhard and Fleischer2006; Belo et al. Reference Belo, Passos, Junior, Goulart, Sherlock and Braga2009), some regions and environments in South America remain poorly studied. For example, little is known about the prevalence of blood parasites of wild birds in Venezuela, particularly from the arid Caribbean coast. Habitat quality can affect vector density and the composition of the bird communities (Reiter and Lapointe, Reference Reiter and Lapointe2009; Ramírez-Albores, Reference Ramírez-Albores2010), and one might expect parasite-host relationships to differ between arid and humid environments. Current land practices in Venezuelan arid zones are causing severe environmental change that threatens the long-term survival of the habitat-specialist birds restricted to these areas (Rodriguez-Ferraro and Blake, Reference Rodríguez-Ferraro and Blake2008). The abundance of some common bird species in dry desert scrub and dense thorny thickets along the Caribbean coasts of Venezuela and Colombia differs considerably among areas as a consequence of anthropogenic factors, such as habitat modification and poaching. Habitat alteration resulting from climate change also might influence the distribution and abundance of wildlife species and thus may be a major driver of change in the ecology of pathogen transmission.
Information on the influence of precipitation on haemoparasite diversity, distribution, and prevalence in tropical environments could help to clarify the effect of habitat and landscape on parasite-host interactions in wild populations, including how parasite species distribution and abundance might respond to climate change in the future.
In this study, we report on the presence and distribution of the haematozoa of birds in the arid zones of northern Venezuela. This region is especially well suited for the study of avian malaria in an arid environment: the area belongs to the ‘peri-Caribbean arid belt’, which is one of 6 Neotropical arid zones and considered an Endemic Bird Area (EBA; Stattersfield et al. Reference Stattersfield, Crosby, Long and Wege1998) because of the occurrence of restricted-range and habitat-specialist bird species.
We tested the null hypothesis that haemosporidian parasite lineages do not differ between the arid regions of Venezuela. We assume that competent vectors occur in every arid region and that generalist host species with wide distributions, such as Mimus gilvus, are susceptible to most of the parasite lineages present in the arid lineages of Northern Venezuela. We also tested the hypothesis that some parasite lineages are unique to the arid regions of Venezuela owing to the presence of arid habitat endemic birds. Thus, the avian malaria parasites of this region might potentially reflect the unique avifauna of this arid climate zone.
MATERIALS AND METHODS
Study areas
Sampling was conducted in the arid zones of northern Venezuela, that are characterized by thorn scrubs dominated by species belonging to the Cactaceae, Mimoseae (Fabaceae), and Capparaceae (Sarmiento, Reference Sarmiento1972). Mean annual temperature is 28°C, and annual rainfall ranges between 300 and 700 mm, with a long and severe dry season punctuated by 2 brief rainy peaks in July–August and December (Sarmiento, Reference Sarmiento and Goodall1976).
Samples were obtained from 6 arid areas in northern Venezuela (Rodríguez-Ferraro and Blake, Reference Rodríguez-Ferraro and Blake2008) (Fig. 1). (1) Paraguaná Peninsula (PP) is located in northwestern Venezuela. It was an island during the Pliocene and extends over 2500 km2. (2 and 3) Falcon (FL) and Lara (LL) lowlands represent the most extensive arid zone in Venezuela, and are located in the Western region, with an approximate area of 16 000 km2. (4) The Clarines-Piritu region (CP) extends over about 4500 km2 in northeastern Venezuela and covers the Unare Depression, between the central and eastern portions of the Coastal Mountain Range. (5) Araya Peninsula (AP) occupies 900 km2 in northeastern Venezuela and comprises the lowlands to the north of the eastern Coastal Mountain Range. (6) Macanao Peninsula (MP), about 300 km2, comprises the westernmost portion of Margarita Island. We divided these areas between a western group (PP, FL, LL) and an eastern group (MP, CP, AP).

Fig. 1. Location of arid zones (shaded) in northern Venezuela, and areas studied. Study areas of Western region include the following: PP, Paraguaná Peninsula; FL, Falcón Lowlands; LL, Lara Lowlands and MP, Macanao Peninsula. Areas studied of Eastern region: CP, Clarines-Piritu; AP, Araya Peninsula.
