INTRODUCTION
Fasciolosis, an infection by the liver flukes Fasciola hepatica Linn. 1758 or Fasciola gigantica Cobbold, 1856, is the major trematode infection of ruminants and a significant zoonosis. The disease has been increasing its range over the past decade (Mas-Coma et al. Reference Mas-Coma, Bargues and Valero2005) and it has been proposed that a contributing factor in this increase has been the environmental changes associated with a warming climate (Mitchell, Reference Mitchell2002) that has promoted an expansion of populations of the lymnaeid snails that provide the liver fluke with its intermediate host. The potential severity of this situation has been compounded by the selection of anthelmintic-resistant isolates of F. hepatica (Wolstenholme et al. Reference Wolstenholme, Fairweather, Prichard, von Samson-Himmelstjerna and Sangster2004).
In temperate regions, F. hepatica is the more common cause of fasciolosis than F. gigantica whereas in tropical regions of Eurasia, the reverse is generally true. This is most likely associated with the occurrence of permissive lymnaeid species. In Europe for example, F. hepatica utilizes Lymnaea (Galba) truncatula (Müller, 1774) as its intermediate host whilst F. gigantica in Africa uses Lymnaea (Radix) natalensis (Krauss, 1848). In some locations (Japan, Korea, China, and Iran) molecular and morphometric studies (Itagaki and Tsutsumi, Reference Itagaki and Tsutsumi1998; Agatsuma et al. Reference Agatsuma, Arakawa, Iwagami, Honzako, Cahyaningsih, Kang and Hong2000; Huang et al. Reference Huang, He, Wang and Zhu2004; Ashrafi et al. Reference Ashrafi, Valero, Panova, Periago, Massoud and Mas-Coma2006) have shown that intermediate forms of the liver fluke can be found, suggesting that the 2 species may hybridize. Such hybrids might be better adapted to the changing environmental conditions associated with climate change and thus would enjoy a selective advantage (Seehausen, Reference Seehausen2004). This potential increase in ‘fitness’ could be countered by the need to be able to infect and multiply in the local lymnaeid species. The efficiency with which different isolates of F. hepatica reproduce in their intermediate host has been shown to vary greatly (Walker et al. Reference Walker, Fletcher, Hoey, Fairweather and Trudgett2006) and infrapopulations of F. hepatica can be genetically very diverse (Walker et al. Reference Walker, Prodöhl, Fletcher, Hanna, Kantzoura, Hoey and Trudgett2007) – in contrast to local populations of lymnaeids which are thought to be largely clonal or have low genetic diversity (Stothard et al. Reference Stothard, Bremond, Andriamaro, Loxton, Sellin and Rollinson2000; Trouvé et al. Reference Trouvé, Degen, Renaud and Goudet2003). It may be assumed that those F. hepatica/F. gigantica hybrids which may have arisen are likely to be the product of a relatively rare event such that individuals within the founding population will be relatively homogenous, lacking populational genetic diversity. The establishment of intermediate/hybrid fasciolids requires the presence of a permissive intermediate host. Whilst L. natalensis does not seem to be capable of supporting the asexual multiplicative stages of F. hepatica (Boray, Reference Boray, Gaafar, Howard and Marsh1985), there have been conflicting reports in the literature concerning the ability of L. truncatula to serve as an intermediate host for F. gigantica (Spithill and Dalton, Reference Spithill and Dalton1998). More recently Dar and colleagues have demonstrated efficient production of F. gigantica cercariae from L. truncatula in the laboratory and ascribed the apparent non-permissiveness of the F. gigantica/L. truncatula combination reported by others to allopatric differences between the parasite and its host (Dar et al. Reference Dar, Rondelaud and Dreyfuss2004).
