INTRODUCTION
The eukaryotic ribosome is a complex structure composed of about 80 different ribosomal proteins (RP) and 4 structural rRNAs, which are assembled into a small and large subunit. Together these subunits form the translational machinery of a cell. Ribosomes are essential for a cell and its constituents are highly expressed in rapidly dividing cells. Expression of RP and rRNA genes in eukaryotes is coordinately regulated in response to stress and growth stimuli, which permits the cell to adjust the number of ribosomes and overall protein synthetic capacity to environmental conditions (Pearson and Haber, 1980; Ju and Warner, 1994; Meyuhas, 2000). For higher eukaryotes, ribosome biosynthesis is regulated at the level of translation (Meyuhas, 2000). In contrast, it is primarily transcriptionally regulated in Saccharomyces cerevisiae, where transcription is switched off during stress (e.g. nutrient deficiency) and spore formation (Warner, 1999). Transcription of RP genes is then coordinately regulated by a combination of transcription factors and specific DNA elements present in their promoters. At the moment, ribosome biosynthesis has scarcely been studied in parasites. Recently, we demonstrated that coccidian parasites also transcriptionally regulate de novo ribosome biosynthesis (Schaap et al. 2005). For example, in Eimeria tenella rRNA and RP genes are abundantly transcribed in rapidly growing merozoite stages and almost not transcribed in dormant oocyst stages. Similarly, Toxoplasma gondii showed a 100-fold difference in RP transcription, when rapidly growing tachyzoites were compared with oocyst stages (Schaap et al. 2005). Thus, Coccidia can transcriptionally regulate ribosome biosynthesis, dependent on their life-cycle stage.
Since transcription of RP genes was simultaneously regulated in Coccidia, we anticipate that this is coordinated for the complete set of RP genes by an underlying control mechanism. Therefore, we made a start to analyse the transcriptional regulation of the complete set of RP genes as a whole in T. gondii. By clustering of publicly available expressed sequence tags (ESTs), all cytoplasmic T. gondii RPs were deduced. Although a large number of ESTs is available for this parasite and EST clusters were published (Ajioka et al. 1998; Li et al. 2003), these proteins were not identified previously. To determine if and how transcription of all T. gondii RP genes may be coordinately regulated, these genes were studied for their genomic organization. Moreover, their upstream sequences were analysed for the presence of conserved DNA elements as well as promoter strength. The identification of 2 novel conserved elements in upstream sequences of the complete set of T. gondii RP genes allows us to study transcriptional control of RP genes.
MATERIALS AND METHODS
Identification of the complete set of T. gondii cytoplasmic RPs
For the identification of the coding sequences of T. gondii cytoplasmic RPs publicly accessible ESTs annotated for RPs (2192 ESTs, dd. 24 April 2003) were collected from NCBI/Nucleotide (http://www.ncbi.nlm.nih.gov). Clustering of ESTs into contigs was performed with Sequencher 4.1.4® software, after which the consensus of each contig was exported to CloneManager 6.0 to determine open reading frames (ORFs). Protein predictions were verified for the consensus of each contig by BLAST search against the database of T. gondii Twinscan2 predicted proteins on www.toxodb.org. BLAST searches for similarity to RPs in other organisms were performed at NCBI/BLAST (swissprot db). Pfam domain searches were performed at http://www.sanger.ac.uk/Software/Pfam and were used for the annotation of each T. gondii RP-specific ORF. The coding sequences of the complete set of T. gondii RPs reported in this paper are available in the Third Party Annotation Section of the DDBJ/EMBL/GenBank databases under the accession numbers TPA: BK004896-BK004974 (protein sequences at DAA04986-DAA05064).
Comparative analysis of RP upstream sequences
A database was made consisting of the upstream sequences of 79 T. gondii RP genes. About 1000 bp of genomic sequence upstream of the start of each RP EST contig were collected, assuming that the start of each RP EST contig roughly corresponds to the presumptive transcriptional start site of the RP gene. These preliminary genomic sequence data were obtained from The Institute for Genomic Research (TIGR) website at http://www.tigr.org by blastn using each cDNA consensus from the final EST database as query. Searches for conserved motifs were carried out by comparison of these RP upstream sequences using the program Multiple Em for Motif Elicitation (MEME) (http://meme.sdsc.edu). MEME was run under the following settings: a motif width ranging from 6 to 50 bases, a discovery limit of 10 motifs and the total number found per site ranging from 2 to 150.