Field sampling
Bird samples were collected during bimonthly trips to each area between September 2004 and August 2005. Birds were captured using mist nets (12 m×2·8 m×36 mm mesh). Blood was collected using heparinized microcapillary tubes following venipuncture of the brachial vein with a sterile syringe needle (Gaunt and Oring, Reference Gaunt and Oring1997) and stored in lysis buffer.
DNA extraction
Approximately 20 μl of blood sample was stored at room temperature (22–25°C) in cell lysis solution (Quiagen Inc, Valencia, California) before processing in the laboratory at the University of Missouri-St Louis. DNA from blood samples was extracted using a PureGene® commercial kit according to the manufacturer's protocol (Gentra systems, Minneapolis, MN, USA). The DNA pellet was re-suspended in 100 μl of hydration solution and kept at −20°C until use.
Screening
We used screening primers designed to amplify a 154-nucleotide segment of RNA-coding mitochondrial DNA (Fallon et al. Reference Fallon, Ricklefs, Swanson and Bermingham2003). PCR reactions were run in 10 μl volumes that contained the following final concentrations: 0·4 mM of each primer, 200 mM of each dNTP (PROMEGA), 10 mM Tris–HCl, pH 8·5, 50 mM KCl, and 1 U of Taq DNA polymerase (PHONEUTRIA, Minas Gerais, Brazil). Thermal cycling conditions were as follows: initial denaturation of 2 min at 94°C followed by 35 cycles with 1 min denaturation at 94°C, 1 min annealing at 62 °C, and extension at 72°C for 1 min 10 sec. This was followed by a final extension of 3 min at 72°C. PCR products were screened on 1·5% agarose gels, stained with ethidium bromide and visualized with a UV light source.
Cytochrome b amplification and sequencing
From samples in which we detected hematozoan infection (mitochondrial DNA amplification), we amplified a fragment of 591 bp of the cyt b gene (Perkins and Schall, Reference Perkins and Schall2002) under the following conditions: with 1 μl of genomic DNA was subjected to an initial denaturation of 4 min at 94°C, followed by 35 cycles of 94°C for 20 sec, 49°C for 10 sec, and 68°C for 45 sec, and a final extension at 68°C for 3 min. For most samples, a 0·5-μl aliquot of this product was used as a template for a nested reaction with primers described by Ricklefs et al. (Reference Ricklefs, Swanson, Fallon, Martinez, Scheuerlein and Latta2005) under initial denaturation of 94°C for 1 min and 28 cycles of 94°C for 20 sec, 52°C for 10 sec, and 68°C for 50 sec and then 68°C for 7 min. PCR products were screened on 1% agarose gels, stained with ethidium bromide, and visualized with a UV light source.
Positive PCR products were purified for cycle sequence reactions using ExoSAP-IT® (USB Corporation, Cleveland, Ohio) following the manufacturer's instructions. Bi-directional sequencing using primers 413F and 926R with dye-terminator fluorescent labelling was performed in an ABI Prism 3100 automated sequencer ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). We sequenced approximately 500 base pairs of the cyt b gene for Plasmodium spp. and Haemoproteus spp.
Phylogenetic analysis
DNA sequences were aligned using CLUSTALX (Thompson et al. Reference Thompson, Gibson and Plewniak1997) and edited using Seq Man II version 4 (DNASTAR Inc.) and are available through GenBank (Accession numbers JN819517- JN819533). Sequences were compared for identification to their closest matches in GenBank using the NCBI nucleotide Blast search, and to unpublished sequences using a local blast search in the laboratory of R. E. Ricklefs. We used MODELTEST version 3.6 (Posada and Crandall, Reference Posada and Crandall1998) to determine the most appropriate evolutionary model for our data. A maximum likelihood phylogenetic tree was produced for the parasite sequences using RAxML (Stamataxis, 2006) with the GTR + gamma model of nucleotide evolution and 100 bootstrap replications.