The topography of the Southern Highlands of Tanzania provides an area in which the interaction between F. hepatica, F. gigantica and their hosts may be studied. The provinces of Mbeya and Iringa cover an area of approximately 100 000 square kilometres with altitudes varying from 500 to 3000 metres. Fasciolosis is a significant problem (Hammond, Reference Hammond1965; Botcher, Reference Botcher1967; Mahlau, Reference Mahlau1970; Ecmovic and Mahlau Reference Ecmovic and Mahlau1973; Msanga, Reference Msanga1985; Keyyu et al. Reference Keyyu, Kassuku, Kyvsgaard and Monrad2005) with prevalences almost universal in some areas. Elsewhere in East Africa, F. gigantica is generally the causative helminth but there have been reports of F. hepatica in cattle from highland regions of Kenya, Ethiopia, Uganda and Zaire (Ogambo-Ongama, Reference Ogambo-Ongama1969, Reference Ogambo-Ongoma1972; Yilma and Malone, Reference Yilma and Malone1998; Malone et al. Reference Malone, Gimmes, Hansen, Yilma, Slingenberg, Snijders, Nachtergaele and Ataman1998). In Ethiopia it has been shown that there is a marked difference in the distribution of Fasciola species according to altitude with F. gigantica dominating at altitudes below 1800 m whilst F. hepatica is limited to altitudes above 1200 m. Much of the data relating to fasciolosis in East Africa was obtained before the introduction of molecular methods capable of distinguishing between F. hepatica, F. gigantica and possible hybrids (Itagaki et al. Reference Itagaki and Tsutsumi1998) or the development of molecular markers for the speciation of lymnaeid snails. In this study we have collected samples of lymnaeid snails and flukes from cattle raised within a range of altitudes in the Southern Highlands of Tanzania. These have been initially identified using morphometric data followed by molecular techniques in order to provide answers to the following questions. What are the intermediate hosts for Fasciola spp. at different altitudes? Is F. hepatica found at high altitude in East Africa? Is there any evidence of F. hepatica/F. gigantica hybridization in our samples?
MATERIALS AND METHODS
Snail survey
The freshwater snail survey was carried out between 2001 and 2003 and sample sites were revisited in 2006. The sampling sites (Fig. 1 and Table 1) were within the Southern Highlands of Tanzania, ranging from Iringa Town, Iringa Region to the Kitulo Plateau of the Mbeya Region. These sample sites range in altitude from 500 m to 2850 m above sea level. Snails were collected from several habitats consisting of streams, small ponds, rivers and swamps. Each habitat was visited on at least 3 occasions: at the beginning of the rainy season (November–December), at the end of the rainy season (May–June), and during the dry season (August–September). In places where snails were found the perimeter of the ponds was sampled at metre intervals and, where possible, the bottoms of shallow ponds were sampled at random points. Between 100 and 200 individuals were collected, and the type of habitat and geo-reference information was recorded. All snails were classified in the field using appropriate keys and field guides (Kristensen, Reference Kristensen1987).

Fig. 1. Map of sampling sites at which lymnaeid snails were found. Locations numbered as in Table 1
Table 1. Sampling sites with altitude
(Numbers in parentheses indicate location on map.)

* Lymnaea (Radix) natalensis present.
** Lymnaea (Galba) truncatula present.
Identification of digenean larvae harboured within snails
Snails were screened for trematode infections by placing each into a 10 ml glass beaker with 5 ml of pond water and placed under strong artificial light for between 4 and 6 h to stimulate them to shed cercariae. Digenean larvae species were identified using the cercarial identification key as supplied within the African freshwater snails field guide as described previously by Frandsen and Christensen (Reference Frandsen and Christensen1984).
DNA extraction and storage
Snails were initially washed in distilled water and then stored in 99% ethanol. DNA extractions were performed using a Nucleon® Genomic DNA Extraction Kit (Tepnel Life Sciences) as per the manufacturer's instruction. Extracted DNA was resuspended in 50 μl of Tris-EDTA buffer. Only lymnaeid snails collected in June 2006 were subjected to the later molecular investigation.