Parasite strain and culturing
T. gondii RHΔHXGPRT (Donald et al. 1996) tachyzoites were maintained in culture at 5% CO2 and 37 °C by serial passage in Vero cells or human foreskin fibroblasts (HFF), grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% heat-inactivated fetal calf serum and 2 mM L-glutamine.
Molecular techniques
Ten different LacZ expression vectors were made by placing a LacZ reporter gene under the control of either the T. gondii alpha-tubulin promoter (TUB) or upstream sequences of a T. gondii RP gene (pRPS3, pRPS10, pRPS13, pRPS25, pRPS29, pRPL9, pRPL13 and pRPL38). The different LacZ constructs were based on the pCAT-GFP plasmid (Striepen et al. 1998), which contains a fusion of chloramphenicol acetyltransferase gene (CAT) to green fluorescent protein (GFP) gene driven by the dihydrofolate reductase (DHFR) promoter and flanked by the DHFR 3′ untranslated region (UTR). First, GFP was replaced with the LacZ gene, which was derived from genomic DNA of Escherichia coli BL21 and amplified by polymerase chain reaction (PCR) using the primers (restriction sites are underlined, start- and stopcodons are in bold); LacZ-AvrII (fw): 5′-CGATCCTAGGATGACCATGATTACGGATTCACTGGCCGTCGTTTTACAACGTCGTG-3′ and LacZ-PstI (rv): 5′-CGATCTGCAGTTATTTTTGACACCAGACCAACTGG-3′. The PCR product was digested with AvrII/PstI and inserted in pCAT-GFP (AvrII/PstI digested) resulting in pCAT-LacZ. The TUB and RP upstream sequences (except pRPS13) and their complete 5′ UTR were PCR amplified from T. gondii RHΔHXGPRT genomic DNA using the primers depicted in Table 1. The PCR products were digested with HindIII/AvrII or KpnI/AvrII and inserted in pCAT-LacZ by replacing the pCAT sequence. The resulting constructs were named pTUB[AvrII]LacZ or pRP(xx)LacZ (xx refers to the name of RP used).
Table 1. List of primers used to generate pRPLacZ reporter constructs (The restriction sites HindIII, KpnI, AvrII and BglII are underlined.)

Since RPS13 upstream sequences contain an internal AvrII restriction site, the construct pRPS13LacZ was made in a different way together with a pTUBLacZ construct. These constructs were also based on pCAT-GFP replacing CAT-GFP with the LacZ gene. LacZ was PCR amplified using LacZ-BglII (fw): 5′-CGATAGATCTATGACCATGAT TACGGATTCACTG-3′ and LacZ-PstI (rv), digested with BglII/PstI and inserted in pCAT-GFP (BglII/PstI digested). The resulting construct was named pDHFRLacZ. The pRPS13 and pTUB were PCR amplified as described above for the other promoters. The PCR products were digested with HindIII/BglII and inserted in pDHFRLacZ by replacing the DHFR promoter (HindIII/BglII digested). The constructs were named pTUB[BglII]LacZ and pRPS13LacZ.
Electroporation of T. gondii RHΔHXGPRT tachyzoites and CPRG assay
Transient transfections were carried out by electroporation as described previously (Soldati and Boothroyd, 1993) using freshly harvested T. gondii RHΔHXGPRT tachyzoites (107) and 20 μg sterilized circular plasmid DNA (QIAGEN Plasmid maxi kit, Qiagen) in a 2 mm gap cuvette (BTX electroporator; 1·8 kV, 100 Ω, 25 μF) in a total volume of 400 μl electroporation buffer, which was composed of 120 mM KCl, 0·15 mM CaCl2, 10 mM K2HPO4/KH2PO4 pH 7·6, 25 mM HEPES, 2 mM EDTA, 5 mM MgCl2. Immediately prior to use, fresh 2 mM ATP and 5 mM glutathione (GSH) were supplemented to the buffer and sterilized by filtration through a 0·22 μm filter. After electroporation triplicates (50 μl) of each sample were added to separate wells (24-well plates) containing a confluent monolayer of HFF cells. Parasites were cultured overnight (37 °C, 5% CO2) and LacZ activity was determined 16–24 h post-infection by a chlorophenol red-β-D-galactopyranoside (CPRG) assay as described previously (Seeber and Boothroyd, 1996). Briefly, infected monolayers were lysed by adding 200 μl of assay buffer (100 mM HEPES pH 8·0, 1 mM MgSO4, 1% Triton X-100, 5 mM dithiothreitol) per well and were then incubated for 1 h at 50 °C. An aliquot (50 μl) of the cleared lysate was diluted into assay buffer (100 μl final volume) and mixed with an equal volume of assay buffer containing 2 mM CPRG (Roche) as substrate. Substrate conversion was carried out at 30 °C (~4 h) and measured at 570 nm using a microplate reader.
