Introduction
Rhipicephalus microplus is a monoxenous tick that infests preferentially bovines, and causes great economic damage to livestock (Willadsen, Reference Willadsen2006). Acaricides are the main control method against ticks, but its intensive use can contaminate milk, meat and environment (Willadsen, Reference Willadsen2004; de la Fuente et al. Reference de la Fuente, Almazán, Canales, de la Lastra, Kocan and Willadsen2007). Moreover, its high cost and occurrence of resistant populations encourages the development of anti-tick vaccines (Lovis et al. Reference Lovis, Reggi, Berggoetz, Betschart and Sager2013; Mendes et al. Reference Mendes, Mendes and Sato2013). Commercial vaccines present variation in efficiency according to tick species (de Vos et al. Reference de Vos, Zeinstra, Taoufik, Willadsen and Jongejan2001), strain (Fragoso et al. Reference Fragoso, Rad, Ortiz, Rodríguez, Redondo, Herrera and de la Fuente1998) and geographic localization (de la Fuente et al. Reference de la Fuente, Rodríguez, Montero, Redondo, García-García, Méndez, Serrano, Valdés, Enríquez, Canales, Ramos, Boué, Machado and Lleonart1999); thus the search for new protective antigens remains (de la Fuente et al. Reference de la Fuente, Kopácek, Lew-Tabor and Maritz-Oliver2016).
Various salivary proteins have been isolated and characterized, since saliva of hematophagous arthropods contains molecules capable of inhibiting responses associated with the homeostasis of the host's physiological system. Salivary molecules of these organisms can act as modulators of the host immune system and hence help in maintaining hematophagy (Reck et al. Reference Reck, Berger, Marks, Zingali, Canal, Ferreira, Guimarães and Termignoni2009; Kotál et al. Reference Kotál, Langhansová, Lieskovská, Andersen, Francischetti, Chavakis, Kopecký, Pedra, Kotsyfakis and Chmelař2015). Thus, vaccines derived from salivary antigens could block the attachment and feeding, by elicitation of a host's immune response and neutralization of secreted molecules. In addition, host resistance can be naturally acquired when, after successive tick infestations, the host assembles an immune response against antigens exposed to its immune system, presumably against molecules secreted in the tick saliva (Trager, Reference Trager1939; de Castro and Newson, Reference de Castro and Newson1993). The level of ticks that complete the life cycle thus tend to decrease gradually along infestations (Fielden et al. Reference Fielden, Rechav and Bryson1992), but does not prevent that some individuals in the tick population complete the parasite cycle (Roberts, Reference Roberts1968; Ribeiro, Reference Ribeiro1987). Different proteins may be able to elicit the same response following artificial immunization and, after vaccinated, recurrent contacts with the tick should enhance the immunity of the host (as a booster vaccine) without expenditures with additional doses of antigen (Shapiro et al. Reference Shapiro, Voigt and Ellis1989; Nuttall et al. Reference Nuttall, Trimnell, Kazimirova and Labuda2006).
Among the salivary molecules being studied, glycine-rich proteins (GRPs) are considered potential antigens against ticks. Vaccination trials using GRPs as antigen were already described against R. haemaphisaloides (Zhou et al. Reference Zhou, Gong, Zhou, Xuan and Fujisaki2006) and Haemaphysalis longicornis (Mulenga et al. Reference Mulenga, Sugimoto, Ingram, Ohashi and Onuma1999), while the protein 64P, a GRP of R. appendiculatus, was patented and, since it presents cross-reactivity with different tick species, suggested as a possible broad-spectrum vaccine (Trimnell et al. Reference Trimnell, Hails and Nuttall2002, Reference Trimnell, Davies, Lissina, Hails and Nuttall2005). Concerning R. microplus, a four-antigen cocktail vaccination trial containing a salivary GRP conferred a 72·3% overall efficacy (Maruyama et al. Reference Maruyama, Garcia, Teixeira, Brandão, Anderson, Ribeiro, Valenzuela, Horackova, Veríssimo, Katiki, Banin, Zangirolamo, Gardinasse, Ferreira and de Miranda-Santos2017). GRPs have been identified in proteomes and transcriptomes of a variety of tick species, but little is known about their roles (Ribeiro et al. Reference Ribeiro, Alarcon-Chaidez, Francischetti, Mans, Mather, Valenzuela and Wikel2006; Garcia et al. Reference Garcia, Gardinassi, Ribeiro, Anatriello, Ferreira, Moreira, Mafra, Martins, Szabó, de Miranda-Santos and Maruyama2014; Tirloni et al. Reference Tirloni, Reck, Terra, Martins, Mulenga, Sherman, Fox, Yates, Termignoni, Pinto and da Silva Vaz2014; Karim and Ribeiro, Reference Karim and Ribeiro2015). In this context, the aim of this study was to describe a GRP from R. microplus (named RmGRP), characterize the levels of gene expression and protein presence in various tick tissues/developmental stages, evaluate the effect of the corresponding gene (rmgrp) silencing by dsRNA treatment in partially engorged females, as well as analyse the recombinant RmGRP (rRmGRP) recognition by antisera raised against tick tissues and sera from naturally and experimentally infested bovines.
Material and methods
Ticks
Ticks were collected from experimentally infested bovines Bos taurus (Hereford) housed in individual pens on slatted floors in Faculdade de Veterinária of the Universidade Federal do Rio Grande do Sul (Porto Alegre, RS – Brazil). Partially engorged females and males (Porto Alegre strain) were collected directly on infested bovines and fully engorged females were collected on the floor after detachment/dropping from the cattle. Ticks were kept at 28 °C under 85% relative humidity. All procedures involving animals were in accordance with the Ethics Committee on Animal Use (CEUA/UFRGS) under the protocol number 27559.
Rmgrp cDNA isolation
A cDNA library synthesized from different developmental stages of R. microplus life cycle (larvae, nymphs, non-engorged females, adult males and partially engorged females) (Renard et al. Reference Renard, Garcia, Cardoso, Richter, Sanakari, Ozaki, Termignoni and Masuda2000) was screened on nitrocellulose membranes (Schleicher and Schüll, USA) with cDNA synthesized from partially engorged adult females poly-A RNA labelled with [α−32P]dATP by random priming (Sambrook et al. Reference Sambrook, Fritsch and Maniatis1989). Hybridization was carried out overnight at 65 °C in hybridization buffer (6X SSC) containing 5% (w/v) cow non-fat dry milk, 200 mg mL−1 denatured salmon sperm DNA and cDNA probe. Filters were then washed for 30 min each in 6× SSC at room temperature, followed by 2× SSC, 1X SSC, 0·5× SSC and 0·1× SSC at 65 °C. One thousand and 600 pfu were screened and positive clones were isolated and excised into plasmid. One positive clone showing high similarity to GRP sequences from ticks after DNA sequencing and DNA database comparison (BLASTX, NCBI) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was selected and the respective putative protein named RmGRP (GenBank accession number KY271084).
RmGRP sequence analysis
Similar amino acid sequences to RmGRP were obtained in protein databases using BLASTP (NCBI) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and multiple alignment was made with CLUSTALW (http://embnet.vital-it.ch/software/ClustalW.html; Thompson et al. Reference Thompson, Higgins and Gibson1994). BLASTP was also used to search a R. microplus transcriptome database (Rm-INCT-EM) produced using Illumina Sequencing technology (BioProject ID PRJNA232001 at Transcriptome Shotgun Assembly database – GenBank). Molecular weight and isoelectric point (pI) were determined using Compute MW/pI (http://web.expasy.org/cgi-bin/compute_pi/pi_tool; Gasteiger et al. Reference Gasteiger, Hoogland, Gattiker, Duvaud, Wilkins, Appel, Bairoch and Walker2005) and amino acid composition was analysed by ProtParam (http://web.expasy.org/protparam/; Gasteiger et al. Reference Gasteiger, Hoogland, Gattiker, Duvaud, Wilkins, Appel, Bairoch and Walker2005). Secondary structure and signal peptide prediction were performed by GOR4 IV (https://npsa-prabi.ibcp.fr/cgi-bin/secpred_gor4.pl; Garnier et al. Reference Garnier, Gibrat, Robson and Doolittle1996) and SignalP 4·1 (http://www.cbs.dtu.dk/services/SignalP/; Petersen et al. Reference Petersen, Brunak, Heijne and Nielsen2011). N-glycosylation, O-glycosylation and Ser, Thr and Tyr phosphorylation sites were predicted using NetNGlyc (http://www.cbs.dtu.dk/services/NetNGlyc/), NetOGlyc (http://www.cbs.dtu.dk/services/NetOGlyc/; Steentoft et al. Reference Steentoft, Vakhrushev, Joshi, Kong, Vester-Christensen, Schjoldager, Lavrsen, Dabelsteen, Pedersen, Marcos-Silva, Gupta, Bennett, Mandel, Brunak, Wandall, Levery and Clausen2013) and NetPhos (http://www.cbs.dtu.dk/services/NetPhos/; Blom et al. Reference Blom, Gammeltoft and Brunak1999), respectively.
cDNA synthesis and qRT–PCR
RNA extraction was performed using TRIzol (Invitrogen), as recommended by the manufacturer, from the following tissue samples: guts, salivary glands, ovaries and fat bodies of partially and fully engorged adult females; whole adult males; 1-, 5-, 10- and 15-day-old larvae; 3-, 6-, 12-, 18- and 21-day-old eggs. All dissections and tissue macerations were performed as previously described (Leal et al. Reference Leal, Seixas, Mattos, Coutinho, Masuda, da Silva Vaz and Ferreira2013). RNA quantification and quality evaluation were determined by spectrophotometry at A280 nm and by the ratio A260/A280 nm. Reverse transcription was performed using 500 ng of RNA in each reaction and Superscript III reverse transcriptase (Invitrogen), following the manufacturer's recommendations. cDNA synthesized from partially and fully engorged females’ tissues, eggs, larvae and adult males RNA were submitted to qPCR. The primers targeting a 207-bp amplicon from the GRP's cDNA sequence (5′-CTTGCCCCCTGAGTCCTACA-3′ and 5′-TTCCACAGTGCAGGCTTCAA-3′) used in qRT–PCR were designed with the PrimerExpress 3·0 software (Applied Biosystems, Foster City, USA). Gene expression corresponding to rmgrp was normalized by the expression of the 40S ribosomal protein gene, which was amplified using the primers 5′-GGACGACCGATGGCTACCT-3′ and 5′-TGAGTTGATTGGCGCACTTCT-3′ (Pohl et al. Reference Pohl, Sorgine, Leal, Logullo, Oliveira, da Silva Vaz and Masuda2008). The qPCR reactions were performed in duplicates with Applied Biosystems Step One Plus thermocycler using Platinum® SYBR® Green qPCR SuperMix kit (Invitrogen), 10 pm primers and 100 ng cDNA. Relative Expression Software Tool (REST) was used for data analyses (Pfaffl et al. Reference Pfaffl, Horgan and Dempfle2002). The qPCR experiment was done in accordance with the MIQE Guidelines (Bustin et al. Reference Bustin, Beaulieu, Huggett, Jaggi, Kibenge, Olsvik, Penning and Toegel2010).
dsRNA synthesis and RNA interference
The RNA interference experiment was adapted from Pohl et al. (Reference Pohl, Klafke, Carvalho, Martins, Daffre, da Silva Vaz and Masuda2011). Forward and reverse primers (5′-GGATCCTAATACGACTCACTATAGGATGAAATTGTTGCTGGTCGC-3′ and 5′-GGATCCTAATACGACTCACTATAGGTGAAATGCGTTTGGAAGAT-3′, respectively), containing a T7 promotor sequence (underlined), were synthesized to amplify a 704 bp sequence of rmgrp. Primers were designed using the IDT RNAi oligo design tool (https://www.idtdna.com/site/order/designtool/index/DSIRNA_CUSTOM), indicating potential dicer recognition sites in the target sequence. The specificity of the designed double-stranded RNA (dsRNA) was verified by BLAST searches against the GenBank database and R. microplus transcriptome sequences. Elongase Enzyme Mix (Invitrogen), 0·6 pm primers and 100 ng cDNA were used in the PCR reactions. cDNA was purified with Geneclean II kit (MP Biomedicals) and used as template to double-stranded RNA (dsRNA) synthesis with Ribomax Express T7 kit (Promega), according to the manufacturer's instructions. The dsRNA quantification was spectrophotometrically determined using A260 nm and diluted in Milli-Q water to 8 µg µL−1. The dsRNA corresponding to Plasmodium falciparum MSP1 gene was also equally synthesized to be used as negative control. Forward (5′-TAATACGACTCACTATAGGCTGATGCAAGCGATTCAGAT-3′) and reverse (5′-TAATACGACTCACTATAGGGTGTATTTCCAGAATTGGCC-3′) primers used to amplify msp1 were supplied by Dr Gerhard Wunderlich (Departamento de Parasitologia – Universidade de São Paulo, SP-Brazil).