Molecular identification of field-collected snail species
Field-based identification of lymnaeid snail species was confirmed using available phylogenetic markers (Stothard et al. Reference Stothard, Bremond, Andriamaro, Loxton, Sellin and Rollinson2000) based on sequence data of Bargues and Mas-Coma (Reference Bargues and Mas-Coma1997) to amplify 18S rDNA nucleotide sequences coding for the variable V1 and V2 regions within the 18S helical 6 and E10-1 structures. Primers were as used by Stothard et al. (Reference Stothard, Bremond, Andriamaro, Loxton, Sellin and Rollinson2000) except the GC clamp region, placed on the 5′ end for subsequent denaturing gradient gel electrophoresis was removed. Amplification reactions were performed in a total reaction volume of 25 μl containing: 100 mm Tris-HCL, pH 8·8, 1·5 mm MgCl2, 0·2 mm dNTPs, 10 pmols of each primer, 50 ng genomic DNA and 1·0 U Taq polymerase (Invitrogen). After optimization, the following thermal cycling conditions were adopted: an initial denaturation of 94°C for 2 min then 40 cycles of 94°C for 1 min, 68°C for 1 min, 72°C for 1 min, then a final extension of 72°C for 10 min before cooling to 4°C. PCR products from 16 to 24 individual snails from each sampling site were cleaned (GenElute™ PCR Clean-up Kit; Sigma) and then commercially sequenced in both directions by Macrogen Inc. Resultant sequences were assembled and aligned against those from 61 other gastropods downloaded from GenBank using Clustal W (Thompson et al. Reference Thompson, Higgins and Gibson1994) within BioEdit (Hall, Reference Hall1999). To provide support for the morphological and molecular identification of snail species, phylogenetic analysis was carried out within PAUP (Swofford, Reference Swofford1991) using parsimony. Group support was provided by 1000 bootstrap replicates.
Fasciolid infections within lymnaeid snails
Lymnaeids collected in the field were screened for infection with fasciolids with the aim of ascertaining which species, F. gigantica or F. hepatica, was present. Markers available in the scientific press were employed to screen snails for F. hepatica (Cucher et al. Reference Cucher, Carnevale, Prepelitchi, Labbé and Wisnivesky-Coli2006) and F. gigantica (Mostafa et al. Reference Mostafa, Taha and Ramadan2003; Velusamy et al. Reference Velusamy, Singh and Raina2004) infections. Fasciolid species were confirmed by analysis of the 618 bp sequence of the 28S rRNA gene as described by Marcilla et al. (Reference Marcilla, Bargues and Mas-Coma2002).
Morphology and size of eggs and adult liver fluke determination from the high altitude zone
Faecal samples were randomly collected from 92 out of 1000 dairy cattle at Kitulo dairy farm. The faecal samples were transported under cool conditions to the laboratory at Sokoine University, Morogoro. At the laboratory the detection of Fasciola spp. eggs was carried out using the Malachite green sedimentation technique described previously by Dinnik and Dinnik (Reference Dinnik and Dinnik1959). The eggs of all positive animals were mixed and 100 eggs were randomly picked with a pipette; their width and length were measured using a calibrated compound microscope. From the same farm, 16 adult flukes were collected from the bile duct of a Friesian bull calf (approx. 12 months old) that had been identified as harbouring mature flukes from analysis of faecal samples, as previously described. The flukes were preserved in 99% ethanol. At the laboratory, the size of the flukes (length and width) was recorded.
Morphology and size of eggs and adult liver fluke determination from the low altitude zone
In the low altitude zone, lengths and widths of a total 100 eggs of liver fluke randomly drawn from a mixture of gallbladder contents of 12 infected cattle slaughtered at Iringa abattoir were measured. In the same low altitude zone lengths and widths of 50 and 48 adult flukes from 10 and 14 cattle slaughtered at Mazizini (Dar es Salaam) and Babati (Manyara) respectively were measured.
Data analysis
Morphological data were analysed by Statistix® windows version 7 statistical software package. Descriptive statistics procedure was used to summarize the data in frequency distribution and histogram charts. T-test procedures were used to compare the sizes of the eggs/adults between the two zones.