Three independent assays were carried out as described above using HFF cells infected with untransfected T. gondii RHΔHXGPRT tachyzoites as a negative control. Within an individual assay all data were subtracted by the LacZ level of this control sample, after which the data were converted to percentages. For conversion into percentages, the average of the pTUBLacZ[BglII] triplicate was set on 100%, after which all samples were related to this sample. All triplicates of the 3 independent assays were used to calculate the average of each tested construct with its standard deviation. Since equal concentrations of construct DNA were transfected, the obtained data were converted to equimolar amounts.
RESULTS
The complete set of T. gondii cytoplasmic RPs
To identify the complete set of RP coding sequences, T. gondii RH tachyzoite ESTs were initially collected, which had been annotated as RP. This resulted in a dataset containing 2192 ESTs which were assembled into 84 contigs. These contigs were checked for sequence inconsistencies and the consensus of each contig was used to deduce ORFs. These ORFs showed high similarity to 31 eukaryotic small subunit RPs (containing 2 partial contigs similar to RPS23 which could not yet be merged), 43 eukaryotic large subunit RPs and the eukaryotic ribosomal proteins SA, P0, P1 and P2, as determined by standard protein-protein BLAST. In addition, ORFs of 5 contigs showed similarity with prokaryotic RPs, namely RPS17, RPL13, RPL22, RPL24 and RPL28. These proteins are probably components of the plastid or mitochondrion and for that reason they were excluded from the dataset. Subsequently, the consensus of each contig was used in a new search at NCBI (tblastn; est_others db) to identify additional ESTs which had either not been annotated or falsely annotated. A total of 588 additional T. gondii RP ESTs were identified which were added to the initial dataset, resulting in a final dataset consisting of 2780 ESTs assembled in 78 contigs. Compared to the complete set of RPs present in human or yeast (Mager et al. 1997; Kenmochi et al. 1998; Planta and Mager, 1998; Yoshihama et al. 2002), only the small RPL41 of T. gondii was missing in the dataset. Using human RPL41 (GenBank Accession no. P28751) as query, 18 homologous T. gondii ESTs were obtained which clustered into 1 contig. With the RPL41 contig included, 79 different contigs were generated each containing a full-length ORF similar to a eukaryotic RP. Protein predictions were confirmed by BLAST search against the database of T. gondii Twinscan2 predicted proteins. In general, coding sequences of these putative T. gondii RPs were highly similar to their human and yeast homologue (shown in Table 2), except for the putative T. gondii RPL28 (8·00E-03). T. gondii RPL28 was 21% identical with human RPL28 and 44% identical with E. tenella RPL28 (which was identified from 10 assembled ESTs, results not shown).
Table 2. Toxoplasma gondii cytoplasmic ribosomal proteins (Shown from the left to the right are: identification number (ID), the name for each RP protein in T. gondii (TgRP) and features for each T. gondii RP such as GenBank Accession number of the coding sequence (acc nr), its calculated molecular weight (MW), total number of amino acids (AA) and the number of ESTs present per RP contig. This is followed by Twinscan protein predictions including T. gondii chromosome (Chr.) location, and followed by the Pfam domains, which showed homology to parts of each T. gondii RP together with the E-value. Furthermore, the homology of T. gondii RPs with the human and yeast RPs are given together with the E-value. (a) Region of the T. gondii RP protein in which the Pfam domain is present. (b) The complete protein consists of an N-terminal fusion of ubiquitin to the RP. (c) ESTs showing similarity to ubiquitin were excluded. (d) Designated previously as ribosomal protein S37, now as ribosomal protein S31 (Mager et al. 1997). (e) Instead of standard blastp, protein BLAST search for short nearly exact matches was used for RPS30 and RPL41. (f) Incomplete coding sequence; missing part (25 amino acids) at the C-terminus was determined using T. gondii genomic DNA sequence data (TIGR) according to similarity with human and yeast RPL3 protein sequences. (g) Ambiguous start of translation; human and yeast RPL4 consists of 140 additional amino acids at the N-terminus. Although their accompanying coding sequences were present on the T. gondii genome, no ESTs were detected for that extension. Therefore, the largest ORF as deduced from assembled ESTs is given. (h) These proteins are smaller in size compared to their human homologues, however, similar in size compared to homologues in yeast and Eimeria tenella.)