Partially engorged females were collected from an infested bovine and 28 ticks between 20 and 45 mg were then selected. All individuals were placed with the dorsal region down on a flat surface using a double-sided tape. Artificial feeding was performed according to the method of Fabres et al. (Reference Fabres, de Andrade, Guizzo, Sorgine, Paiva-Silva, Masuda, da Silva Vaz Silva and Logullo2010). Briefly, a glass capillary with cattle whole blood and citrate (anticoagulant) was inserted into the mouthparts of each tick, which were fed for 1 h under 28 °C with 85% relative humidity. The partially engorged females were distributed in two groups of 14 ticks and carefully injected with 1 µL (8 µg) of dsRNA corresponding to rmgrp or msp1 (non-related gene as negative control) using a Hamilton syringe. Following dsRNA injection, ticks were artificially fed until the next day. After 24 h of dsRNA administration, four ticks of each group were dissected, being ovaries and salivary glands removed and stored in TRIzol at −70 °C. The remaining 10 females were reweighed, fixed using a double-sided tape with the dorsal region down in Petri dishes, and kept for seven days in a culture chamber at 28 °C with 85% relative humidity to oviposition. Eggs were weighted and placed in microtubes with the lid open, closed with cotton and incubated in the same conditions mentioned above until hatching. Larvae were weighed after separation from eggshells and unhatched eggs.
Ovaries from partially engorged females, isolated 24 h after dsRNA injection, were processed individually for RNA extraction, whereas the four salivary glands were combined in a pool for protein extraction. Proteins and RNAs were extracted using TRIzol (Invitrogen). RNAs were treated with DNAse I (Invitrogen) before cDNA synthesis, which was performed as already described in the item cDNA synthesis and qRT–PCR. Gene expression knockdown was confirmed by qRT–PCR using Rotor-Gene Q thermocycler (Qiagen). Reactions were performed in duplicate using Go-Taq qPCR Master Mix (Promega), 300 ng cDNA and 10 pm primers. Data were analysed by REST as described in the item cDNA synthesis and qRT–PCR. RmGRP presence in salivary glands of silenced ticks was analysed by Western blot.
Cloning, expression and purification of the rRmGRP
The DNA sequence encoding RmGRP was amplified by PCR using the isolated cDNA library plasmid DNA as template, and forward (5′-TTTTTGCATATGAAATTGTTGCTGGTCGCAG-3′) and reverse (5′-TTTTTGCTCGAGTTAGTGGTGGTGGTGGTGGTGATTGTTCCTACGGTTGCCG-3′) primers containing restrictions sites for NdeI and XhoI (underlined). Reverse primer was designed with a 6× Histidine tag encoding sequence (bold). The PCR product was cloned into PCR-Blunt vector using Zero Blunt PCR Cloning kit (Invitrogen) and the recombinant plasmid was cleaved with the restriction enzymes. The resulting fragment was inserted into the vector pET 23a (Novagen) using T4 DNA ligase (Fermentas) and the construction was electroporated into Escherichia coli BL21 Star strain cells.
One colony of BL21/pET 23a-RmGRP was grown in 5 mL of Luria–Bertani (LB) medium with 50 µg mL−1 ampicillin overnight at 37 °C with shaking at 170 rpm. The culture was added in 1000 mL of the LB medium with 50 µg mL−1 ampicillin and 1 mm Isopropyl β-D-1-thiogalactopyranoside (IPTG). Protein expression was performed during 5 h at 37 °C in a shaker at 170 rpm. Cells were centrifuged at 5000 × g for 5 min at 4 °C. The pellet was resuspended in 100 mL of PBS and disrupted in a French press. The culture was centrifuged at 18 000 × g for 20 min at 4 °C. rRmGRP was detected only in insoluble fraction, thus the supernatant was discarded and the pellet was resuspended in 50 mL of buffer containing 8 M urea, 10 mm Tris–HCl, 1 mm dithiothreitol, pH 8·0. The resuspended pellet was incubated for 1 h at room temperature with stirring and filtered using a 0·45 µm membrane filter (adapted from Jiz et al. Reference Jiz, Wu, Meng, Pond-Tor, Reynolds, Friedman, Olveda, Acosta and Kurtis2008).
rRmGRP was purified by affinity chromatography using a HisTrap column (GE Healthcare) according to the manufacturer's instructions. Urea 8 M was added to the binding and elution buffers. Protein purity was monitored by 15% SDS–PAGE stained with Coomassie Brilliant Blue R (Sigma-Aldrich). Recombinant protein identity was verified by Western blot using anti-polyhistidine alkaline phosphatase-conjugated IgG (Sigma-Aldrich). Recombinant protein concentration was estimated by spectrophotometry at A280 nm.
Antigen preparation and rabbit immunization
Fully engorged female ticks were obtained after spontaneous detachment from the host while males and partially engorged females (25–150 mg) of R. microplus were manually removed from cattle and washed with PBS. Salivary glands from partially and fully engorged females, guts and ovaries from fully engorged females were dissected using a scalpel blade, separated of other tissues with a fined-tipped forceps and washed with PBS. Five-days-old larvae and eggs collected in the 15th and 18th day after oviposition were homogenized with a tissue grinder as described by da Silva Vaz et al. (Reference da Silva Vaz, Logullo, Sergine, Veloso, Rosa de Lima, Gonzales, Masuda, Oliveira and Masuda1998). Protein extracts were prepared according to the method of da Silva Vaz et al. (Reference da Silva Vaz, Ozaki and Masuda1994). Briefly, tissues were sonicated (Ultrasonicator Cole Parmer 4710, 500 W, 4 and 20% duty cycle) for 30 s four times in an ice bath and solubilized in extraction buffer (0·5% sodium deoxicolate, 0·1% pepstatin A, 0·1% leupeptin, 0·1 mm N-tosyl-L-phenylalanine chloromethyl ketone, 10 mm tris buffer, pH 8·2). After incubation for 15 min in an ice bath, the material was centrifuged at 32 000 × g for 40 min and supernatants stored at−70 °C. Salivation was induced by injection of 5 µL of 2% pilocarpine solution in a wet chamber and saliva was collected for a period of 2 h directly from tick mouthparts. Recombinant Boophilus yolk pro-cathepsin (rBYC) was produced according to the method of Leal et al. (Reference Leal, Pohl, Ferreira, Nascimento-Silva, Sorgine, Logullo, Oliveira, Farias, da Silva Vaz and Masuda2006).
Rabbits were immunized nine times with varying volumes (200 µL to 1 mL) of saliva from partially and fully engorged females, while rabbit immunizations with protein extracts of salivary glands from partially engorged females, eggs and guts from fully engorged females were performed according to the method of Canal et al. (Reference Canal, Maia, da Silva Vaz, Chies, Farias, Masuda, Gonzales, Ozaki and Dewes1995). Briefly, protein extracts emulsified with Freund's complete adjuvant and additional boosters with antigen emulsified in Freund's incomplete adjuvant were inoculated in rabbits. Sera were collected fifteen days after the last booster. The anti-rRmGRP antiserum was generated by the immunization of a rabbit seven times at 14-days intervals with 100 µg of rRmGRP emulsified in Freund's incomplete adjuvant. Serum was collected 15 days after the last booster. Anti-rBYC serum was raised in rabbit as already described (Leal et al. Reference Leal, Pohl, Ferreira, Nascimento-Silva, Sorgine, Logullo, Oliveira, Farias, da Silva Vaz and Masuda2006).
Sera of infested bovines
Sera from six naturally infested and a non-infested Bos indicus (Nelore) were provided by the Departamento de Veterinária Preventiva, at the Universidade Federal de Pelotas (Brazil). Sera of three bovines B. taurus (Hereford) from an area free of R. microplus submitted to twelve successive experimental infestations were also used (Cruz et al. Reference Cruz, Silva, Mattos, da Silva Vaz, Masuda and Ferreira2008). Heavy infestations were performed with each calf being infested once a month for 6 months with 18 000 R. microplus larvae along the back, followed for 6 consecutive monthly light infestations of 800 R. microplus larvae. Sera were collected after each infestation. rBYC (used as an unrelated control protein; Supplementary Fig. 1A–C) and rRmGRP recognition by sera from naturally and experimentally infested bovines were performed by Western-blot.
Western blot
rRmGRP (1·7 mg cm−2), rBYC (1·7 mg cm−2) or tick/tissue extracts (50 µg well−1) were resuspended in sample buffer containing 62·5 mm Tris-HCl, pH 8, 0·001% bromophenol blue, 10% glycerol, 5% β-mercaptoethanol, 2% SDS and 8 M urea, separated by 12 or 15% SDS–PAGE and transferred to nitrocellulose membrane at 70 V for 1 h, according to the method of Dunn (Reference Dunn1986). Nitrocellulose membranes and strips of 4 mm were blocked for 2 h at room temperature with blocking buffer (cow non-fat dry milk 5% in PBS with 0·05% Tween 20). Sera were diluted in blocking buffer and adsorbed for 1 h at room temperature with E. coli BL21 strain lysate expressing pET 23a vector. The E. coli BL21 strain-pET 23a lysate was prepared according to the method of Rott et al. (Reference Rott, Fernández, Farias, Ceni, Ferreira, Haag and Zaha2000). Nitrocellulose membranes and strips were incubated with the sera overnight at 4 °C. After five washes for 10 min with blocking buffer, strips were incubated for 1 h with anti-IgG (bovine or rabbit) conjugated to peroxidase or alkaline phosphatase (Sigma-Aldrich) diluted 1:4000 in blocking buffer. After five washes with PBS 0·05% tween 20, ECL Western Blotting Substrate kit (Abcam) was used in membranes incubated with anti-IgG (bovine or rabbit) conjugated to peroxidase, while NBT and BCIP were used as substrate in membranes incubated with anti-rabbit IgG conjugated to alkaline phosphatase. The bands intensities were analysed using a Carestream Gel Logic 2200 PRO Imaging System and the associated Image Analysis Software.