Molecular identification of fasciolids using PCR and PCR-RFLP analysis
Fasciolids were detected in snails using PCR primers reported to be species specific (Mostafa et al. Reference Mostafa, Taha and Ramadan2003; Velusamy et al. Reference Velusamy, Singh and Raina2004; Cucher et al. Reference Cucher, Carnevale, Prepelitchi, Labbé and Wisnivesky-Coli2006). Species identification of adult fasciolids collected was undertaken by employing the PCR-RFLP protocol designed by Marcilla and colleagues (Reference Marcilla, Bargues and Mas-Coma2002).
Mitochondrial DNA analysis of adult fasciolids
Using the region of the mitochondrial genome identified as being most informative (Walker et al. Reference Walker, Prodöhl, Fletcher, Hanna, Kantzoura, Hoey and Trudgett2007) we have produced 1250 bp amplicons of representative flukes from each population. These were commercially sequenced in forward and reverse directions and the resulting sequences submitted for analysis using BLAST (Altschul et al. Reference Altschul, Madden, Schaffer, Zhang, Zhang, Miller and Lipman1997) software.
RESULTS
Distribution of lymnaeids
With regard to altitude, the first lymnaeid snail species (L. natalensis by morphology) was found at a location with an altitude of 1090 m having been absent at one lower sample site at 517 m (Table 1). Lymnaea natalensis was found at 15 of the 24 sites visited whereas snails identified as L. truncatula on morphological characteristics were only found at 2 adjacent sites (2712 m and 2720 m) on the Kitulo Plateau. These ponds were approximately 25 m apart, draining into a shared stream. Lymnaea natalensis was not found above an altitude of 1890 m.
Digenean larvae
Both L. natalensis and L. truncatula shed cercariae which were classified as belonging to the Gymnocephalous group indicating the presence of either fasciolid or paramphistome infections within these snails. Lymnaea natalensis was also found to harbour cercariae belonging to Schistosomatidae.
Molecular identification of field-collected snail species
