INTRODUCTION
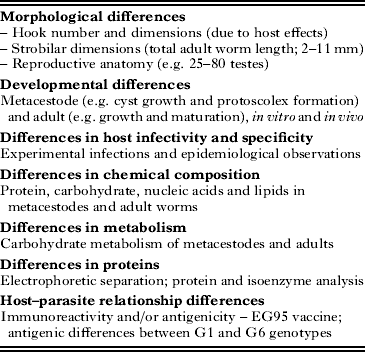
The classification of Echinococcus has long been controversial because of a lack of phenotypic characters, inadequate taxonomic descriptors and lack of evidence for geographical or ecological segregation (reviewed in Thompson et al. Reference Thompson, Lymbery and Constantine1995; Thompson, Reference Thompson, Gillespie and Pearson2001; Thompson and McManus, Reference Thompson and McManus2002). Based mainly on host-parasite specificity characteristics, many species and subspecies of Echinococcus were described originally (Ortlepp, Reference Ortlepp1934; Lopez-Neyra and Soler Planas, Reference Lopez-Neyra and Soler Planas1943; Williams and Sweatman, Reference Williams and Sweatman1963; Verster, Reference Verster1965). However, most of these taxa were regarded as synonyms for Echinococcus granulosus (Rausch, Reference Rausch1967), and subsequent taxonomic revisions recognised only four valid species: E. granulosus, E. multilocularis, E. oligarthrus and E. vogeli (Rausch and Bernstein, Reference Rausch and Bernstein1972). Opposing views on the breeding system and population genetics of Echinococcus further complicated discussion on the taxonomy (Thompson and Lymbery, Reference Thompson and Lymbery1988). Over the past 50 years, however, field and laboratory observations have revealed considerable phenotypic variability among isolates of Echinococcus. This variation has largely been observed in E. granulosus, and between isolates of the parasite from different species of intermediate host (Table 1).
Table 1. Some of the phenotypic variation observed in Echinococcus granulosus (Modified from Thompson and McManus, Reference Thompson and McManus2002)

Note: DNA differences reflect this phenotypic variation.
Based mainly on mitochondrial DNA-based studies, it has been shown that E. granulosus comprises 10 genotypes (G1 to G10), which have been elevated to distinct species, comprising E. granulosus sensu stricto (G1, G2 and G3); E. equinus (G4); and E. ortleppi (G5) and its sister species, E. canadensis (G6, G7, G8, G9, G10) (Nakao et al. Reference Nakao, McManus, Schantz, Craig and Ito2007). Recently, the lion strain has been proposed as another new species, E. felidis positioned as a sister taxon of E. granulosus s.s. (Hüttner et al. Reference Hüttner, Nakao, Wassermann, Siefert, Boomker, Dinkel, Sako, Mackenstedt, Romig and Ito2008). Echinococcus shiquicus, a sister species to E. multilocularis, has been found in Tibet (Xiao et al. Reference Xiao, Qiu, Nakao, Li, Yang, Chen, Schantz, Craig and Ito2005; Reference Xiao, Qiu, Nakao, Li, Yang, Chen, Schantz, Craig and Ito2006). This article provides an overview of some of the phenotypic variation observed in isolates of Echinococcus, why the concept of a ‘strain’ was developed, and how molecular and other data have reinforced the need to revise the taxonomic status so that up to 9 species are now recognised.