Statistical analysis
Statistical analyses were performed using GraphPad Prism version 7.00 for Windows (GraphPad Software, La Jolla, California, USA) by one-way ANOVA and Tukey's post-test or Student's unpaired t-test. Values with significance of P < 0·05 were considered statistically different. Analyses were performed using two-tailed test.
Results
RmGRP sequence analysis
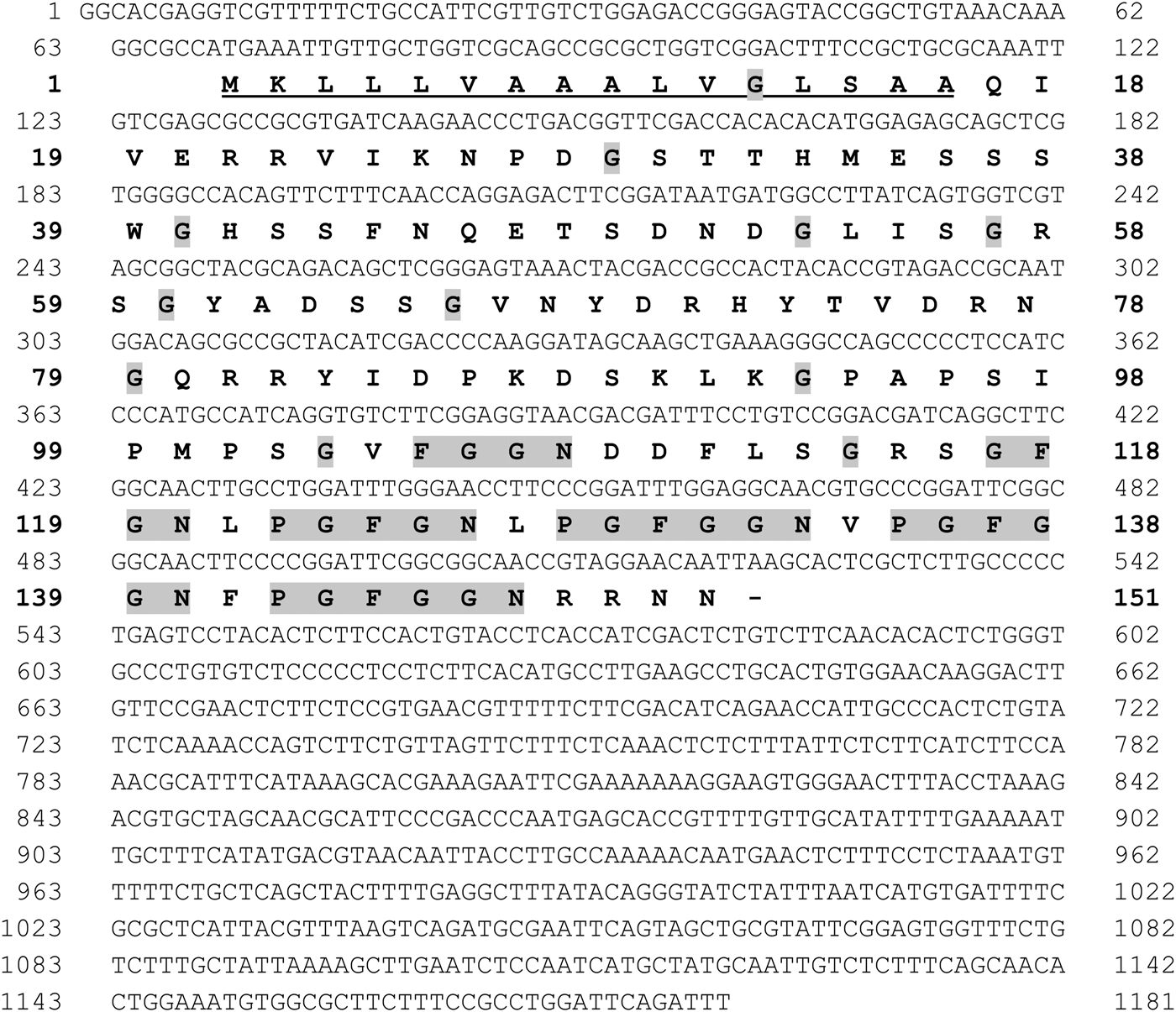
The cDNA sequence encoding RmGRP (1181 bp) was analysed showing the presence of an open reading frame of 456 bp (151 amino acids), including a putative signal peptide composed by 16 amino acids (see Fig. 1). The protein was predicted to have a molecular weight of 15·9 kDa and pI 9·1, consisting mainly of random coil (70%), and presenting 25 potential sites for phosphorylation and 21 sites for o-glycosylation. The amino acid composition shows high presence of glycine (17%), serine (11%) and asparagine (8%), but do not present cysteines. The distribution of prolines and glycines shows distinct profiles in the two moieties of the protein. The N-terminal half (amino acids 1–79) does not present proline and the glycine residues are distributed not forming any distinct pattern, whereas the C-terminal half (amino acid 80 to 151) shows a high proportion of prolines (12·5%) and most glycines are present in repeats, preceded or not by proline, as: FGGN, GFGN (two copies) and GFGGN (three copies) (Fig. 1). Alignment of RmGRP with GRPs showed high similarity with proteins of R. appendiculatus, R. puchellus, Amblyomma maculatum, A. cajennense, A. americanum and Ixodes scapularis, which present identities of 91, 91, 89, 89, 87 and 72%, respectively. The aligned amino acid sequences indicated five conserved repeats, whereas the FGGN sequence (residues 105–8) was not present in the other GRPs and two repeats PGFGGN (residues 128–133 and 135–140) were incomplete in I. scapularis, forming a single ‘PGFGN’ sequence (Fig. 2).
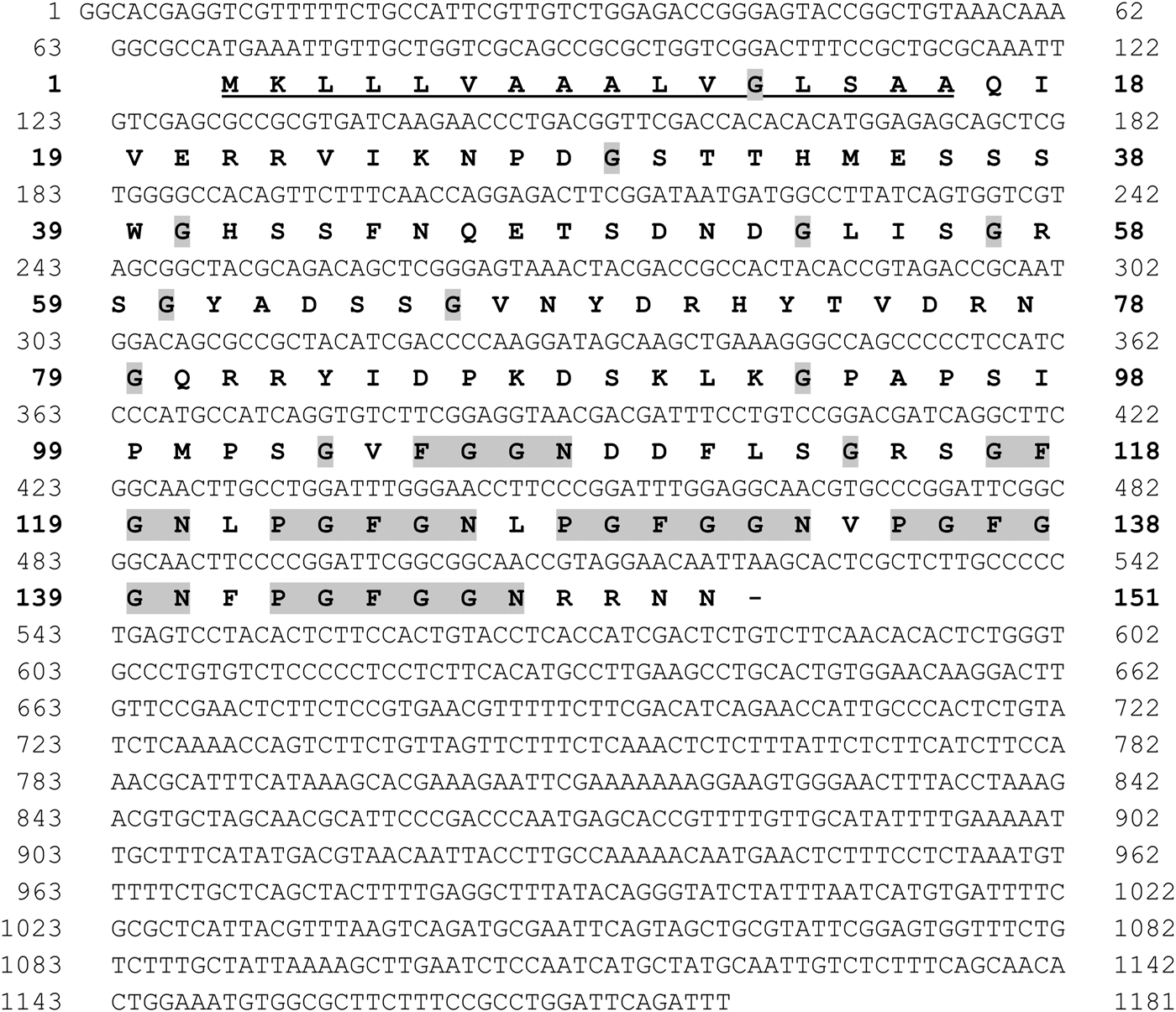
Fig. 1. Nucleotide and predicted amino acid sequence of the cDNA of a glycine-rich protein of Rhipicephalus microplus (RmGRP). Signal peptide is underlined. Glycine residues and the repeats FGGN, GFGN and GFGGN, preceded or not by P (Proline), are highlighted in gray.

Fig. 2. Multiple sequence alignment analysis. ClustalW was used to align RmGRP with glycine-rich proteins from four tick species. Repeats are highlighted in gray boxes. Accession numbers: Ixodes scapularis, AAY66550·1; Rhipicephalus microplus, KY271084; Rhipicephalus appendiculatus, JAP85432·1; Rhipicephalus pulchellus, JAA64016·1; Amblyomma americanum, EL593292·1; Amblyomma maculatum, AEO33043·1; Amblyomma cajennense, JAC24024·1.