Statistics
Contingency table analyses were used to detect interactions between location, sex (where distinguished), season, and parasite prevalence. Statistics were carried out using Prism 5.0 for Mac OS (GraphPad Software, Inc.). The estimator Mao Tau (Colwell et al. Reference Colwell, Mao and Chang2004) was used to compare parasite species richness between two regions using the scores of 200 randomizations produced by EstimateS 8.2 (Colwell, Reference Colwell2008) to estimate parasite lineage richness in each study site.
RESULTS
Samples from 527 wild birds (11 families and 20 species) were screened for haemosporidian parasites. Of these, 41% were infected with Plasmodium spp. (13 individuals) or Haemoproteus spp. (148 individuals), using PCR as the diagnostic method; 55 individuals that presented mixed infections were not considered in this study (data not shown).
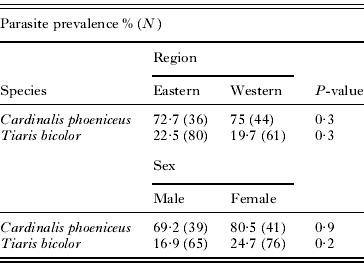
The species with the highest prevalence of infection were Icterus nigrogularis (n=19, 100%), Mimus gilvus (n=51, 84·3%), Melanerpes rubricapillus (n=14, 78·6%), and Cardinalis phoeniceus (n=80, 73·8%). The overall frequency of haemosporidian infection did not differ between the western (45%; n=252) and eastern (37·5%; n=275) regions (P=0·1). Homogeneity between western and eastern regions was also apparent in the two most abundant species in our sample, Cardinalis phoeniceus and Tiaris bicolor (Table 1). Haemosporidian prevalence was not heterogeneous with respect to dry versus rainy seasons, either in the total sample (P=0·07) or in C. phoeniceus and T. bicolor (P>0·5). Blood parasite prevalence did not differ between males and females of C. phoeniceus and T. bicolor (P=0·3) (Table 1).
Table 1. Prevalence of Plasmodium/Haemoproteus according to regions studied and the sex in two species wild birds

| Parasite prevalence % (N) | ||||
| Species | Region | P-value | ||
| Eastern | Western | |||
| Cardinalis phoeniceus | 72·7 (36) | 75 (44) | 0·3 | |
| Tiaris bicolor | 22·5 (80) | 19·7 (61) | 0·3 | |
| Sex | ||||
| Male | Female | |||
| Cardinalis phoeniceus | 69·2 (39) | 80·5 (41) | 0·9 | |
| Tiaris bicolor | 16·9 (65) | 24·7 (76) | 0·2 | |
General absence of regionalization of parasite lineages
Cytochrome b sequences revealed 7 Plasmodium lineages and 10 Haemoproteus lineages in the entire sample (Fig. 2). Of the lineages recovered from 5 or more host individuals, only 1 was restricted to a single region (Plasmodium Ven01 in the east). Thus, there is little evidence of regionalization in the parasite faunas across the arid northern coast of Venezuela.

Fig. 2. Phylogram of Plasmodium spp. and Haemoproteus spp. lineages in community birds. Phylogenetic relationships of the 17 haemosporidian parasite lineages found in 2 different regions, based on cyt b sequences. Numbers located on the top of the branches indicate ML bootstrap support (100 replications, only values above 50% are shown). The presence of each parasite lineage in the two areas studied is indicated by: western region (•) and eastern region (△). Parasite lineages described in others studies (*). The values of ‘N’ are the samples size of each lineage.
Ven01 was restricted to the host species Mimus gilvus, which had similar representation in our samples from 2 regions (east, n=28; west, n=23). Four of 5 examples of the Haemoproteus lineage Ven13 were restricted to Mimus gilvus in the western region (the other example was from Xiphorhynchus picus in the east). This appears to be the only potential case of partitioning of host individuals among parasite lineages on a regional basis. Two other lineages recovered from M. gilvus (Ven04 and Ven05) occurred in both regions.