PHENOTYPIC VARIATION IN ECHINOCOCCUS
Early physiological and biochemical studies
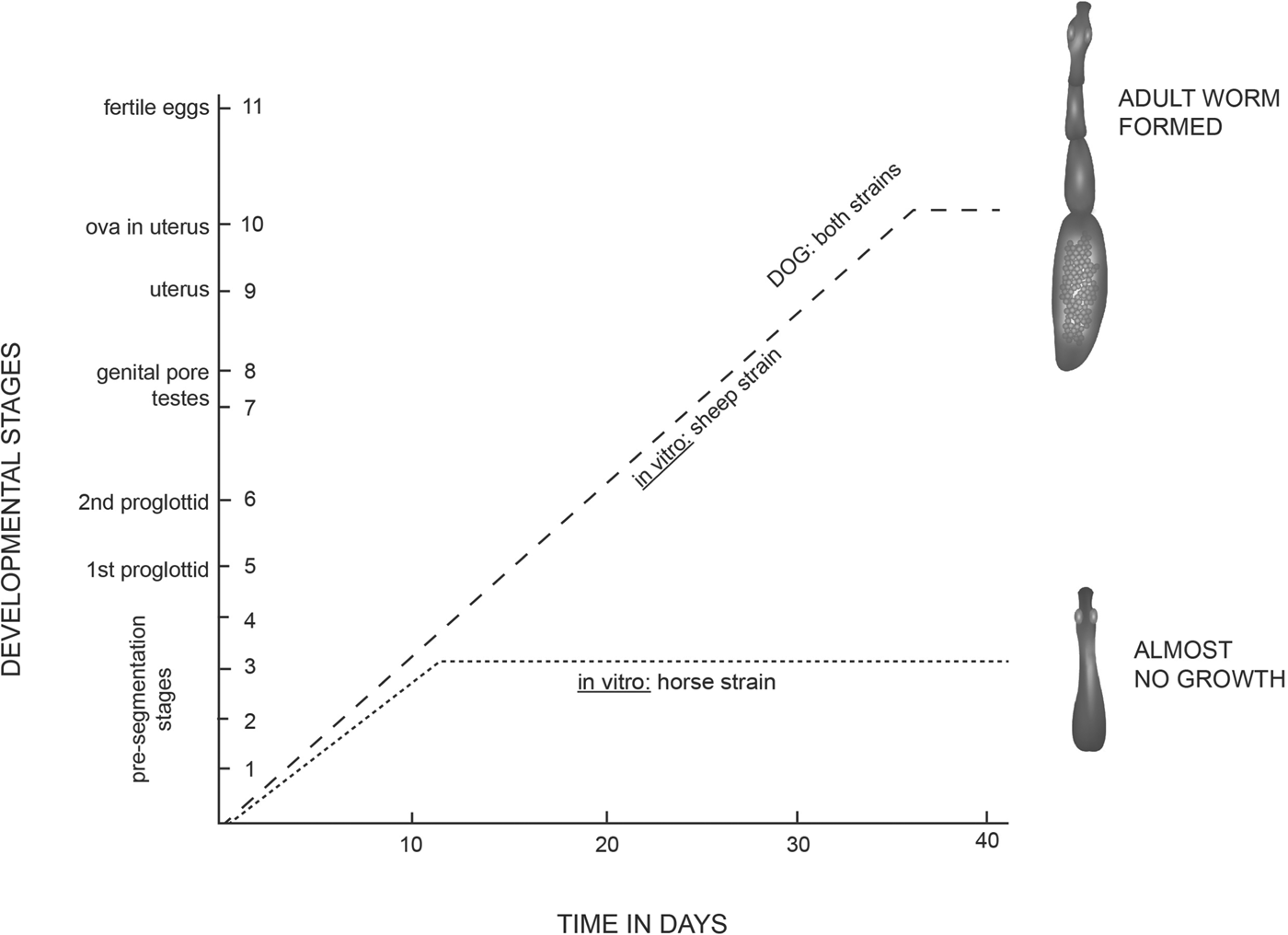
Smyth and Smyth (Reference Smyth and Smyth1964) were the first to point out that the Echinococcus organisms have a mode of reproduction which favours the expression of mutants, with the result that new parasite variants or strains can readily arise. However, the key to our understanding of the phenomenon of strain variation came in the early 1970s with the seminal studies of James Desmond Smyth on the strobilar development of Echinococcus in vitro where he demonstrated that in media which supported the development and maturation of E. granulosus of sheep origin, isolates of the parasite from horses failed to develop or mature (Smyth and Davies, Reference Smyth and Davies1974) (Fig. 1). This was at first attributed to suspected faults in technique or media components but when, after two years of experiments involving some 200 cultures, horse material failed to strobilate, it was realised that this result represented a new phenomenon, and it was concluded that isolates of E. granulosus from horses represented a different ‘strain’ from that of sheep with some unique (possibly nutritional) factor or culture condition for sexual differentiation (Smyth and Davies, Reference Smyth and Davies1974) which even today has not been identified. This appears to be some unusual requirement, for isolates of buffalo, camel, cattle, goat and human origin were subsequently shown to differentiate sexually in the (sheep) in vitro system (Macpherson and Smyth, Reference Macpherson and Smyth1985). In addition to the in vitro growth differences, epidemiological data (Hatch and Smyth, Reference Hatch and Smyth1975; Smyth, Reference Smyth1977) suggested that the horse-dog form may be a different strain with possibly no, or only low infectivity to humans. Indeed, all subsequent studies have reinforced this view.