Transcription profiling of rmgrp
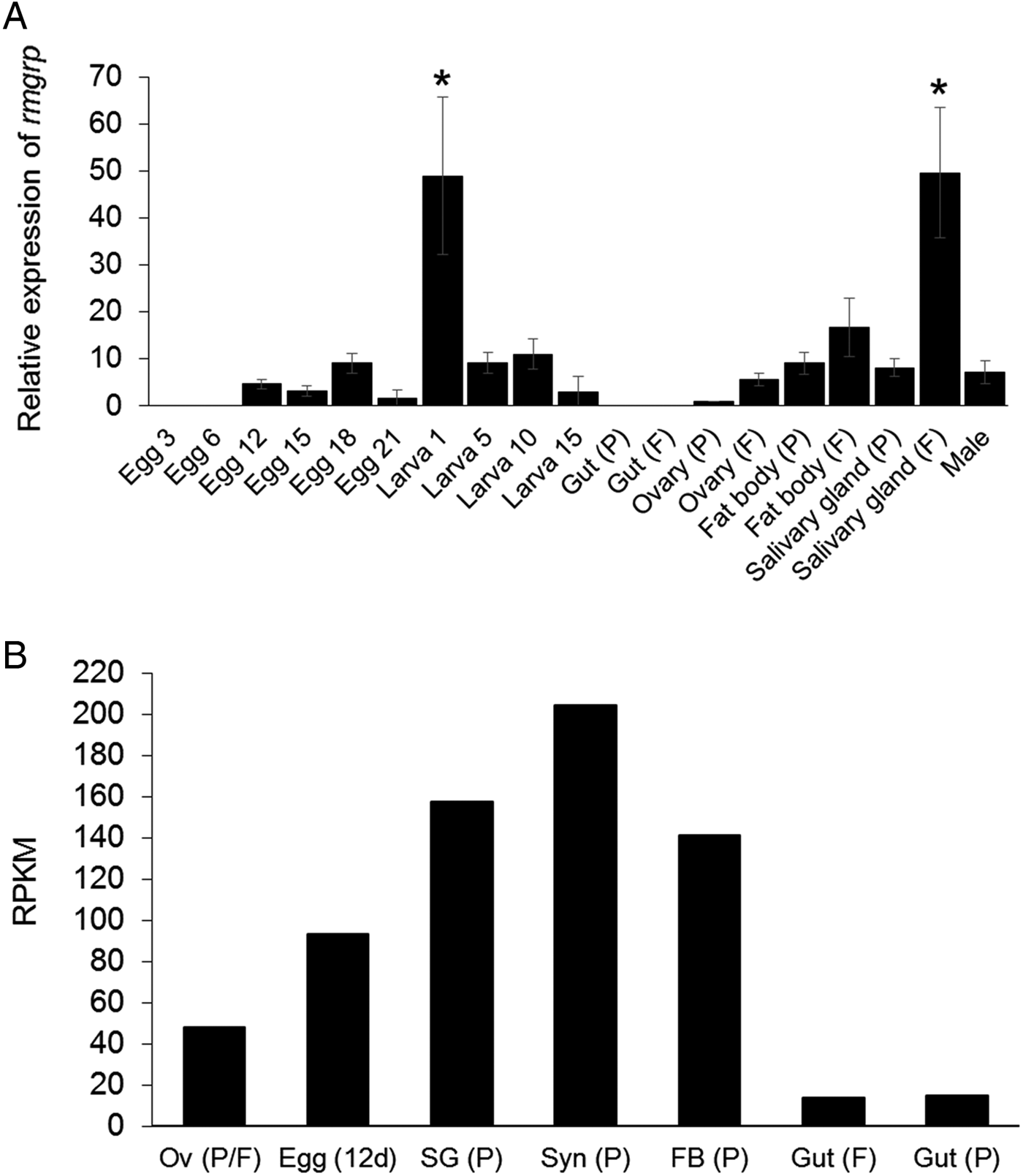
The transcription of rmgrp was detected by qRT–PCR in all tick developmental stages: eggs, larvae, adult females and males. However, rmgrp transcription was not detected in 3- and 6-day-old eggs and in the guts of partially and fully engorged females. The highest relative transcription levels were identified in 1-day-old larvae and salivary glands of fully engorged females (P < 0·01). rmgrp transcription levels showed to be higher in salivary glands (P = 0·0004), fat bodies and ovaries of fully engorged comparing to partially engorged females (Fig. 3A), although the differences concerning the last two organs did not reach statistical significance.
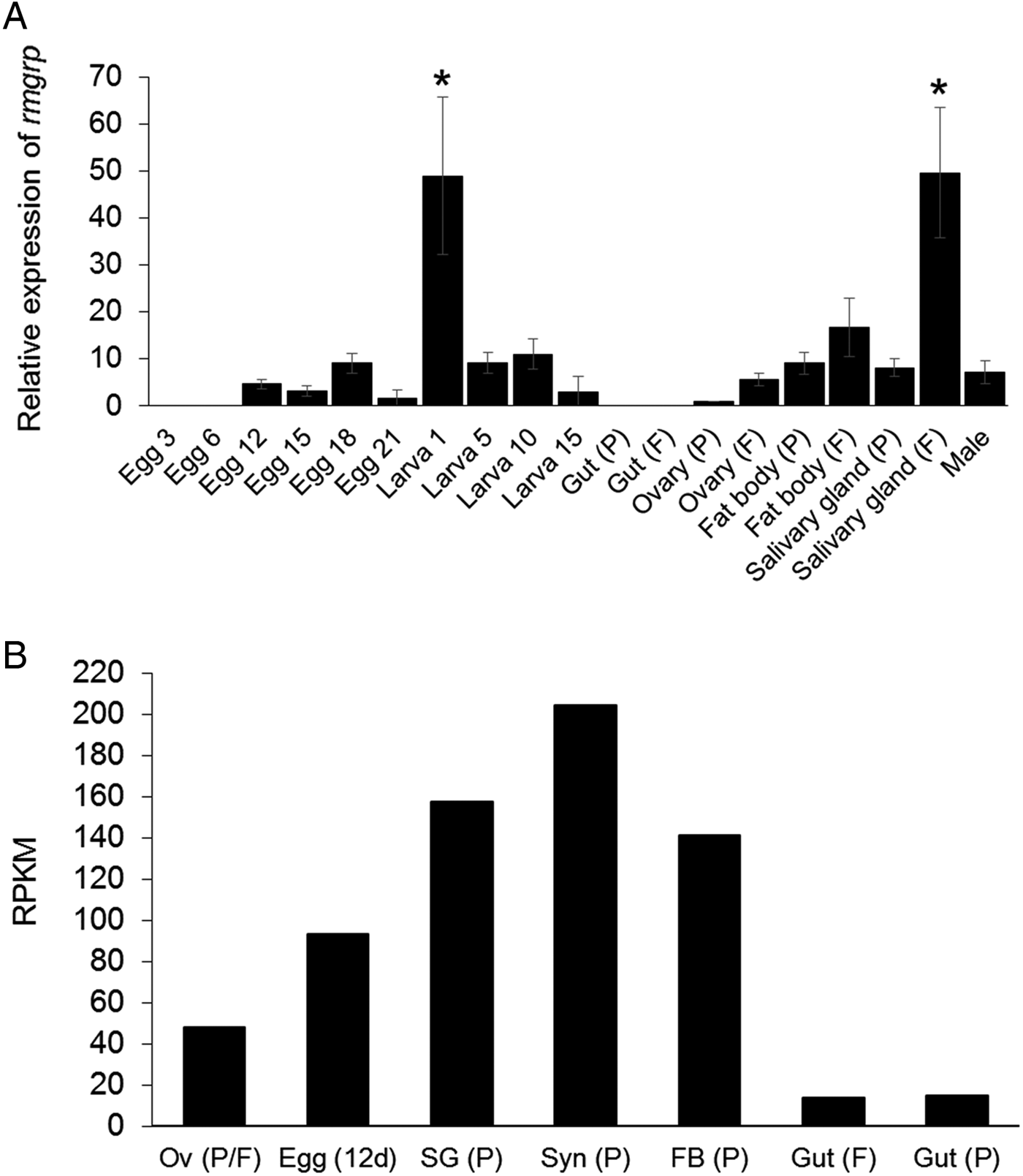
Fig. 3. Transcription profiling of rmgrp. (A) Analysis of relative rmgrp transcript levels in eggs, larvae, partially and fully engorged female tissues and males of Rhipicephalus microplus by qRT–PCR. P: Partially engorged females; F: Fully engorged females. Eggs and larvae are represented by days after laying and hatching, respectively. rmgrp transcription was normalized using the 40S ribosomal gene. Data are expressed as average ± s.d. The asterisks (*) indicate means statically different from others. Statistical analyses were performed by one-way ANOVA and Tukey post-test (P < 0·01). (B) Analysis of relative rmgrp transcript levels by Blastp comparison of the RmGRP sequence with a transcriptome database of different adult tissues/developmental stages of R. microplus (Rm-INCT-EM) produced using Illumina Sequencing technology (BioProject ID PRJNA232001 at Transcriptome Shotgun Assembly database – GenBank). Data are expressed as Reads Per Kilobase Million (RPKM). Messenger RNA of each transcriptome was obtained from: Ov (P/F), ovaries (mixed of partially and fully engorged females); Egg (12d), eggs 12 days after oviposition; Gut (F), guts of fully engorged females; and Syn (P), synganglia, SG (P), salivary glands, FB (P), fat bodies and Gut (P), guts of partially engorged females. Partially engorged female ticks were manually removed from cattle, while fully engorged female ticks were obtained after spontaneous detachment from the host.
In addition to the qRT–PCR analysis, a database from tick protein sequences deduced from contig assembly of a R. microplus transcriptome from ovaries (mixed of partially and fully engorged females), guts of fully engorged females, synganglia, salivary glands, fat bodies and guts of partially engorged females and 12-day-old eggs was used to quantify rmgrp transcripts (Fig. 3B). All tissues analysed showed the presence of the rmgrp sequence, including guts from partially and fully engorged females, although these tissues presented lower transcript expression levels, as showed by RPKM normalization (reads per kilo base per million mapped reads) (Mortazavi et al. Reference Mortazavi, Williams, McCue, Schaeffer and Wold2008).
Cloning, expression and purification of RmGRP
Integrity of the RmGRP coding nucleotide sequence insertion in the pET23a vector was confirmed by DNA sequencing and respective protein expression was confirmed by 15% SDS–PAGE and Western blot (Fig. 4A and B). The relative migration of rRmGRP indicated a molecular mass of about 19 kDa, while the predicted size was 16·7 kDa (15·9 kDa of RmGRP plus 0·8 kDa of 6× His tag). The Western-blot analysis also shows that the anti-His tag antibody recognized equally the recombinant protein in purified and non-purified forms.