Eight of the lineages found in this study had previously been described in other studies (Table 2). Nine lineages are described for the first time in this study: 3 Plasmodium and 6 Haemoproteus, although several of these lineages are additionally known from unpublished work in the laboratory of R. E. Ricklefs (see Discussion section).
Table 2. Relation of parasite lineages to those previously reported in other areas

| Parasite lineage | GenBank number Identical Lineages | Source |
| Ven_01 Plas | AF465559 | Ricklefs and Fallon (Reference Ricklefs and Fallon2002) |
| Ven_03 Haem | GQ395658 | Levin et al. (Reference Levin, Outlaw, Hernan Vargas and Parker2009) |
| Ven_07 Plas | DQ838997 | Beadell et al. (Reference Beadell, Ishtiaq, Covas, Melo, Warren, Atkinson, Bensch, Graves, Jhala, Peirce, Rahmani, Fonseca and Fleischer2006) |
| Ven_11 Haem | AF465568 | Ricklefs and Fallon (Reference Ricklefs and Fallon2002) |
| Ven_13 Haem | HM222483 | Ricklefs and Outlaw (Reference Ricklefs and Outlaw2010) |
| Ven_14 Haem | GQ395647 | Levin et al. (Reference Levin, Outlaw, Hernan Vargas and Parker2009) |
| Ven_15 Plas | GQ395654 | Levin et al. (Reference Levin, Outlaw, Hernan Vargas and Parker2009) |
| Ven_17 Plas | EU627831 | Ishak et al. (Reference Ishak, Dumbacher, Anderson, Keane, Valkiūnas, Haig, Tell and Sehgal2008) |
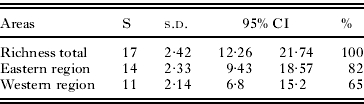
Parasite diversity was higher in the eastern region (82% of the combined diversity) than in the western region (65%), but the differences were not statistically significant (P>0·05) (Table 3). The rarefaction curve and Mao Tau estimator indicated that likely we have not sampled all of the lineages expected to occur in 2 regions in arid zones of Venezuela, suggesting the need for additional sampling (Fig. 3).

Fig. 3. Mao Tau species accumulation curves for the parasite lineages of 2 regions. (a) Total richness of parasite lineages, (--) is the 95% CI and (−) is Total richness. (b) Richness of parasite lineages in the eastern and western regions separately (the 95% confidence intervals for the two regions overlap; see Table 3).
Table 3. Parasite lineage richness as estimated by Mao Tau (S) in Western and Eastern regions in arid zones of Venezuela
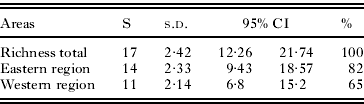
| Areas | S | s.d. | 95% CI | % | |
| Richness total | 17 | 2·42 | 12·26 | 21·74 | 100 |
| Eastern region | 14 | 2·33 | 9·43 | 18·57 | 82 |
| Western region | 11 | 2·14 | 6·8 | 15·2 | 65 |
The Table 4 lists the parasite lineages and host species and families parasitized.