Fig. 1. Development of the protoscoleces of Echinococcus granulosus of sheep and horse origin under identical conditions in vitro. The sheep strain formed proglottids and grew to sexual maturity; the horse strain failed even to segment and did not develop beyond stage 3. In the most successful cultures, the rate of development in vitro of the sheep strain lagged only a few days behind that in the dog in vivo. (Modified from Smyth and Davies, Reference Smyth and Davies1974).
That sheep and horse hydatids represent different strains was confirmed by the demonstration of biochemical differences between them (McManus and Smyth, Reference McManus and Smyth1978, Reference McManus and Smyth1982). A comparison of the basic biochemical composition and carbohydrate metabolism of protoscolex larvae showed marked differences, providing a striking example of a phenomenon described for other helminths such as Hymenolepis diminuta and Haemonchus contortus. Protoscoleces of the horse strain contained less protein and RNA and more lipid than those of the sheep strain. Under aerobic conditions, the sheep strain used more oxygen and glycogen and produced more succinic and acetic acids but less lactic acid than the horse strain. Anaerobically, there was little difference in glycogen utilisation, but the sheep strain produced less succinic and lactic acids, and more acetic acid and ethanol. In other words, both aerobically and anaerobically, the major end-products of the sheep strain were mainly acetic acid with some succinic acid, whereas those of the horse strain were mainly lactic acid with some succinic acid. A subsequent, wider ranging study carried out on E. granulosus from Kenya involving protoscoleces from five different host species (sheep, goats, camels, cattle and humans) and adults, also indicated metabolic variability (McManus, Reference McManus1981) although the significance of the variability was, at the time, unclear.
In other early work, isoenzyme markers were used to discriminate species and strains of Echinococcus (Le Riche and Sewell, Reference Le Riche and Sewell1978; McManus and Smyth, Reference McManus and Smyth1979; Macpherson and McManus, Reference Macpherson and McManus1982). In one study, extracts of protoscoleces of the horse and sheep strains of E. granulosus and E. multilocularis were compared on the basis of their isoenzyme patterns for 10 enzymes (acid phosphatase, lactate dehydrogenase, malate dehydrogenase, malic enzyme, glucose phosphate isomerase (GPI), phosphoglucomutase (PGM), isocitrate dehydrogenase, adenylate kinase, aldolase and alpha-glycerophosphate dehydrogenase) by means of isoelectric focusing (IEF) in polyacrylamide gels; interspecific and intraspecific differences were apparent in the isoenzyme profiles of all the enzymes except adenylate kinase, whose pattern and activity was identical for both strains of E. granulosus (McManus and Smyth, Reference McManus and Smyth1979).