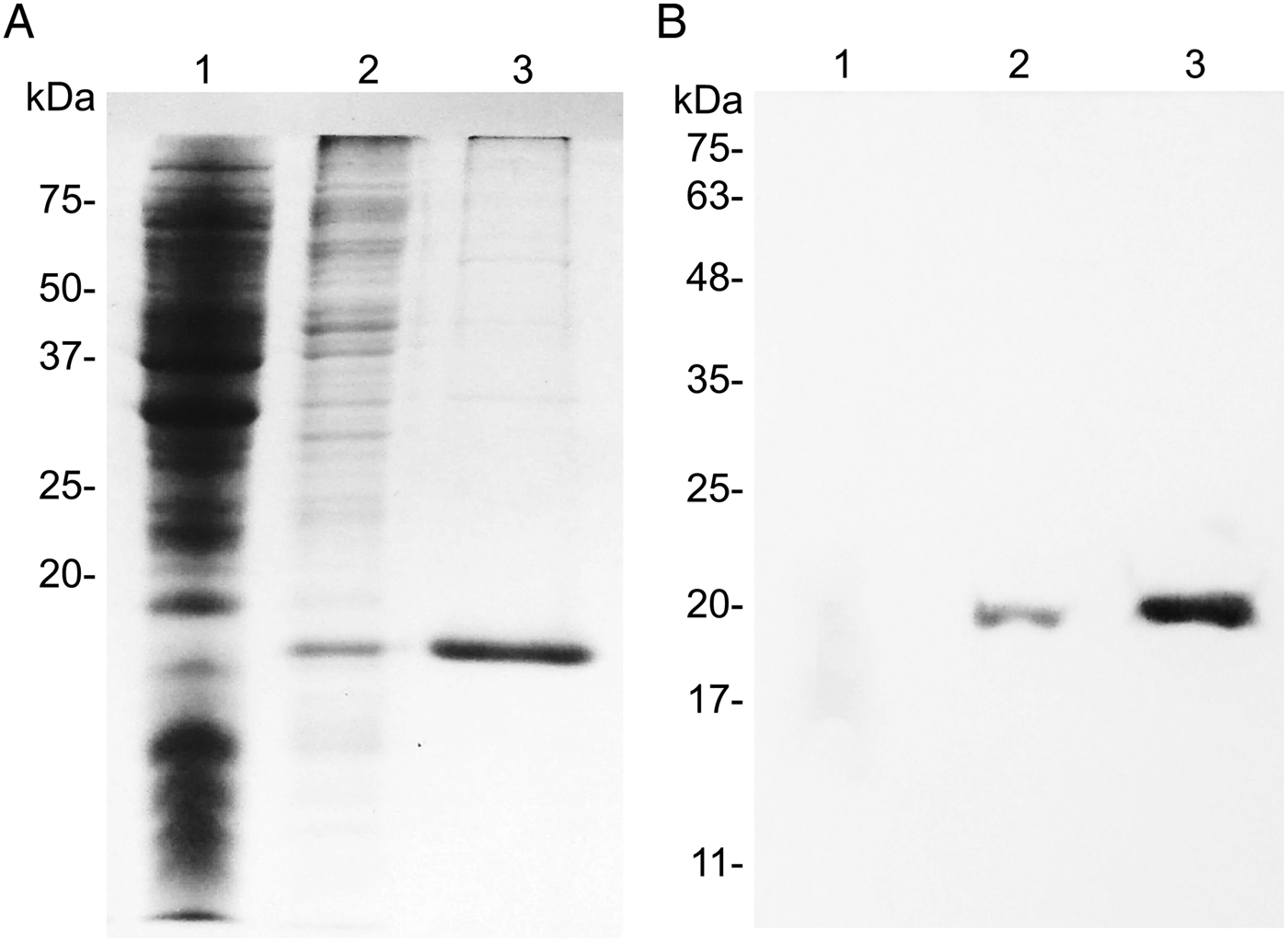
Fig. 4. SDS–PAGE (A) and Western blot (B) analysis of recombinant RmGRP. 1: E. coli BL21/pET 23a; 2: RmGRP expressed in E. coli BL21; 3: rRmGRP purified. Western blot was performed using Anti-PolyHistidine Alkaline Phosphatase (Sigma-Aldrich) diluted 1:4000. Precision Plus Protein Standards (Bio-Rad) and BLUeye Prestained Protein Ladder (Sigma-Aldrich) were used as molecular weight markers in (A) and (B), respectively.
RmGRP gene silencing
RNAi experiment was performed separating ticks in two groups: injected with dsRmGRP or dsMSP1 (control). After 24 h of the treatment, ticks of each group were dissected, being ovaries and salivary glands individually separated. RNA of ovaries was used to evaluate the rmgrp silencing level by qRT–PCR and protein extract of pooled salivary glands was used to verify RmGRP depletion in silenced ticks by Western blot. The remaining ticks of each group were placed to oviposition. Eggs and larvae weight, as well as weight gain of females 24 h after treatment, were measured and compared between groups. The fraction of eggs hatched in larvae (hatching rate) was determined as the ratio of total weight of hatched larvae by total weight of eggs laid. Gene silencing was observed in three of four ticks treated with dsRmGRP, which showed a reduction of 12, 70 and 96% in rmgrp expression (Fig. 5A). RmGRP depletion was observed at the protein level, with molecules of ~24 and ~16 KDa showing decreased recognition in salivary glands from silenced ticks (Fig. 5B). The weight gain of partially engorged females 24 h after dsRNA injection and eggs weight were not statistically different between groups (injected with dsRmGRP or dsMSP1), but the weight of hatched larvae was lower in the group treated with dsRmGRP in comparison with the control (P = 0·0007). Hatching rate was 55·8% in the control group (treated with dsMSP1) and 8·6% in the group injected with dsRmGRP, representing a larvae weight reduction of 91% (Table 1). Tick mortality was not observed after dsRNA treatment (data not shown).
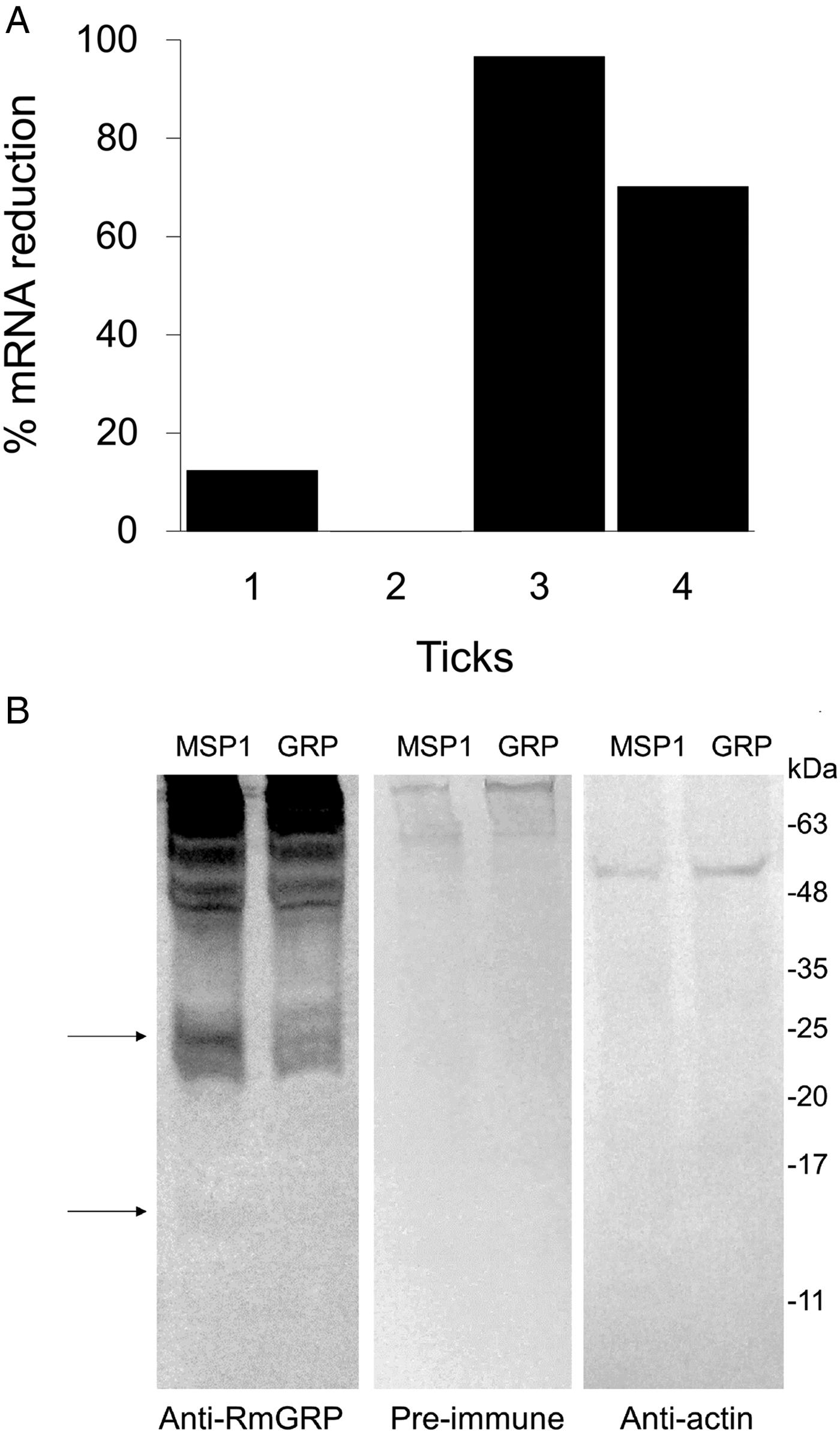
Fig. 5. RNAi-mediated rmgrp silencing in ovary (A) and salivary gland (B) of Rhipicephalus microplus adult females injected with dsRmGRP or dsMSP1 (control). Ovaries extracted 24 h after treatment were used to determine the suppression of mRNA corresponding to RmGRP of each treated tick (1, 2, 3 and 4). mRNA expression was analysed by qRT–PCR using the comparative Ct method, which was normalized using the 40S ribosomal gene. Percentage of mRNA reduction was calculated by comparison of rmgrp transcription from ticks treated with dsMSP-1 or dsRmGRP (A). Salivary glands extracted 24 h after dsMSP1 or dsRmGRP injection were used to verify RmGRP presence after rmgrp silencing. Western blot was performed using anti-RmGRP rabbit serum (dilution 1:4000), pre-immune serum (1:4000) and polyclonal antibody anti-actin (1:2000). Arrows indicate bands showing reduced recognition in silenced ticks (B).
Table 1. Effect of gene silencing by treatment of partially engorged females with dsRNAs of genes coding for a glycine-rich protein of Rhipicephalus microplus (RmGRP)

*Merozoite surface protein-1 of Plasmodium falciparum (negative control).
**Statistically significant (Statistical analyses were performed by unpaired t-test (P < 0·001)).
Weight gain (mg), weight gain (%), egg weight (mg) and larvae weight (mg) are presented as average ± s.d.
a final weight – initial weight (before artificial feeding).
b weight gain (mg) × 100/initial weight.
c (100-(egg weight of each tick treated with dsRmGRP × 100/egg weight mean of dsMSP-1 group)).
d [100-(larvae weight of each tick treated with dsRmGRP × 100/larvae weight mean of dsMSP-1 group)].
e (total larvae weight/total eggs weight) × 100.
Identification of RmGRP within tick tissues and developmental stages
The antiserum against rRmGRP was used in a Western-blot analysis with tick saliva and different adult tissues/developmental stages, as can be seen in Fig. 6. The number of molecules recognized suggests the presence of cross-reactivity to other proteins, possibly other GRPs. The molecule of ~24 kDa depleted in the RNAi experiment is present in salivary glands from fully engorged females and saliva from partially engorged females, indicating the RmGRP secretion into the host. Molecules of ~21 kDa compatible with RmGRP were also detected in ovaries and males.