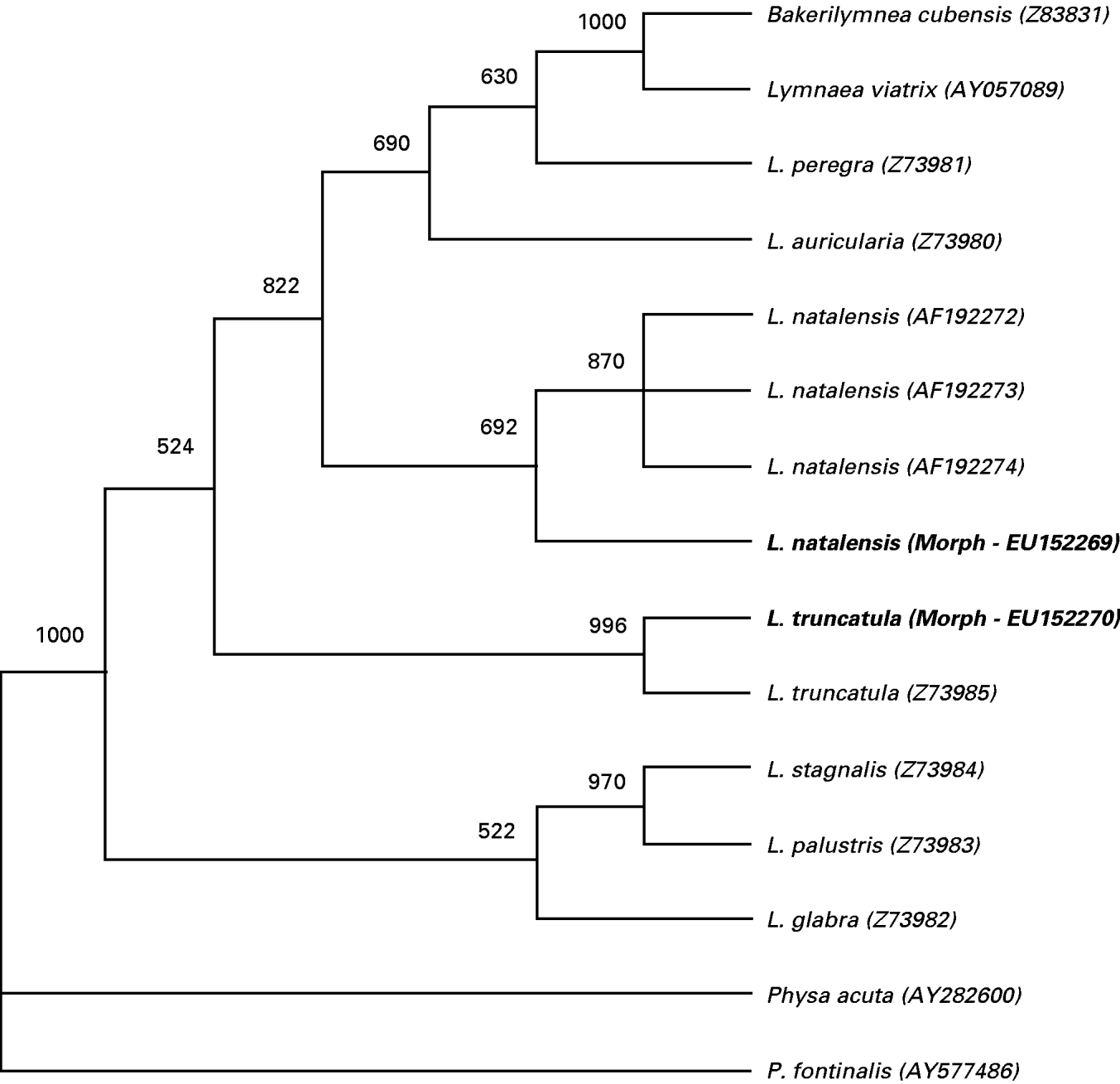
Fig. 2 shows an extract from the phylogenetic analysis illustrating the relationships within the lymnaeids and containing the unknown Tanzanian morphs. The snails from the Kitulo Plateau were clustered with the L. truncatula GenBank sequence (Z73985) with bootstrapping support of 99·6%. The Iringa, Mbeya and Njombe samples were clustered with the L. natalensis GenBank sequences (AF192272-4) with bootstrapping support of 69%. These results confirm our hypothesis with regard to the existence of L. truncatula at Kitulo (GenBank Accession number EU152270) and L. natalensis (GenBank Accession number EU152269) at all sites below this altitude.

Fig. 2. Parsimony majority rule consensus tree based on 318 bp from the variable V1 and V2 regions within the 18S helical 6 and E10-1 structures of the 18S small subunit. Bootstrap support values based on 1000 replicates are shown at nodes. GenBank identification numbers follow species names. AF192272, Lymnaea natalensis from South Africa; AF192273, L. natalensis from Madagascar; AF192274, L. natalensis from Madagascar; Z73985, L. truncatula from Corsica, France.
Identification of fasciolids – morphology
The mean size of the fluke eggs from the low zone was 155 μm (S.E.±9·56 μm) in length by 89·6 μm (S.E. ±8·19 μm) in width with ranges of 130–180 μm whilst that from cattle grazing the Kitulo Plateau was 127 μm (s.e.±7·38 μm) by 63·42 μm (s.e.±4·19 μm) with ranges of 115–150 μm. Welch's approximate t-test gave a value for t of 2·318 with 196 degrees of freedom indicating that they were from significantly different populations (P=0·0215). The flukes from the low altitude zone were larger than those from Kitulo and their mean body length to body width ratio was 2·85 whereas that for the flukes from Kitulo was 1·59. Statistical analysis of the data using Welch's approximate ‘t’-test gave a value of t of 4·226 (N=107) which gives a probability of P<0·0001 that they were drawn from different populations.