The isoenzyme patterns for GPI and PGM were later compared by IEF for soluble enzyme extracts from protoscoleces obtained from hydatid cysts of human, camel, cattle, sheep and goat origin in Kenya (Macpherson and McManus, Reference Macpherson and McManus1982). Consistent GPI and PGM isoenzyme patterns were obtained for larvae of human, camel and sheep material, with the camel material exhibiting distinct profiles for both enzymes. Two isoenzyme patterns were evident in the goat material; the more common goat patterns were similar to those of human, cattle and sheep material. The more rare goat patterns were similar to those obtained for the camel samples.
The molecular era and the genetic basis for the observed phenotypic variation in E. granulosus
In the 1980s the new techniques in molecular biology offered great potential for providing a new approach to studies on the taxonomy, genetics and population biology of parasites. The sensitivity and specificity of DNA analysis was ideal for ascertaining the extent of strain variation in E. granulosus. Indeed, the application of molecular tools for characterizing isolates of Echinococcus has had a major impact on our understanding of the population genetics, epidemiology and taxonomy of the parasite and the genetic basis of the phenotypic variation evident.
Having isolated high molecular weight and pure DNA from E. granulosus and E. multilocularis that was cleavable by restriction enzymes (McManus et al. Reference McManus, Knight and Simpson1985), McManus and Simpson (Reference McManus and Simpson1985) used cloned DNA fragments of the ribosomal RNA gene of Schistosoma mansoni to discriminate between isolates of E. granulosus from UK horses and sheep as well as E. granulosus from E. multilocularis used restriction fragment length polymorphism (RFLP) analysis involving a Southern blot hybridization approach. Subsequently, a recombinant plasmid (coded pEG18) with a 2·3 kb DNA fragment unique for E. granulosus was cloned and used, with other cloned probes, in RFLP analysis to independently and reproducibly discriminate between the UK horse and sheep strains of E. granulosus and to characterize a large number of isolates from different host species from various geographical areas (McManus and Rishi, Reference McManus and Rishi1989).
The study did not demonstrate any significant genetic variation within the United Kingdom horse/dog or sheep/dog strains but confirmed the distinctiveness of the two strains shown previously. The sheep/dog strain was shown to be cosmopolitan in its distribution and fertile bovine material originating from the United Kingdom, Kenya, Spain and India conformed to this strain by DNA hybridization. In contrast, cattle isolates from Holland produced markedly different DNA hybridization banding profiles indicating that cattle could harbour more than one strain of E. granulosus. Similarly, it was shown that goats could harbour two different strains of E. granulosus, the sheep/dog strain and a form which infects camels. The analysis showed that the strain of E. granulosus infecting equines in Spain and Ireland was genetically identical to that found in horses in the United Kingdom. There was also a different strain infecting pigs in Poland and Yugoslavia. This pig/dog strain appeared to be very similar genetically to the forms of E. granulosus which use camels and goats as intermediate hosts and was similar, though not identical, to the variant infecting Dutch cattle.
Building on this work, a procedure was developed that linked the polymerase chain reaction (PCR) with RFLP analysis of the ITS-1 region of ribosomal DNA of E. granulosus; this method (PCR-RFLP analysis) was used successfully to study variation in E. granulosus isolates from several regions (Bowles and McManus, Reference Bowles and McManus1993). An example of a study where the procedure proved useful was that of Wachira et al. (Reference Wachira, Bowles, Zeyhle and McManus1993) who examined 208 larval isolates and 40 worm samples of E. granulosus from various hosts in Kenya, confirming the existence of the camel and sheep strains there and showing that the distribution of the camel strain appeared to be restricted to the Turkana region, where camels are kept as livestock. The study also showed that although the life-cycle patterns of the two strains overlap both geographically and in intermediate and definitive hosts, the strains maintain their homogeneous genetic identity.