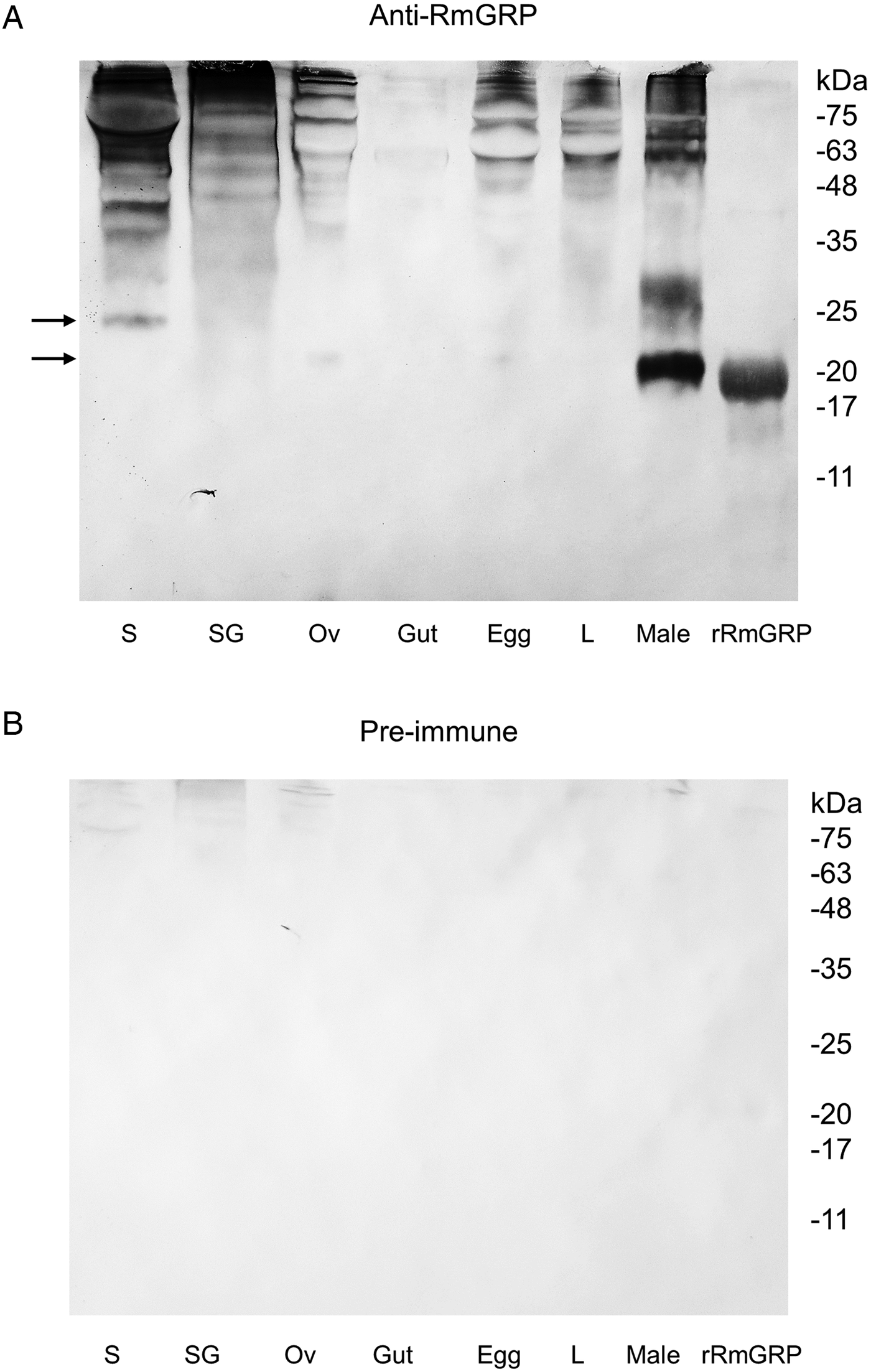
Fig. 6. Western-blot of R. microplus tissues and developmental stages extracts, saliva and purified rRmGRP. Anti-RmGRP rabbit serum (dilution 1:4000) (A) and pre-immune serum (1:4000) (B) were used as probe. Tissues and developmental stages analysed: saliva (S) of partially engorged females, salivary gland (SG), ovary (Ov) and gut of fully engorged females, 18-day-old egg, 5-day-old larva (L) and male extracts. rRmGRP was used as positive control. Arrows indicate molecules of ~21 and ~24 kDa. BLUeye Prestained Protein Ladder (Sigma-Aldrich) was used as molecular weight marker.
rRmGRP antigenicity evaluation
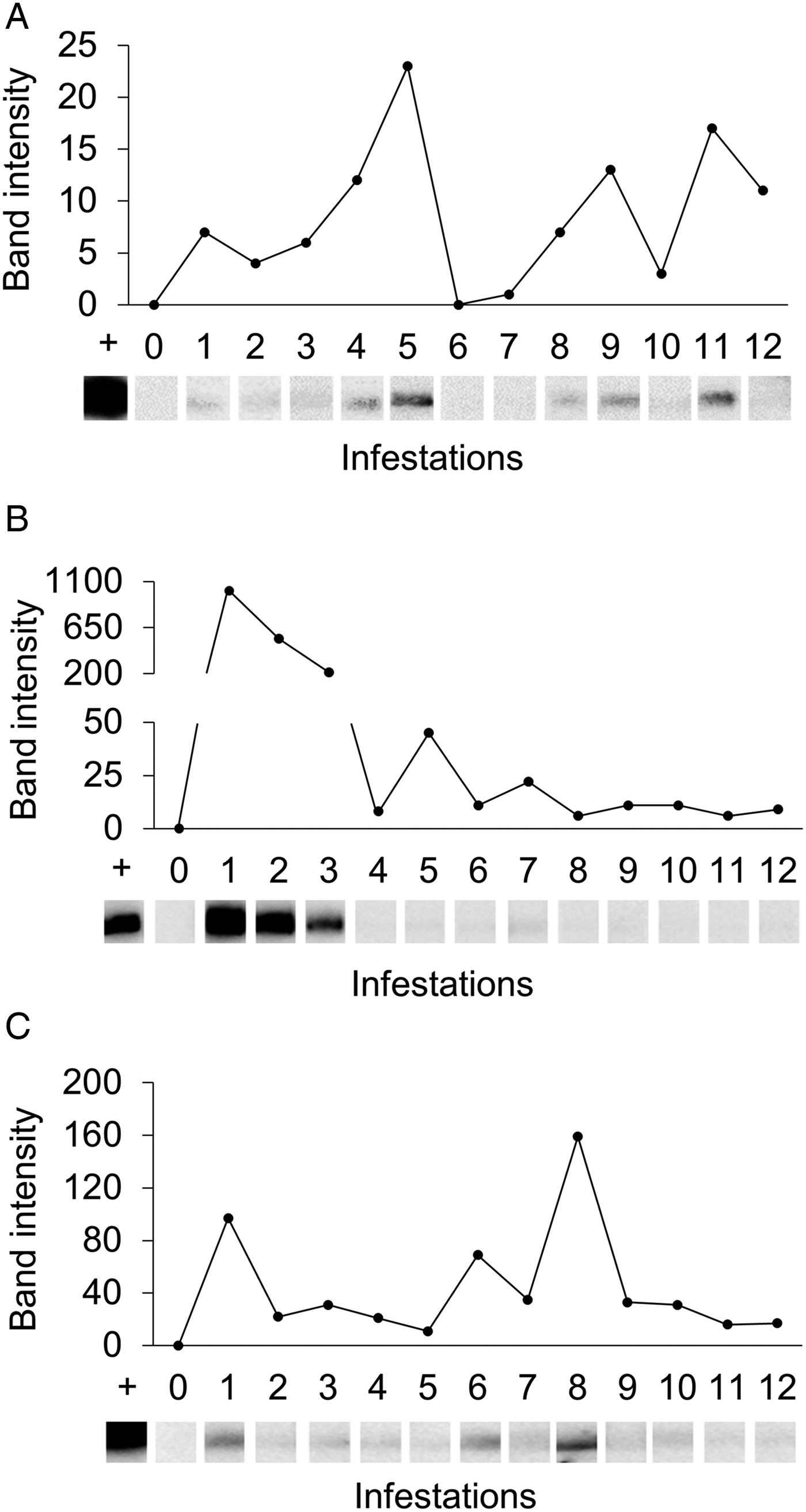
Western-blot analyses were performed in order to evaluate the antigenicity of RmGRP under natural and experimental infestations, as well as after rabbit immunization with protein extracts of instars, adult tissues or saliva. Fig. 7 shows the recognition of rRmGRP by immune rabbit sera raised against 15-day-old eggs, salivary glands of partially engorged females and saliva of partially and fully engorged females, while sera against fully engorged female guts did not. rRmGRP antibody recognition was also tested using sera from three B. taurus bovines submitted to twelve experimental successive infestations (six heavy infestations followed by six light infestations). All sera from bovines 2 and 3 recognized rRmGRP, while bovine 1 presented sera from infestations 6 (corresponding to the last heavy infestation) and 7 (corresponding to the first light infestation) that did not recognize rRmGRP under the conditions tested. The intensity of recognition also varied greatly among the sera of bovine 2, showing up to a 10-fold difference, with the highest intensity following infestation 1 and a gradual decline until infestation 3, with a major decline in the 4th serum infestation, which level of recognition was maintained in the remaining sera (Fig. 8B). The rRmGRP recognition by sera from bovines 1 and 3 did not exhibit a similar intensity difference of magnitude, although in bovine 3 sera a two-fold difference is present (Fig. 8C) and in bovine 1 is possible to perceive a slight gradual increase of band intensities from infestation 2 to 5 sera, without rRmGRP recognition after infestation 6 (Fig. 8A). Thus, the experimentally infested bovines presented heterogeneous rRmGRP recognition profiles by sera collected after both light and heavy infestations. rRmGRP was also recognized by sera from six naturally infested bovines B. indicus, presenting also variation in the recognition levels between individuals (Fig. 9).