Molecular techniques – fasciolid infections within lymnaeid snails
Using PCR primers to detect the presence of fasciolid infections a representative sample of snails from 4 out of the 5 lymnaeid populations suggested the presence of these trematodes in both L. truncatula and L. natalensis populations. However, when appropriate controls were employed it was evident that the molecular approach used was not able to discriminate between F. hepatica and F. gigantica species (results not shown). PCR-RFLP analysis of 28S rDNA from adult fasciolids produced fragments that were diagnostic for F. hepatica and F. gigantica and indicated that the flukes in all of the infected snails sampled from below 2000 m were F. gigantica. The restriction pattern with the PCR products from the snails from the Kitulo plateau was consistent with F. hepatica (Fig. 3).

Fig. 3. Identification of fasciolid species by PCR-RFLP analysis of a 618 bp fragment amplified from 28S rRNA gene and digested with AvaII (3a – lanes 1–3 Fasciola hepatica, lanes 4–8 F. gigantica, lane 9 1 Kb DNA ladder) and DraII (3b – lanes 1–3 F. hepatica, lanes 4–8 F. gigantica, lane 9 1 Kb DNA ladder)) (Marcilla et al. Reference Marcilla, Bargues and Mas-Coma2002).
Molecular techniques – BLAST analysis of fasciolid mtDNA
The sequences from the putative F. hepatica flukes (Accession number EU282862) gave 97% homology with the appropriate region of the complete mitochondrial sequence of F. hepatica (Accession number AF216697) whereas those from the putative F. gigantica flukes (Accession numbers EU282859, EU282860, EU282861) gave 90% sequence homology with the appropriate region of the complete mitochondrial sequence of F. hepatica. (Sequences for F. gigantica homologous to this region of its mtDNA are not available in GenBank.) These were the highest scoring homologues in each case.
DISCUSSION
The present findings validate earlier reports that L. truncatula exists in the East African tropical highlands including Tanzania (Dinnik and Dinnik, Reference Dinnik and Dinnik1959; Kendall, Reference Kendall1965). The occurrence of this temperate snail in tropical highlands may be either the result of inadvertent introduction from other areas or the presence of a relict native population. Migratory birds from temperate countries have been implicated as possible vectors for freshwater pulmonate snails to such tropical regions (Brown, Reference Brown1994). The Kitulo Plateau provides breeding grounds for several species of migratory birds from temperate regions such as the Blue Swallow (Hirundo xantholopus Sundevall, 1850), Pygmy Kingfisher (Ceyx picta Boddaert, 1783), and Denham's Bustard (Neotis denhami Children and Vigors, 1826). The alternative possibility, that the L. truncatula populations of the Kitulo Plateau are an indigenous species, is a question which could be addressed by further sampling and the development and deployment of highly informative phylogenetic markers for this species (Bargues and Mas-Coma, Reference Bargues and Mas-Coma1997; Stothard et al. Reference Stothard, Bremond, Andriamaro, Loxton, Sellin and Rollinson2000; Bargues et al. Reference Bargues, Vigo, Horak, Dvorak, Patzner, Pointier, Jackiewicz, Meier-Brook and Mas-Coma2001). Lymnaea natalensis 18S rDNA sequences were monomorphical across all populations sampled in this study although limited interspecific variation was detected. These changes occurred in the variable V2 region of the predicted RNA secondary structure (E10-1 helix) as reported by Stothard et al. (Reference Stothard, Bremond, Andriamaro, Loxton, Sellin and Rollinson2000) for this species of snail. Likewise, the observed variation at the species level between L. natalensis and L. truncatula within the nucleotide sites 233 to 253 supports similar findings by other authors (Bargues and Mas-Coma, Reference Bargues and Mas-Coma1997).