Since the nomenclature of RPs is often ambiguous and differing between species, all T. gondii RP-specific ORFs were classified by their similarity to human RPs. This classification is in agreement with the annotations for conserved eukaryotic RP domains as defined in the Pfam database (Sanger). As is obvious from Table 2, this annotation sometimes differs from yeast annotated RPs. In summary, the complete set of T. gondii cytoplasmic RPs was identified and consists of 31 small subunit RPs, 44 large subunit RPs and the proteins SA, P0, P1 and P2, being highly similar to RPs present in human and yeast.
Clustering of RP genes on the T. gondii genome
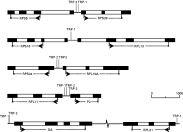
Previously, we showed that RP transcription is coordinately regulated in E. tenella which was also suggested for T. gondii (Schaap et al. 2005). Furthermore, genes encoding proteins which display similar functions or are required in specific tissues are sometimes clustered on the genome to allow coordinate regulated transcription (van Driel et al. 2003). Therefore, it was investigated whether RP genes are clustered on the T. gondii genome. Coding sequences of all T. gondii RPs were used as a query in a BLAST search against the Twinscan database. All RP coding sequences were detected within the Twinscan database except for RPL41. Analysis of their positions revealed that 10 RP genes were paired on the genome, being (1) RPS5 with RPS29, (2) RPS16 with RPL13, (3) RPS24 with RPL10A, (4) RPL11 with P2, and (5) SA with RPL31 (Fig. 1). In the first 4 pairs, RP genes were arranged in a head-to-head orientation with an intergenic region ranging from 280 bp to 380 bp. If a promoter is limited to the intergenic region, this would indicate that these RP putative promoters are at most 280 to 380 bps long. Moreover, these intergenic regions then contain either 2 small promoters or one bidirectional promoter. In pair 5, the genes were arranged in a tail-to-tail orientation being spaced 3764 bp apart. Apart from the above-described gene clusters, all other T. gondii RP genes were spaced more than 10 kb apart. Thus, most RP genes are randomly distributed as individual genes over the genome indicating that their transcription is individually regulated.

Fig. 1. Pairs of RP genes on the Toxoplasma gondii genome. Shown are 5 clusters of paired T. gondii RP genes on scale, with the exception of the intergenic region between the genes SA and RPL31 (being 3764 bp). Exons are shown as black boxes and introns as open boxes. Intergenic regions are shown as solid lines and TRP elements are indicated as vertical lines at their position within the sequence. Arrows depict start codons and X depicts stop codons. For RPS5, RPS16 and RPL13 the start codon is preceded by an intron. For RPS5 the last 119 bps of the 3′ UTR are not depicted and for ribosomal protein P2 the last 26 bps of the 3′ UTR are not shown. The RP genes which are arranged in a head to head orientation contain intergenic regions between 280–380 bp. The genomic coordinates of these clusters on the T. gondii chromosomes are as follows; RPS5/RPS29: LG14 – 1·95 Mb; RPS16/RPL13: VI – 0·85 Mb; RPS24/RPL10A: X – 6·91 Mb; RPL11/P2: XI – 0·82 Mb; SA/RPL31: IX – 1·31 Mb.
Generation of a T. gondii database containing RP upstream sequences
Since T. gondii RP genes are not clustered on the genome, transcription of these genes must be individually regulated though in a concerted manner. Similarly to S. cerevisiae, we investigated if transcription of the complete set of RP genes is coordinately regulated in T. gondii and whether conserved promoter elements are present in this set of T. gondii genes. Therefore, a RP putative promoter database was made consisting of genomic sequences immediately upstream of the presumptive transcriptional start site of all T. gondii RP genes. It should be noted that transcriptional start sites of the 79 RP genes were not experimentally determined, but were based on the start of each RP EST contig. Since these contigs contain many ESTs (35 ESTs on average), the true transcriptional start sites will likely be close to the presumptive transcriptional start sites we used. For 2 RP genes (RPS13 and RPL9) the transcriptional start sites were experimentally determined, which indeed correlated well to the start of the RP EST contigs (van Poppel, manuscript submitted). The T. gondii genomic sequences immediately upstream of the presumptive transcriptional start site of all RP genes were obtained by blastn (TIGR) using the consensus of each RP EST contig as query. If available, 1000 bp of genomic upstream sequences were collected for each RP; for 69 RPs 915–1000 bp were collected while for the remaining 10 RPs between 160 and 844 bp of upstream sequences were obtained. Although the lengths of promoter regions for RP genes are not determined, the upstream sequences present in this database were considered as the putative promoters.