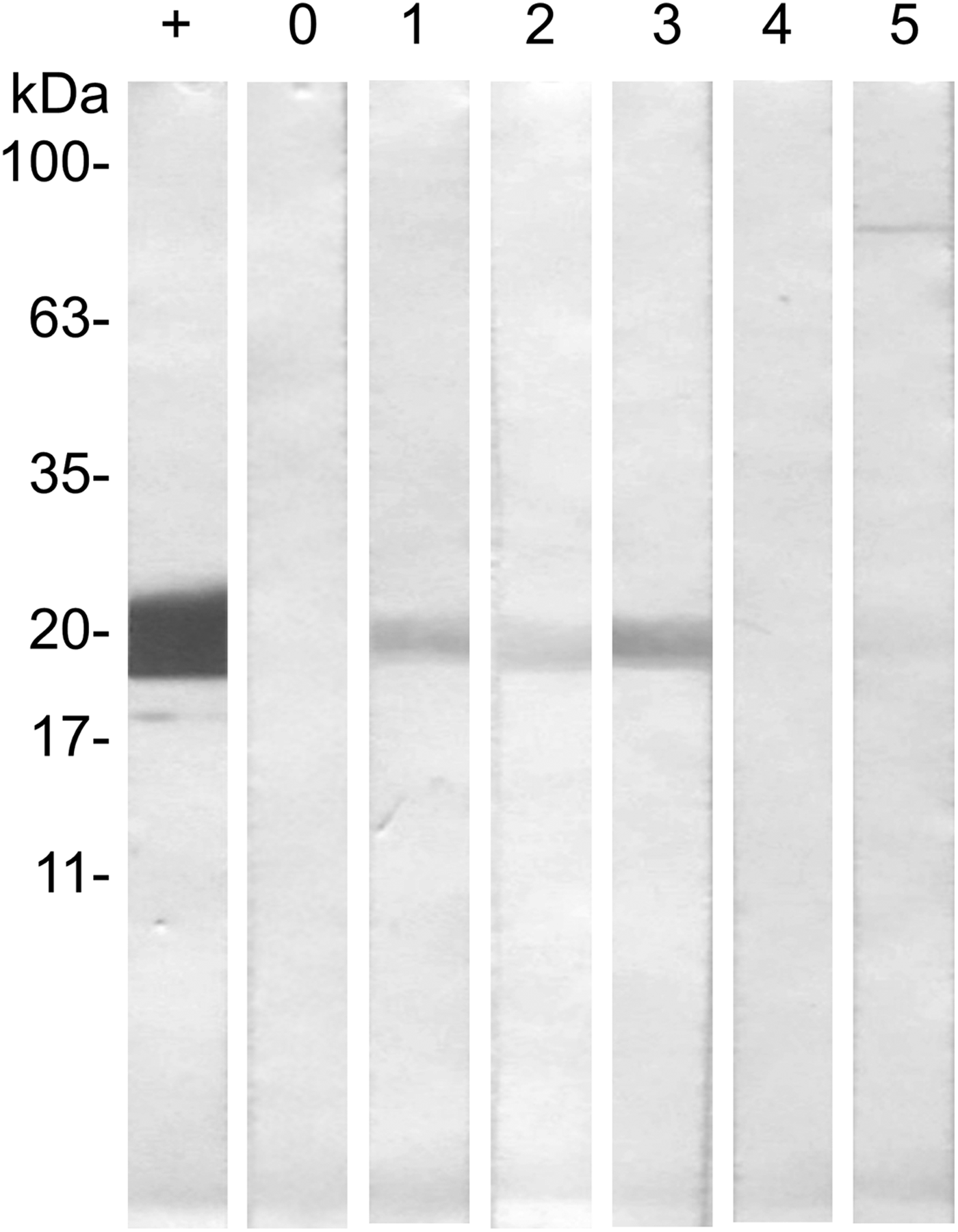
Fig. 7. Western blot of rRmGRP immunogenicity analysis by rabbit sera. rRmGRP was run on a 15% SDS–PAGE and transferred to a nitrocellulose membrane. Strips were probed against: +, Monoclonal Anti-polyHistidine conjugated with alkaline phosphatase (Sigma-Aldrich) (dilution 1:4000); 0, non-immune serum; 1, anti-saliva of partially engorged females serum; 2, anti-salivary glands of partially engorged females serum; 3, anti-saliva of fully engorged females serum; 4, anti-gut serum; 5, anti-egg serum. Sera were diluted 1:200. BLUeye Prestained Protein Ladder (Sigma-Aldrich) was used as molecular weight marker.
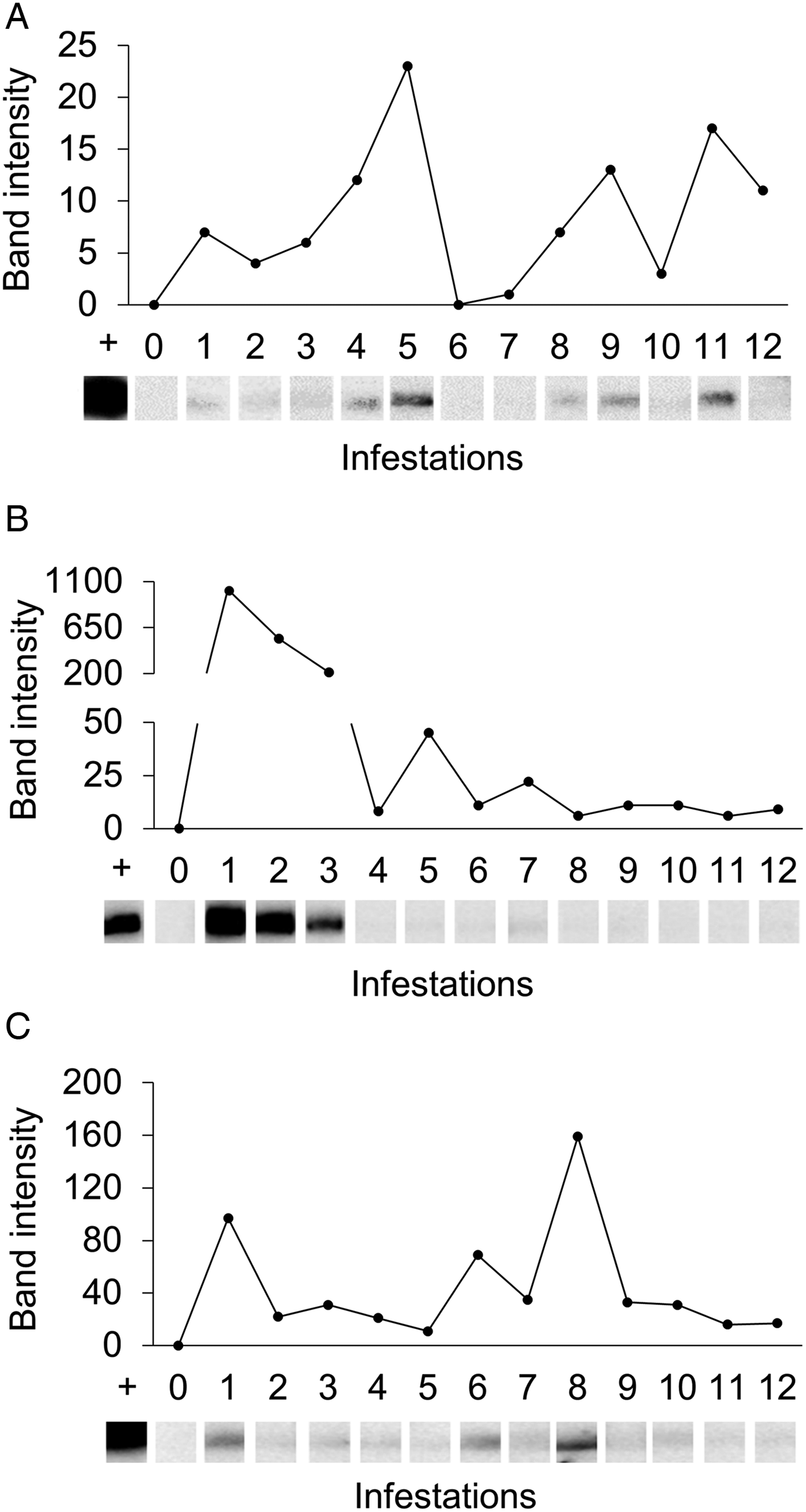
Fig. 8. Recognition of rRmGRP by sera of three experimentally infested Bos taurus. rRmGRP was run on a 15% SDS–PAGE and transferred to a nitrocellulose membrane. Strips were probed by the sera respective to each consecutive experimental infestation. Relative band intensities (infested serum value minus pre-infested serum value) are indicated graphically. +: positive control (anti-R. microplus saliva rabbit serum); 0: pre-infestation bovine sera; 1–12: bovine sera from twelve successive infestations (six heavy infestations followed by six light infestations). Sera of bovines 1 (A), 2 (B) and 3 (C) were diluted 1:250, 1:250 and 1:200, respectively. BLUeye Prestained Protein Ladder (Sigma-Aldrich) was used as molecular weight marker.
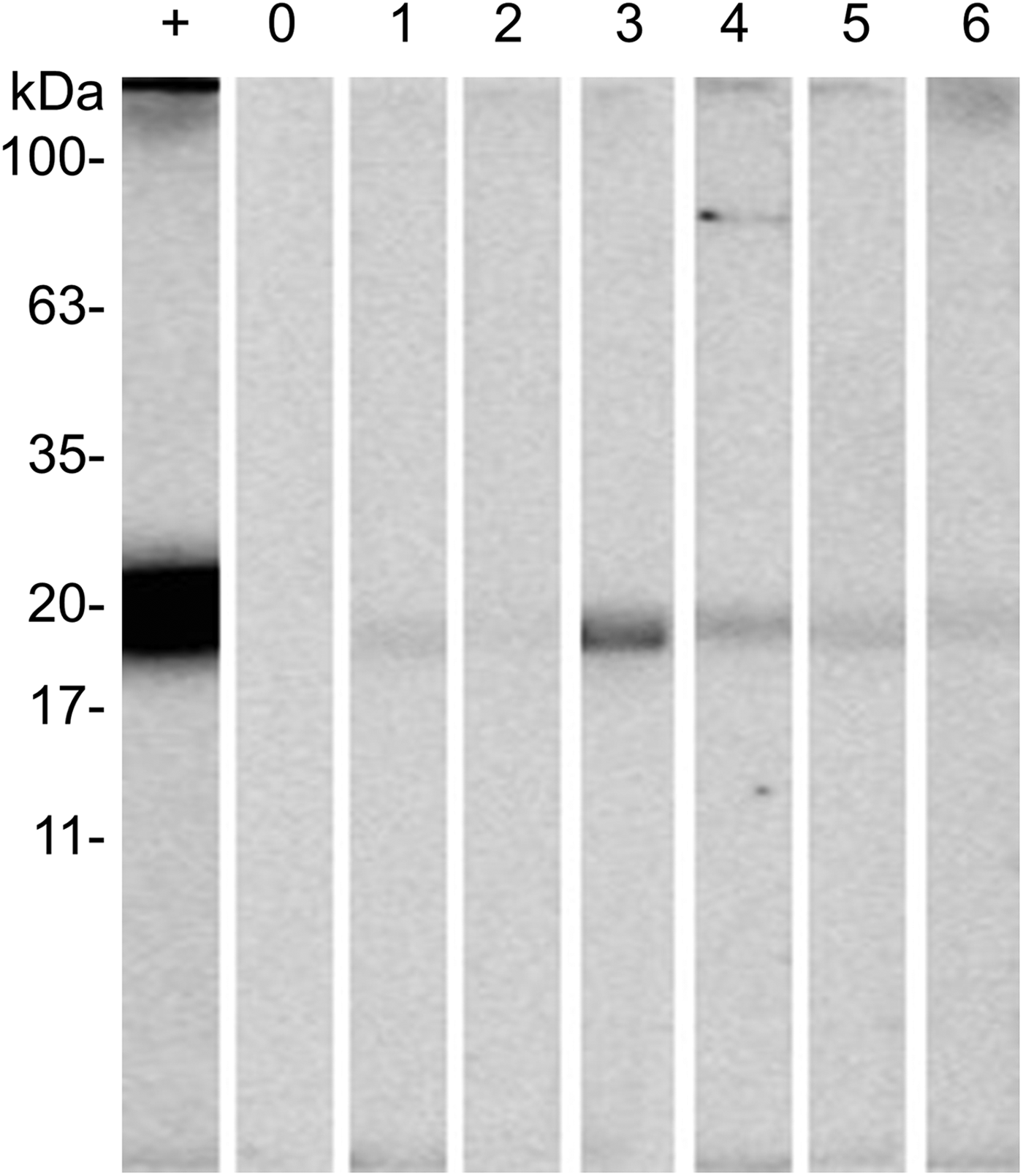
Fig. 9. Recognition of rRmGRP by naturally infested Bos indicus sera. rRmGRP was run on a 15% SDS–PAGE and transferred to a nitrocellulose membrane. Strips were probed against: +, positive control (anti-R. microplus saliva rabbit serum); 0, non-infested bovine serum; 1–6, naturally infested bovine sera. All sera were diluted 1:200. BLUeye prestained protein ladder (Sigma-Aldrich) was used as molecular weight marker.