Table 4. Parasite lineages and host species in arid northern Venezuela

| Parasite lineages | Parasite lineage names used previously (see Table 2 for GenBank numbers) | Haemosporidian genus | Host family | Host species |
| Ven01 | Plasmodium sp. haplotype 64 | Plasmodium sp | Mimidae | Mimus gilvus |
| Ven02 | Haemoproteus sp | Cardinalidae | Cardinalis phoeniceus | |
| Icteridae | Icterus nigrogularis | |||
| Picidae | Melanerpes rubricapillus | |||
| Psittacidae | Aratinga pertinax | |||
| Furnariidae | Xiphorhynchus picus | |||
| Thamnophilidae | Sakesphorus canadensis | |||
| Cardinalidae | Saltator coerulescens | |||
| Thraupidae | Tiaris bicolor | |||
| Thraupis glaucocolpa | ||||
| Ven03 | Haemoproteus sp. LA07GD29 | Haemoproteus sp | Icteridae | Icterus nigrogularis |
| Ven04 | Haemoproteus sp | Mimidae | Mimus gilvus | |
| Psittacidae | Aratinga pertinax | |||
| Ven05 | Haemoproteus sp | Mimidae | Mimus gilvus | |
| Ven06 | Haemoproteus sp | Thraupidae | Coryphospingus pileatus | |
| Tiaris bicolor | ||||
| Ven07 | Plasmodium sp. P11 | Plasmodium sp | Tyrannidae | Sublegatus arenarum |
| Ven08 | Plasmodium sp | Cardinalidae | Cardinalis phoeniceus | |
| Thraupidae | Thraupis glaucocolpa | |||
| Ven09 | Plasmodium sp | Thraupidae | Thraupis glaucocolpa | |
| Icteridae | Icterus nigrogularis | |||
| Ven10 | Plasmodium sp | Cardinalidae | Cardinalis phoeniceus | |
| Ven11 | Haemoproteus sp. haplotype 7 | Haemoproteus sp | Thraupidae | Tiaris bicolor |
| Ven12 | Haemoproteus sp | Cardinalidae | Cardinalis phoeniceus | |
| Ven13 | Haemoproteus sp. YU2MEX510 | Haemoproteus sp | Mimidae | Mimus gilvus |
| Furnariidae | Xiphorhynchus picus | |||
| Ven14 | Haemoproteus sp. GD1GD24 | Haemoproteus sp | Furnariidae | Xiphorhynchus picus |
| Ven15 | Plasmodium sp. JA7J725 | Plasmodium sp | Furnariidae | Xiphorhynchus picus |
| Ven16 | Haemoproteus sp | Cardinalidae | Saltator orenocensis | |
| Thamnophilidae | Formicivora intermedia | |||
| Picidae | Melanerpes rubricapillus | |||
| Ven17 | Plasmodium sp. GHOW3489 | Plasmodium sp | Tyrannidae | Sublegatus arenarum |
DISCUSSION
In this study we observed high prevalence (41%) of malaria parasites in arid zones of coastal northern Venezuela, representing 17 mitochondrial lineages: 7 of Plasmodium and 10 of Haemoproteus. Of those lineages, 8 have been described in other studies, while 9 lineages are described for the first time in this study. We found some lineages in northern Venezuela that also were observed in different localities in the Americas. However, other lineages, including some that are relatively common (e.g. Ven05 and Ven06), have not been reported from elsewhere. The prevalence and diversity of parasite lineages did not differ between the isolated eastern and western regions of arid coastal habitat in Venezuela.
Other studies in Neotropical regions that used molecular analysis have reported high prevalence values consistent with this study (Belo et al. Reference Belo, Passos, Junior, Goulart, Sherlock and Braga2009; Santiago-Alarcon et al. Reference Santiago-Alarcon, Outlaw, Ricklefs and Parker2010; Belo et al. Reference Belo, Pinheiro, Reis, Ricklefs and Braga2011). In contrast, several studies that have screened haematozoan infections by scanning blood smears have typically reported low overall prevalence in the Neotropics (approximately 10%), significantly less than in any other zoogeographical region (Greiner et al. Reference Greiner, Bennett, White and Coombs1975; Winchell, Reference Winchell1978; White et al. Reference White, Greiner, Bennett and Herman1978; Bennett et al. Reference Bennett, Earle, Peirce, Huchzermeyer and Squires-Parsons1991; Valkiūnas et al. Reference Valkiūnas, Salaman and Iezhova2003; Rodriguez and Matta, Reference Rodríguez and Matta2001). More studies of blood parasites in the Neotropical region, directly comparing association PCR and microscopy for diagnosis, are required to fully characterize the distribution of haemosporidian parasites in this region.
The higher occurrence of Haemoproteus infections compared to Plasmodium infections in the arid environments considered in this study could be due to the relative abundance of vectors of Haemoproteus in this region. Biting midges (Ceratopogonidae) and mosquitoes (Culicidae), the primary vectors known for Haemoproteus spp. and Plasmodium spp., respectively, are common in the Neotropics (Garnham, Reference Garnham1966; Linley, Reference Linley1985) but their relative abundance in the arid coastal zones of northern Venezuela is not known.