T. gondii RP upstream sequences contain two highly conserved and localized DNA elements
The complete set of T. gondii RP upstream sequences present in the database was used to search for conserved DNA elements. The upstream sequences of all T. gondii RP genes were compared with each other in multiple searches using the program MEME. This program indicates which sequence elements are overrepresented in a database. Three specific DNA elements were identified which were highly enriched within the RP putative promoter database, being TCGGCTTATATTCGG, [T/C]GCATGC[G/A] and polypyrimidine tracts.
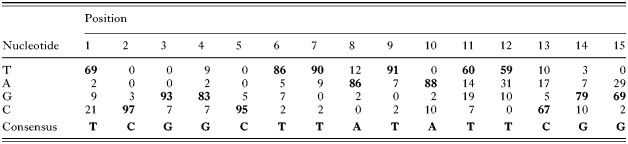
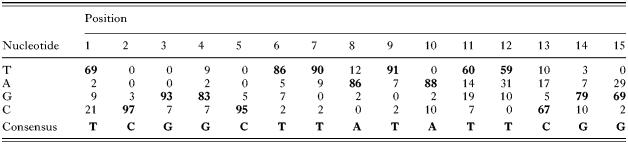
The first sequence element, TCGGCTTATATTCGG (15 bp), was named Toxoplasma Ribosomal Protein-1 element (TRP-1). This conserved and novel element was identified 58 times in the T. gondii RP putative promoter database. TRP-1 elements showed limited variation (see Table 3), and the percentages of conservation per nucleotide are as follows T69979383958690869188T60T59C6779G69 (bold and underlined indicates a conservation [ges ]79%). TRP-1 was present in both orientations and mostly once per RP upstream sequence. TRP-1 was observed twice in the upstream sequences of RPS4, RPL6, RPL35 and ubiquitin-RPL40, and 3 times in the upstream sequence of RPL30. The second identified sequence element, [T/C]GCATGC[G/A], was also novel, containing a reversible sequence of 8 nucleotides and was named Toxoplasma Ribosomal Protein-2 element (TRP-2). The TRP-2 element was found 73 times in the database, mostly once per RP upstream sequence. Two TRP-2 elements were detected in the upstream sequences of RPS11, RPS13, RPS24, RPS28, RPL10A, RPL11, RPL18A, RPL32, RPL41 and P2. Three elements were observed in the upstream sequences of RPS18, RPS27 and RPL27. The third identified sequence element was a polypyrimidine tract consisting of T/C stretches of 5–30 nucleotides. Polypyrimidine tracts have previously been shown to be present in eukaryotic RP genes, but in the 5′ UTR instead of the upstream sequences (Meyuhas, 2000). These tracts were observed 73 times in the total database.
Table 3. Conservation of TRP-1 consensus in Toxoplasma gondii RP upstream sequences (Summarized in this Table are the 58 TRP-1 elements which have been identified in T. gondii RP upstream sequences. For each position, frequency of each individual nucleotide (T, A, G or C) is given in percentages. The most prevalent nucleotide is indicated in bold. On the bottom the consensus obtained from the 58 TRP-1 elements is shown.)
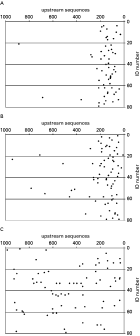
To further characterize these elements, their positions in relation to the start of transcription were analysed. TRP-1 and TRP-2 elements were almost all highly localized to a specific region, 10–330 bp upstream of the presumptive transcriptional start site (Fig. 2A,B). In contrast, the polypyrimidine tracts were randomly distributed over the RP upstream sequences and no specific common localization was observed (Fig. 2C). TRP-1 and/or TRP-2 were present in 95% of all RP upstream sequences; 41 contained 1 TRP element, 34 contained both TRP elements and for only 4 no TRP element was identified. No co-localization of both types of TRP elements was observed.