A modification of the procedure, involving a specific and sensitive PCR/semi-nested PCR system, was later developed by Dinkel et al. (Reference Dinkel, Njoroge, Zimmermann, Wälz, Zeyhle, Elmahdi, Mackenstedt and Romig2004) for the rapid diagnosis of E. granulosus strains in East Africa. Most recently, a multiplex PCR for the simultaneous detection and typing of E. granulosus strains has been developed that has potential for worldwide application in large-scale molecular epidemiological studies on the Echinococcus genus (Boubaker et al. Reference Boubaker, Macchiaroli, Prada, Cucher, Rosenzvit, Ziadinov, Deplazes, Saarma, Babba, Gottstein and Spiliotis2013).
The seminal in vitro culture studies of Smyth provided a platform of understanding for both earlier and subsequent observations on differences in the development and infectivity between isolates of the parasite from various host species in different parts of the world (Thompson and McManus, Reference Thompson and McManus2002). The realisation that variants of E. granulosus develop at different rates in the definitive host has meant that the timing of anthelmintic administration designed to remove worm burdens before patency should be reconsidered. Further, the fact that some variants appear to be poorly infective to humans has resulted in a reappraisal of the public health significance of Echinococcus in areas where such variants are endemic (Thompson and McManus, Reference Thompson and McManus2002).
Given the epidemiological significance of such intraspecific variation in E. granulosus and the international efforts to establish control programmes in different endemic regions, new nomenclature was needed to reflect the phenotypic variability evident between host-derived isolates of E. granulosus (Thompson and McManus, Reference Thompson and McManus2002). Thus, the concept of a ‘strain’, first coined by Smyth, was further developed as a result of the accumulating genomic information to describe variants that differ from other groups of the same species in gene frequencies or DNA sequences, and in one or more characters of actual or potential significance to the epidemiology and control of echinococcosis (Thompson et al. Reference Thompson, Lymbery and Constantine1995; Thompson and McManus, Reference Thompson and McManus2002; McManus and Thompson, Reference McManus and Thompson2003).
Rapidly evolving mitochondrial (mt) sequences have provided a rich source of information for research in evolutionary biology, population genetics and phylogenetics. They have been used extensively in studies of Echinococcus since Bowles et al. (Reference Bowles, Blair and McManus1992) first used the sequence of a region of the mitochondrial cytochrome c oxidase subunit I (CO1; cox1) as a marker of species and strain identity and as a preliminary indication of evolutionary divergence within the genus. Molecular studies, using mainly mtDNA sequences, have now identified ten distinct genotypes (G1–G10) within E. granulosus, a categorisation which follows closely the pattern of strain variation that has emerged based on biological characters (Eckert and Thompson, Reference Eckert and Thompson1997; Thompson and McManus, Reference Thompson and McManus2002; Lavikainen et al. Reference Lavikainen, Lehtinen, Meri, Hirvelä-Koski and Meri2003; McManus and Thompson, Reference McManus and Thompson2003). These genotypes/strains of E. granulosus comprise: sheep strain (G1), Tasmanian sheep strain (G2), buffalo strain (G3), horse strain (G4), cattle strain (G5), camel strain (G6), pig strain (G7), cervid strain (G8), pig/human strain (G9) and Fennoscandian cervid strain (G10).
PHYLOGENETIC ANALYSIS
An invaluable approach for investigating levels of divergence between taxa is by means of phylogenetic trees, a number of which have been constructed for Echinococcus, based on partial and complete mitochondrial DNA and nuclear DNA sequences (Bowles et al. Reference Bowles, Blair and McManus1995; Thompson et al. Reference Thompson, Lymbery and Constantine1995; Le et al. Reference Le, Pearson, Blair, Dai, Zhang and McManus2002; Nakao et al. Reference Nakao, McManus, Schantz, Craig and Ito2007, Reference Nakao, Yanagida, Okamoto, Knapp, Nkouawa, Sako and Ito2010; Moks et al. Reference Moks, Jõgisalu, Valdmann and Saarma2008; Saarma et al. Reference Saarma, Jõgisalu, Moks, Varcasia, Lavikainen, Oksanen, Simsek, Andresiuk, Denegri, González, Ferrer, Gárate, Rinaldi and Maravilla2009; Knapp et al. Reference Knapp, Nakao, Yanagida, Okamoto, Saarma, Lavikainen and Ito2011).