Parasite prevalence did not vary in relation to time of year in this study. Reproduction is highly seasonal in arid northern Venezuela, and the increased stress of reproduction has been shown to depress immune defences and lead to increased parasite prevalence, particularly in females (Møller and Saino, Reference Møller and Saino1994; Weatherhead et al. Reference Weatherhead, Metz, Bennett and Irwin1993; Zuk, Reference Zuk1996; Zuk and McKean, Reference Zuk and McKean1996; Hughes and Randolph, Reference Hughes and Randolph2001). However, blood parasite prevalence in females and males of the two most abundant species in this study did not differ, consistent with the results of a similar study in North America (Ricklefs et al. Reference Ricklefs, Swanson, Fallon, Martinez, Scheuerlein and Latta2005). Based on 33 studies of blood smears from Europe and North America, McCurdy et al. (Reference McCurdy, Shutler, Mullie and Forbes1998) also failed to find a significant difference in parasitism between the sexes. Recently, studies in blue tits (Cyanistes caeruleus) demonstrated differences in parasite prevalence related to host sex depending on the species of Plasmodium. Male and female blue tits differed in prevalence of P. circumflexum (Lachish et al. Reference Lachish, Knowles, Alves, Wood and Sheldon2011), but not other Plasmodium lineages (Szöllosi et al. Reference Szöllosi, Cichoń, Eens, Hasselquist, Kempenaers, Merino, Nilsson, Rosivall, Rytkönen, Török, Wood and Garamszegi2011).
Prevalence of haemoparasites has been found to vary over the annual cycle and between years (Bennett and Lopes, Reference Bennett and Lopes1980; Woodworth-Lynas et al. Reference Woodworth-Lynas, Caines and Bennet1989). Seasonal differences in the prevalence of haemoparasites occur more frequently in temperate regions (Kirkpatrick and Suthers, Reference Kirkpatrick and Suthers1988, Weatherhead and Bennett, Reference Weatherhead and Bennett1992; Hatchwell et al. Reference Hatchwell, Wood, Anwar and Perrins2000), where climate seasonality is more pronounced, limiting the transmission of the parasites to the warm months of the year (Atkinson, Reference Atkinson1988). However, one study of haematozoan prevalence throughout the year in a seasonally dry forest in Puerto Rico found no significant seasonal variation in the prevalence of infection or predominant parasite lineages (Fallon et al. Reference Fallon, Ricklefs, Latta and Bermingham2004). We also did not find seasonal variation in parasite prevalence in this study. Because the prevalence of different parasite species can vary individually in response to environmental and host factors (e.g. Lachish et al. Reference Lachish, Knowles, Alves, Wood and Sheldon2011), it is important to consider abiotic factors (climate and habitat change), biotic factors (age/sex, host species, population density), and vectors, as well as parasite species, when characterizing parasite prevalence.
In this study, we found 17 parasite lineages in arid zones, of which 9 (3 Plasmodium/6 Haemoproteus) had not been described previously. This is the first study on avian malaria in arid environments in South America, and it is important because it reports the high parasite prevalence and parasite diversity present in this area. Temperature and moisture are fundamental determinants of the distribution and abundance of most vector species. Among mosquito species described in this region of Venezuela (Heinemann and Belkin, Reference Heinemann and Belkin1978), Culex quinquefasciatus and Anopheles strode are likely avian malaria vectors (Valkiūnas, Reference Valkiūnas2005). The high parasite prevalence might be associated with anomalous rainfall, which might have promoted vector breeding and survival. In Botswana, Africa, more than two thirds of the variability observed between years in human malaria incidence during January–May could be explained by variation in rainfall during December–February (Thomson et al. Reference Thomson, Mason and Phindela2005). In Venezuela there are reports of anomalous rainfall, such as a torrential rainfall during December 1999, described by Lyon (Reference Lyon2003) that resulted in devastating floods and landslides along the northern coast of Venezuela. These events that occurred in an area with a predominantly dry climate, took place during what is regionally the dry season, and were preceded by unusually heavy seasonal rainfall. More studies in arid zones must be undertaken to understand the parasite-vector-host relationships in these areas.