Fig. 2. Localization of sequence elements in RP upstream sequences. Shown are the locations of TRP-1 elements (A), TRP-2 elements (B) and polypyrimidine tracts (C) in T. gondii RP upstream sequences. Each dot represents a DNA sequence element. The position of the element within the RP upstream sequence is depicted on the X-axis, where the presumptive transcriptional start site corresponds to position 0 and position 1000 represents 1000 bp upstream from the presumptive transcriptional start site. The ID number of the respective T. gondii RP is indicated on the Y-axis. Of each RP gene 1000 bps of upstream sequence were analysed except for: RPS17 (753 bp), RPS18 (844 bp), RPL3 (839 bp), RPL7 (536 bp), RPL11 (828 bp), RPL36 (556 bp), RPL37 (160 bp), RPL39 (760 bp), RPL41 (390 bp) and P0 (291 bp).
To determine if TRP elements are specifically associated with T. gondii RP upstream sequences, the presence of these elements in RP upstream sequences was compared to their occurrences in the T. gondii genome. Since TRP elements were mainly present within the 330 bp upstream of the presumptive transcriptional start site, their enrichment was determined for this region. For TRP-1 a more restricted consensus was used, being CGGCTTATANNNG, to which 20 TRP-1 elements fully complied. Based on random chance we calculated that a TRP-1 element would be present 0·050 times within a region of 330 bps (2 (sense/antisense)×1/1048576 (random chance)×330 (bp)×79 (RPs)=0·050). Thus, TRP-1 elements were 400-fold enriched (20/0·050=400) in the 330 bp upstream of the presumptive transcriptional start site compared to random chance. Similar calculations were performed for the presence of TRP-2 elements showing a 40-fold enrichment in the 330 bp region of all 79 RP putative promoters. Subsequently, the presence of both TRP elements was determined within the T. gondii genome, using publicly accessible Toxoplasma genomic sequence data (being ~6·4×107 bp). In the genomic sequence data TRP-1 and TRP-2 were respectively present 351 times and 30856 times, being a 3-fold and 8-fold enrichment compared to random chance. Since 7588 genes are identified in the T. gondii genome (Twinscan database) and assuming that each of the identified TRP elements is localized within a single gene promoter, RP putative promoter regions are significantly overrepresented with TRP-1 elements (20/79) compared to the genome (351/7588; χ2-test=64·23; P[Lt ]0·005). However, this could not be concluded for occurrence of TRP-2 elements.
T. gondii RP promoters have variable promoter strength
To determine whether TRP elements could be correlated to gene expression, a study was performed in which 8 different RP genes were randomly selected which contained either TRP-1, TRP-2 or both TRP elements in their putative promoter regions, being RPS3, RPS10, RPS13, RPS25, RPS29, RPL9, RPL13 and RPL38. Of these RP genes, upstream sequences together with their 5′ UTR were cloned immediately upstream of the LacZ reporter gene and compared with the T. gondii TUB promoter with 5′ UTR for their strength to drive expression. Using the LacZ reporter gene as readout, expression levels were determined with a CPRG assay. Three independent assays were performed in which each construct was transiently transfected in the T. gondii strain RHΔHXGPRT. These experiments showed reproducible expression patterns for each individual construct, which are shown in Fig. 3. As a positive control pTUBLacZ was used. This construct was cloned twice in which the 5′ UTR of TUB was separated from the LacZ startcodon by either an AvrII or a BglII restriction site. Although this region is important for translation efficiency (Seeber, 1997), the difference in restriction sites did not affect expression levels (see Fig. 3), allowing us to use both restriction sites in these two constructs for integration of RP sequences. LacZ was expressed at different levels, when driven by different RP promoters with their 5′ UTR. Highest expression was observed for the RPS13 promoter (including 5′ UTR), which was comparable in strength to the strong T. gondii TUB promoter, whereas 15-fold lower expression was obtained with the RPS29 promoter (including 5′ UTR) being the weakest tested RP promoter. These data showed that RP promoters and their 5′ UTR differ in their strength to drive expression of a heterologous gene. No direct correlation could be observed between expression levels and the presence of a TRP element in the upstream sequences for the analysed RPs.