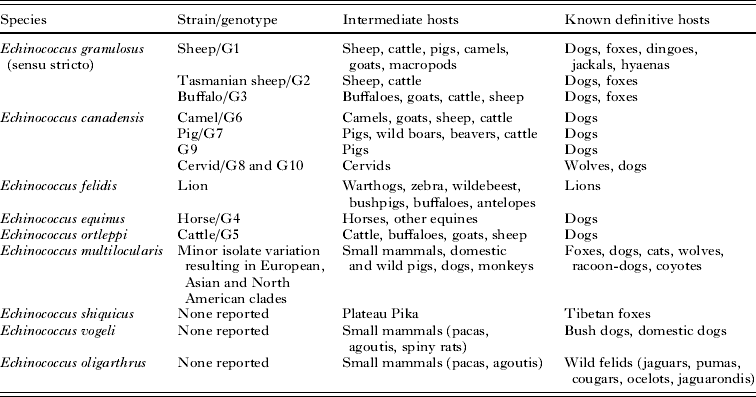
These DNA-based phylogenetic studies have shown an even more pronounced genetic divergence between the ten E. granulosus genotypes than previously recognised and, taking this information and other criteria into account, the taxonomy of Echinococcus has been revised (Thompson and McManus, Reference Thompson and McManus2002; McManus and Thompson, Reference McManus and Thompson2003; Nakao et al. Reference Nakao, McManus, Schantz, Craig and Ito2007; Thompson, Reference Thompson2008; Pednekar et al. Reference Pednekar, Gatne, Thompson and Traub2009; Saarma et al. Reference Saarma, Jõgisalu, Moks, Varcasia, Lavikainen, Oksanen, Simsek, Andresiuk, Denegri, González, Ferrer, Gárate, Rinaldi and Maravilla2009; Knapp et al. Reference Knapp, Nakao, Yanagida, Okamoto, Saarma, Lavikainen and Ito2011) (Table 2). E. granulosus is now considered as a complex consisting of at least four species: E. granulosus s.s. (genotypes G1 to G3), E. equinus (G4) and E. ortleppi (G5), but the species status of genotypes G6 to G10 is still ambiguous (Thompson and McManus, Reference Thompson and McManus2002; McManus and Thompson, Reference McManus and Thompson2003). Taxonomic revision to unify the cervid, camel and pig strains into a single species, E. canadensis, has been suggested (Lavikainen et al. Reference Lavikainen, Lehtinen, Agren and Meri2005; Nakao et al. Reference Nakao, McManus, Schantz, Craig and Ito2007; Moks et al. Reference Moks, Jõgisalu, Valdmann and Saarma2008).
Table 2. Some taxonomic features of the genus Echinococcus

Note: All species, except E. equinus, have been shown to be infective to humans.