The common lineages of Haemoproteus and Plasmodium occurred in both the eastern and western portions of the arid Venezuelan coast. Geographically widespread birds can make the parasite community homogeneous across the arid regions. In this area, habitat degradation occurs at a very local scale and does not promote contact between eastern and western regions, because humid regions between sampling areas act as barriers for arid specialist birds. However, only 2 parasite lineages (Ven01 and Ven13) appeared to be partitioned between the eastern and western regions in a single host species, Mimus gilvus, which is present throughout northern and central Venezuela and uses a variety of arid and mesic habitats. The partitioning might be a sampling effect as we recovered only 5 examples of each of these lineages. Other, more common parasite lineages were distributed across the region.
Some lineages found in this study in northern Venezuela have been observed in different localities in the Americas. Among Haemoproteus lineages previously reported from other species and areas, Ven03 has been recovered from other Icterus spp. in Jamaica and the Yucatan Peninsula of Mexico. Ven04 differs by a single nucleotide from a lineage found in several endemic Mimidae in the Lesser Antilles and in the catbird Dumetella carolinensis in eastern North America. Ven11 had already been described in another study of Tiaris bicolor in Venezuela (Ricklefs and Fallon, Reference Ricklefs and Fallon2002; their lineage H07). Ven13 is also common in M. gilvus in the Mexican state of Yucatan. Ven14 is an abundant parasite of the bananaquit Coereba flaveola throughout the West Indies. Ven16 was found as single infections in 4 species in this study, but also in the flycatcher Hemitriccus margaritaceiventer in Tocantins State, Brazil (Belo et al. Reference Belo, Pinheiro, Reis, Ricklefs and Braga2011, HQ287536).
Four of the 7 Plasmodium lineages observed in this study also occur in other regions. Ven01 infecting Mimus gilvus in our study was obtained from Vireo griseus in Missouri, USA, but has also been recovered from a variety of species in several locations around the Caribbean Basin. Ven07 was observed in this study infecting Sublegatus arenarum, and Beadell et al. (Reference Beadell, Ishtiaq, Covas, Melo, Warren, Atkinson, Bensch, Graves, Jhala, Peirce, Rahmani, Fonseca and Fleischer2006) recovered this parasite lineage from Troglodytes aedon in Uruguay; this lineage is also frequently encountered in a variety of species in the Caribbean Basin. We obtained Ven15 from Xiphorhynchus picus, but it has also been recorded in North Carolina infecting a captive Great Horned Owl Bubo virginianus as well as in many species in the Caribbean Basin and North America. Ven17 was detected in Bubo virginianus in northern California.
In spite of the widespread distributions of some of the parasites found in northern Venezuela, several of the lineages, including some that are relatively common (e.g. Ven05 and Ven06) have not been reported from elsewhere. Clearly, additional studies are needed to characterize the host and geographical distribution of avian malaria parasite lineages, which will provide a foundation for a better understanding of the influence of landscape, vector abundance and diversity, and host identity on haemosporidian parasite diversity and prevalence.
ACKNOWLEDGEMENTS
Methods used for bird capture and blood collection were approved by the Institutional Animal Care and Use Committee at UMSL (Protocol 03-22). Bird surveys and collection of blood samples were conducted under permits issued by Inparques (N° 087) and the Dirección de Fauna at the Ministerio del Ambiente in Venezuela (N° PAA-035.2005). Permits needed for exportation of blood samples, and to conduct genetic analyses, were issued by the Oficina de Permisiones (N° 01-03-03-1298, CITES N° 1705) and the Oficina de Biodiversidad (Oficio N° 41-0262) at the Ministerio del Ambiente in Venezuela, respectively.
FINANCIAL SUPPORT
Laboratory work was supported by funds from the Curators of the University of Missouri to R. E. Ricklefs. The CNPq granted a scholarship to Ms. Nayara O. Belo.