In the present survey, L. truncatula was the only lymnaeid species observed to exist on the Kitulo Plateau at an altitude of approximately 3000 m. Such data support previous findings that this snail species is capable of adaptation to many different environments such as the very high altitudes (over 4000 m) of the Bolivian Altiplano (Jabbour-Zahab et al. Reference Jabbour-Zahab, Pointier, Balzan, Khallayoune and Renaud1997; Mas-Coma et al. Reference Mas-Coma, Funatsu and Bargues2001). The presence of L. truncatula on the Kitulo LMU farm further extends the problem of fasciolosis in domestic ruminants of this area where the presence of L. natalensis is limited by the extreme environmental conditions. Under both natural and experimental conditions, other lymnaeid snails species may be infected with F. hepatica (Soulsby, Reference Soulsby1982) but it is widely accepted that variants of L. truncatula are the fundamental intermediate hosts (Malone, Reference Malone and Boray1994). Studies have also shown that L. natalensis is not a good intermediate host for F. hepatica (Boray, Reference Boray, Gaafar, Howard and Marsh1985). Therefore, as in Ethiopia, it would appear logical that the observed widespread distribution of L. natalensis in the moderately low altitude areas of the Southern Highlands of Tanzania in this present study, is responsible for the serious problem of ruminant fasciolosis due to F. gigantica in this area (Mahlau et al. Reference Mahlau1970; Wilson, Reference Wilson1995; Makundi, Reference Makundi2001) and that L. truncatula is responsible for the recently reported problems with F. hepatica at Kitulo LMU farm (Namuba et al. Reference Namuba, Makundi and Kassuku2003).
The origin of the F. hepatica-like flukes collected on the Kitulo plateau is of interest. The farm was originally stocked with Australian merino sheep and, more recently, dairy cattle of (ultimately) European origin suggesting that the flukes may have been imported with the sheep or cattle and became established due to the pre-existing presence of L. truncatula. We are currently comparing mtDNA sequences from these flukes with those from European specimens of F. hepatica in order to determine their provenance.
Although the flukes from the low altitude sample sites were within the range of lengths reported for F. gigantica, comparison with morphological data from Ashrafi and colleagues suggested that both the highland and lowland populations found in Tanzania were F. hepatica-like, in that the body width to length ratios of the populations were within the ranges quoted for F. hepatica in their extensive study (Ashrafi et al. Reference Ashrafi, Valero, Panova, Periago, Massoud and Mas-Coma2006). However, PCR amplification of a partial fragment of the 28S ribosomal DNA gene from the flukes and digestion with Ava I and Dra II gave a more definitive resolution with restriction patterns consistent with the lowland flukes being F. gigantica-like and those from the Kitulo plateau being F. hepatica-like. We did not observe any RFLP patterns indicative of the presence of hybrid Fasciola spp. in either population. The high level of homology seen with the mitochondrial sequences from these flukes confirms their identification. A lower level of homology to the GenBank F. hepatica mitochondrial genome was seen with the lowland flukes, the relevant region of the F. gigantica mt genome is not present in GenBank.
It has been reported that F. gigantica eggs can embryonate and hatch successfully at 17–22°C (Guralp et al. Reference Guralp, Oxcan and Simms1964). In this study F. gigantica was found at relatively high altitudes (<1800 m) where climatic conditions are within that range and comparable to those experienced by F. hepatica in much of Australia, South America and southern Europe. F. hepatica was only found above 2700 m where night temperature may drop to −6°C. There was concordance between the presence of L. truncatula/F. hepatica-like flukes and L. natalensis/F. gigantica-like flukes, suggesting that the determining factor in the distribution of F. hepatica and F. gigantica is not temperature per se but rather the presence of an intermediate host in which each species can reproduce efficiently.
In conclusion, the distribution of F. hepatica and F. gigantica seen in the Tanzanian Southern Highlands supports the hypothesis that the determining factor in the spread and establishment of either of these species is the pre-existing presence of their permissive intermediate host.
This study was supported by a British Society for Parasitology Ann Bishop Award (2006) to S. M. W. and the DeLiver programme of the EU 6th Framework Initiative.