Fig. 3. LacZ expression driven by different RP promoters. Shown on the left are upstream sequences of different RP genes with their 5′ UTR fused to LacZ, which were tested for expression. The RP upstream sequences are indicated with a solid line, followed by black boxes for exons and open boxes for introns of the respective RP 5′ UTRs (on scale). Roughly 1 kb of RP upstream sequence was used; pRPS29 (1009 bp); pRPL38 (953 bp); pRPS3 (1000 bp); pRPL13 (954 bp); pRPL9 (828 bp); pRPS25 (941 bp); pRPS10 (1000 bp) and pRPS13 (1276 bp). The construct pRPS10LacZ contains 5 codons of RPS10 prior to LacZ. As a control the TUB promoter and its 5′ UTR were used to drive expression. The LacZ ORF is indicated as a grey arrow (not on scale; 3·1 kb). The localization of TRP-1 elements is indicated as open triangles and of TRP-2 elements as open circles. Presumptive transcriptional start sites of RP genes and their flanking genes are indicated by a thin arrow. Each construct was analysed with 3 independent CPRG assays. On the right the LacZ enzyme activity as determined for each construct is related to the average LacZ enzyme activity obtained by pTUBLacZ[BglII] and shown as an average percentage with its standard deviation. Results are shown as equimolar amounts of transfected plasmid DNA.
DISCUSSION
Ribosomes are responsible for protein synthesis and as such are essential for growth in all living organisms (Warner, 1999; Meyuhas, 2000; Schaap et al. 2005). Synthesis of de novo ribosomes requires expression of 4 different rRNAs and a large set of RPs. The 4 different rRNAs were previously described for T. gondii (Guay et al. 1992; Gagnon et al. 1996). Here, the complete set of T. gondii cytoplasmic RPs is identified consisting of 79 different proteins. These proteins are highly similar in both numbers and protein sequence to higher eukaryotes like human, suggesting a conserved ribosome complexity.
Since synthesis of these ribosomal components consumes a large proportion of the cell's energy, it is tightly and coordinately regulated in eukaryotes. In most eukaryotes, synthesis of the set of RPs is primarily regulated at the level of translation (Meyuhas, 2000). In contrast, in S. cerevisiae and Coccidia like T. gondii and E. tenella, synthesis of RPs is regulated at the level of transcription and differs dependent on its life-cycle stage (Warner, 1999; Schaap et al. 2005). For T. gondii we previously showed that the complete set of RP genes is highly transcribed in tachyzoites, whereas transcripts were almost absent in the oocyst stages (Schaap et al. 2005). Since these gene products are functionally linked and also simultaneously transcribed, it was suggested that they are coordinately regulated.
Coordinated transcriptional control of a set of genes is usually regulated by one or more transcription factors binding to specific promoter elements. In addition, gene clusters exist in higher eukaryotes for functionally related genes, such as α-globin genes, β-globin genes, histone genes and Hox genes (van Driel et al. 2003). RPs are also functionally related proteins and in T. gondii 8 RP genes are located paired on the genome. The paired RP genes are arranged in a head to head orientation with small intergenic regions of 280–380 bps. Their small size suggests these regions may operate as bidirectional promoters. However, most RP genes (71 resp.) were randomly distributed in the genome and over the different T. gondii chromosomes suggesting that a conserved promoter structure should control the coordinate expression of these genes.
To study if transcription of the set of individual RP genes is coordinated in T. gondii by a combination of promoter elements and transcription factors, their upstream sequences were compared for conserved DNA elements. Since T. gondii RP promoters have not been defined before, 1000 bps of upstream sequences were selected for all RP genes. By comparative analysis two novel highly conserved DNA elements were identified in the upstream sequences of nearly all T. gondii RP genes, named TRP-1 (consensus TCGGCTTATATTCGG) and TRP-2 ([T/C]GCATGC[G/A]). Both elements were specifically localized in a region 10–330 bps upstream of the presumptive transcriptional start site of these RP genes. In addition, TRP-1 and TRP-2 elements were highly over-represented in these RP upstream regions compared to random chance, respectively 400- and 40-fold. Due to their specific localization these TRP elements likely operate as promoter elements in the coordinated transcriptional control of T. gondii RP genes.