DNA sequencing of mitochondrial genes and nuclear protein coding genes, phylogeny construction and morphological studies have identified E. shiquicus as a new sister species to E. multilocularis (Xiao et al. Reference Xiao, Qiu, Nakao, Li, Yang, Chen, Schantz, Craig and Ito2005, Reference Xiao, Qiu, Nakao, Li, Yang, Chen, Schantz, Craig and Ito2006, Nakao et al. Reference Nakao, McManus, Schantz, Craig and Ito2007; Saarma et al. Reference Saarma, Jõgisalu, Moks, Varcasia, Lavikainen, Oksanen, Simsek, Andresiuk, Denegri, González, Ferrer, Gárate, Rinaldi and Maravilla2009; Knapp et al. Reference Knapp, Nakao, Yanagida, Okamoto, Saarma, Lavikainen and Ito2011). The larval form occurs in the plateau pika, Ochotona curzoniae, found in Shiqu County, in the Qinghai-Tibet plateau region of western Sichuan, China. The adult stage has been isolated from the Tibetan fox, Vulpes ferrilata. The metacestode develops into a unilocular cyst mainly in the liver. Its zoonotic transmission potential is presently unknown. Furthermore, recent mitochondrial DNA studies have identified E. felidis as a sister species to E. granulosus s.s. whose adult stage has been isolated from African lions with warthogs acting as intermediate hosts (Hüttner et al. 2008, 2009). There are no data available on the pathogenicity of E. felidis to humans, but its public health impact may be minimal, as lions are largely restricted to national parks and game reserves where there is little human activity. E. felidis may, however, have an impact on pastoralists in East Africa who coexist with wildlife (Moro and Schantz, Reference Moro and Schantz2009).
FINAL COMMENTS
There is remarkable genetic homogeneity within E. multilocularis specimens isolated from different geographical regions based on the analysis of classical coding and non-coding DNA targets although European, Asian and North American clades have been defined (Bowles et al. Reference Bowles, Blair and McManus1992; Haag et al. Reference Haag, Zaha, Araújo and Gottstein1997; Rinder et al. Reference Rinder, Rausch, Takahashi, Kopp, Thomschke and Löscher1997; Yang et al. Reference Yang, Rosenzvit, Zhang, Zhang and McManus2005; Nakao et al. Reference Nakao, Xiao, Okamoto, Yanagida, Sako and Ito2009). In contrast, a number of distinct genotypes of E. granulosus (designated G1-G10) are now recognised, with the genotype cluster G1-G3 (E. granulosus s.s.) and genotype G6 being responsible for the majority of humans infections.
E. granulosus is now considered as a complex consisting of at least four species having differing life-cycle patterns, host specificity, development rates, antigenicity, transmission dynamics, sensitivity to chemotherapeutic agents and pathology with important implications for the design and development of vaccines, diagnostic assays and drugs impacting on the control of hydatid disease (Thompson and McManus, Reference Thompson and McManus2002). This variability has practical relevance when considering the extensive genetic variability recently reported in the complete coding DNA sequence of the EG95 antigen of E. granulosus which may have a direct influence on host specificity and vaccine efficacy.
The oncosphere-expressed EG95 antigen is the basis of a recombinant vaccine developed for use in livestock to prevent infection with E. granulosus (Gauci et al. Reference Gauci, Heath, Chow and Lightowlers2005). The EG95 antigen was originally cloned from the G1 genotype of E. granulosus and the protein has been found to be encoded by members of a small family of related genes in this genotype. Following on from an earlier study, which revealed substantial nucleotide substitutions (encoding amino acid substitutions) for EG95 in the G6/G7 genotypes (Chow et al. Reference Chow, Gauci, Vural, Jenkins, Heath, Rosenzvit, Harandi and Lightowlers2008), a recent study by Alvarez Rojas et al. (Reference Alvarez Rojas, Gauci, Nolan, Harandi and Lightowlers2012), used genomic DNA cloning techniques to characterize seven eg95-related gene fragments from the G6 genotype of E. granulosus. Three proteins appeared to be encoded by these genes. Considerable differences were found between the EG95 related proteins from the G6 genotype compared with the EG95 protein from the G1 genotype. These differences suggest that the EG95-related proteins from the G6 genotype may have different antigenic epitopes compared with the current vaccine antigen. These data have implications for future vaccine design and provide information that would enable a G6 genotype-specific vaccine to be developed against E. granulosus, should this be considered a desirable addition to the available tools for control of cystic echinococcosis transmission.
ACKNOWLEDGEMENTS
The author would like to thank Madeleine Flynn for producing Fig. 1.
FINANCIAL SUPPORT
Donald P. McManus is a National Health and Medical Research Council of Australia Senior Principal Research Fellow.