Comparative analysis of upstream sequences of RP genes from the coccidian parasite E. tenella showed no enrichment for these TRP elements. Instead a different element was identified, being GGGCTG[T/C]GGGG[G/C][G/T]GC (results not shown) which was similarly positioned to the TRP elements in T. gondii. These findings suggest that a comparable control mechanism may be present for transcription of RP genes in both parasites, but that genus specific DNA elements have evolved.
To determine if TRP elements are exclusively associated with RP genes, known T. gondii promoters were analysed, including GRA1, GRA3, GRA5, GRA6, GRA7, SAG1, TUB1. With one exception, TRP elements were not present in these promoters suggesting that TRP elements are not general promoter elements in T. gondii. In addition, a genome-wide analysis for TRP elements was performed, whereby we did not select for gene promoters only. In total 351 TRP-1 elements were detected within the T. gondii genome, suggesting that this element may operate in transcriptional control of a larger set of gene products, including RP genes. TRP-2 elements were detected 30856 times within the genome. This number is higher than the estimated 7588 genes present in the T. gondii genome (ref. Twinscan) being too high to function as a key element for regulation. Therefore, we expect that, apart from acting in combination with transcription factors, TRP-2 elements may operate in combination with additional gene control such as DNA accessibility. It is well known that chromatin remodelling by histone modifications (acetylation, methylation, phosphorylation) can alter DNA accessibility and thereby influences gene control by the transcription machinery (Horn and Peterson, 2002).
The identification of TRP elements in the set of T. gondii RP genes allowed us to further investigate their role. In a first study, the promoter strength of upstream sequences of 8 different RP genes was analysed and correlated with the presence of TRP-1 and/or TRP-2. No direct correlation could be detected between RP promoter strength and the presence of any of the TRP elements. Remarkably, up to a 15-fold difference in strength was observed between the T. gondii RP promoters. Since ribosome assembly is based on equimolar usage of RPs, another level of regulation must be involved to maintain the stoichiometry among the T. gondii ribosomal components. Such regulation could operate at different levels including mRNA processing and stability, translational efficiency and protein turnover. In this respect it may be relevant that RP genes in T. gondii frequently contain introns in their 5′ UTRs. In S. cerevisiae such introns were suggested to be involved in autoregulation (Warner et al. 1985).
Since S. cerevisiae also transcriptionally regulates RP genes, it is informative to compare T. gondii with S. cerevisiae. In S. cerevisiae RP transcription is dependent on external stimuli such as carbon sources and nutrients, which can trigger signal transduction pathways and thereby activate or inhibit the function of transcription factors. Complicated transcriptional networks are present in S. cerevisiae whereby multiple transcription factors (including Fhl1, Rap1, Yap5, Crf1, Ifh1) bind to different promoter elements, that control transcriptional induction or silencing of S. cerevisiae RP genes (Lee et al. 2002; Martin et al. 2004; Wade et al. 2004). In S. cerevisiae the identified promoter elements are present in many, but not in all RP gene promoters, similar to the presence of TRP elements in T. gondii RP genes. Thus, a set of multiple promoter elements and accompanying transcription factors is required for coordinated regulation of RP transcription. Moreover, in S. cerevisiae the conserved promoter elements in RP genes are also not limited to the select group of RP genes but appeared to be involved in transcriptional control of several other genes as well. No direct homologues of the above-described yeast transcription factors could be identified in the T. gondii genome. However, the identification of TRP elements in T. gondii should allow the characterization of their respective transcription factors.
In summary, the complete set of T. gondii RPs has been identified. Their upstream sequences are enriched with TRP-1 and TRP-2 elements which are specifically localized in front of the transcriptional start sites and therefore expected to be involved in coordinated transcription of RP genes. The identification of these elements creates a basis to further study the underlying mechanism by which RP transcription is controlled in T. gondii. Since ribosome biosynthesis is directly linked to cell growth in all organisms, understanding the transcriptional control of T. gondii RP genes will simultaneously explain how growth is regulated at the molecular level in parasites.
We would like to acknowledge Dr Erik de Vries for help with the genome analysis of TRP elements. EST data were obtained from NCBI at http://www.ncbi.nlm.nih.gov. Preliminary genomic and/or cDNA sequence data was accessed via http://ToxoDB.org and/or http://www.tigr.org/tdb/t_gondii/. Genomic data were provided by The Institute for Genomic Research (supported by the NIH grant no. AI05093), and by the Sanger Center (Wellcome Trust). EST sequences were generated by Washington University (NIH grant no. 1R01AI045806-01A1).