Discussion
GRPs are a superfamily of proteins characterized by the presence of glycines in repeated motifs or isolated along the primary amino acid sequence and were described to present diverse roles. In plants, GRPs were described to act in cell wall structure, cell elongation, signal transduction, defense, RNA binding, as well as present antimicrobial activity (Mangeon et al. Reference Mangeon, Junqueira and Sachetto-Martins2010). In insects, GRPs are involved in cuticle construction (Andersen et al. Reference Andersen, Hojrup and Roepstorff1995; Zhong et al. Reference Zhong, Mita, Shimada and Kawasaki2006) and their expression were shown to be increased in response to stress induced by insecticides, making the cuticle more resistant in face of the new conditions (Zhang et al. Reference Zhang, Goyer and Pelletier2008). Glycine repeats were also detected in proteins from mollusc shells (Sudo et al. Reference Sudo, Fujikawa, Nagakura, Ohkubo, Sakaguchi, Tanaka, Nakashima and Takahashi1997), silkworms chorion proteins (Tsitilout et al. Reference Tsitilout, Rodakis, Alexopoulou, Kafatos, Ito and Iatroul1983) and in silk proteins of spiders (Yamaguchi et al. Reference Yamaguchi, Yamauchi, Gullion and Asakura2009; Yazawa et al. Reference Yazawa, Yamaguchi, Knight and Asakura2011). GRPs may also present distinct patterns of glycine distribution, presence of conserved motifs and signal peptide (Sachetto-Martins et al. Reference Sachetto-Martins, Franco and de Oliveira2000). In ticks, little is known about their functions. The salivary GRPs roles are usually suggested to be linked to the cement cone, which fixes tick mouthparts on the host, helping in its structure formation (Bishop et al. Reference Bishop, Lambson, Wells, Pandit, Osaso, Nikonge, Morzaria, Musoke and Nene2002; Bullard et al. Reference Bullard, Allen, Chao, Douglas, Das, Morgan, Ching and Karim2016b). RmGRP contains a putative signal peptide and is present in female salivary glands and pilocarpine-induced saliva, what indicates that this protein is secreted to the extracellular environment. In addition, RmGRP presents six glycine repeats of FGGN, GFGN or GFGGN, preceded or not by P (proline) in its C-terminal portion. These repeats are conserved in GRPs from R. appendiculatus, R. pulchellus, A. maculatum, A. cajennense, A. americanum, while five are present in I. scapularis. Plant GRPs present very similar repeats, including GGX, GGXXXGG, GXGX and GGX/GXGX (Mangeon et al. Reference Mangeon, Junqueira and Sachetto-Martins2010). Moreover, the amino acid sequence of a spicule matrix protein of Hemicentrotus pulcherrimus micromeres presents repeated regions as QPGFGNQPG[M/V]GG[R/Q/N] (Katoh-Fukui et al. Reference Katoh-Fukui, Noce, Ueda, Fujiwara, Hashimoto, Tanaka and Higashinakagawa1992), therefore including the PGFGN repeat found in RmGRP. Other spicule matrix proteins present the same organization of RmGRP, including a signal peptide and the presence of G[F/M/V]GG repeats preceded by proline in the half end (Livingston et al. Reference Livingston, Shaw, Bailey and Wilt1991), indicating that these glycine repeats can be present in proteins from evolutionary distant organisms. Among the roles suggested for matrix spicule proteins include tissue biomineralization during the sea urchin development (Killian et al. Reference Killian, Croker and Wilt2010). In ticks, a large variation is found in the glycine repeats of GRPs, including a ‘GLX’ triplet repeat domain in the N-terminus and ‘GSP’ in a central repeat block in R. appendiculatus GRPs (Bishop et al. Reference Bishop, Lambson, Wells, Pandit, Osaso, Nikonge, Morzaria, Musoke and Nene2002), ‘GYG’ in R. annulatus (Shahein et al. Reference Shahein, Abouelella, Hussein, Hamed, El-Hakim, Abdel-Shafy and Tork2013) and ‘GLX’ in a R. haemaphysaloides counterpart (Zhou et al. Reference Zhou, Gong, Zhou, Xuan and Fujisaki2006).
Cement proteins are essential to tick fixation and feed on the host, thereby are potential candidates to compose a vaccine against ticks (Mulenga et al. Reference Mulenga, Sugimoto, Ingram, Ohashi and Onuma1999; Zhou et al. Reference Zhou, Gong, Zhou, Xuan and Fujisaki2006). A truncated form of 64P protein (64trp3), isolated from R. appendiculatus, is an example of GRP that provided protection to the immunized host, increasing the mortality rate of adult ticks (Trimnell et al. Reference Trimnell, Hails and Nuttall2002, Reference Trimnell, Davies, Lissina, Hails and Nuttall2005). Havlíková et al. (Reference Havlíková, Roller, Koči, Trimnell, Kazimírová, Klempa and Nuttall2009) found variants in the 64P protein that can be explained by instability of repeated glycine motifs in the C-terminal region, which proved to be highly immunogenic but not protective. It is suggested that the conformation of this protein in saliva remains the C-terminal portion exposed to the host during the tick feeding and, as the glycine repeats are characteristics of vertebrate extracellular matrix proteins, their exposure could result in the evasion of the host's immune system (Trimnell et al. Reference Trimnell, Hails and Nuttall2002). RmGRP presents glycine repeats in its half end, as well as spicule matrix proteins of sea urchin (Katoh-Fukui et al. Reference Katoh-Fukui, Noce, Ueda, Fujiwara, Hashimoto, Higashinakagawa, Killian, Livingston, Wilt, Benson, Sucov and Davidson1991), silk proteins (Beckwitt et al. Reference Beckwitt, Arcidiacono and Stote1998) of spiders and some tick GRPs (Trimnell et al. Reference Trimnell, Hails and Nuttall2002; Ribeiro et al. Reference Ribeiro, Alarcon-Chaidez, Francischetti, Mans, Mather, Valenzuela and Wikel2006; Francischetti et al. Reference Francischetti, Meng, Mans, Guderra, Hall, Veenstra, Pham, Kotsyfakis and Ribeiro2008). Therefore, analogously to 64P, the C-terminal half of RmGRP is possibly immune-dominant and more exposed to the host. Moreover, the high degree of cross-reactivity showed in the Western-blot analysis of tick tissues/developmental stages may also be linked to a higher recognition of glycine repeat motifs. This feature may be especially important in the R. microplus–host relationship since an expressed sequence tags (ESTs) analysis detected a larger quantity of GRPs in R. microplus, when compared to R. sanguineus and A. cajennense (Maruyama et al. Reference Maruyama, Anatriello, Anderson, Ribeiro, Brandão, Valenzuela, Ferreira, Garcia, Szabó, Patel, Bishop and de Miranda-Santos2010). As R. microplus is monoxenous, it spends more days fixed and feeding on a same host than a heteroxenous species (Kemp et al. Reference Kemp, Stone, Binnington, Obenchain and Galun1982). Thereby, the presence of different types of GRPs may be strategic to evade the host's immune system, alternating the expression of these proteins during feeding. In this scenario, it is suggested that the host should mount an immune response against an initial exposed tick GRP, but then the synthesis of the first protein is diminished or turned off while the transcription of a new GRP is activated (Maruyama et al. Reference Maruyama, Anatriello, Anderson, Ribeiro, Brandão, Valenzuela, Ferreira, Garcia, Szabó, Patel, Bishop and de Miranda-Santos2010; Kim et al. Reference Kim, Tirloni, Pinto, Moresco, Yates, da Silva Vaz and Mulenga2016). In addition, it was suggested that Brevirostrata ticks tend to express more GRP genes than Longirostrata species (Maruyama et al. Reference Maruyama, Anatriello, Anderson, Ribeiro, Brandão, Valenzuela, Ferreira, Garcia, Szabó, Patel, Bishop and de Miranda-Santos2010; de Castro et al. Reference de Castro, de Klerk, Pienaar, Latif, Rees and Mans2016).
The RmGRP depletion by RNAi resulted in the diminished recognition of a ~24 kDa molecule in salivary glands, as well as of a ~16 kDa form, pointing to the possible presence of post-translational modifications in the native protein, what is consistent with the RmGRP sequence analysis for phosphorylation and glycosylation putative sites. Interestingly, the ~16 kDa RmGRP was not detected in fully engorged female salivary glands nor saliva. The recognition of the ~24 kDa RmGRP in saliva and salivary glands indicate that this RmGRP form is secreted into the host, in accordance with the presence of the signal peptide and the described secretion of tick salivary GRPs (Garcia et al. Reference Garcia, Gardinassi, Ribeiro, Anatriello, Ferreira, Moreira, Mafra, Martins, Szabó, de Miranda-Santos and Maruyama2014; Tirloni et al. Reference Tirloni, Reck, Terra, Martins, Mulenga, Sherman, Fox, Yates, Termignoni, Pinto and da Silva Vaz2014; Karim and Ribeiro, Reference Karim and Ribeiro2015). In this sense, the relative high transcription of rmgrp in salivary glands was expected, but the fact that rmgrp expression was significantly higher in salivary glands from fully engorged than partially engorged females deserves further attention. Adult female engorgement in R. microplus precedes its drop from the bovine, and, additionally, salivary glands degradation starts while the tick is still fixed to the host (Bennington, Reference Bennington1978; Nunes et al. Reference Nunes, Mathias and Bechara2006). Therefore, the high expression of rmgrp in salivary glands of fully engorged females, as well as in synganglia among partially engorged tick tissues, suggests additional roles for RmGRP possibly not strictly linked to the tick–host interaction. In this sense, Bullard et al. (Reference Bullard, Williams and Karim2016a) analysed mRNA expression levels of nine GRPs of A. americanum in unfed and fed ticks 24–168 h after attachment and verified distinct expression profiles during the blood meal, and mRNA of some proteins were detected in the end of the feeding period. Similarly, Kim et al. (Reference Kim, Tirloni, Pinto, Moresco, Yates, da Silva Vaz and Mulenga2016) identified GRPs in salivary glands of I. scapularis partially fed for 24, 48, 72, 96 and 120 h as well as in fully fed ticks detached and not detached from the host. They found the largest variety of GRPs in fully fed ticks detached from the host and a greater relative abundance of GRPs in fully fed ticks collected on the host. Therefore, different tick species seem to overexpress some GRPs upon blood repletion, as could be seen for rmgrp. High expression levels were also presented by rmgrp in larvae, what may be consistent with a function of RmGRP in the cement cone formation (Harnnoi et al. Reference Harnnoi, Sakaguchi, Xuan and Fujisaki2006; Bullard et al. Reference Bullard, Allen, Chao, Douglas, Das, Morgan, Ching and Karim2016b). Conversely, it must be emphasized that the cement cone tends to be structured until 24 h of fixation (Bennington, Reference Bennington1978), therefore high levels of mRNA transcription to cement proteins are not likely to be present in partially and fully engorged adult females. The particular high rmgrp mRNA levels detected in 1-day-old larvae can also be associated with the results found in the RNAi experiment. Although no effect could be seen in treated ticks in mortality or oviposition, a significantly low hatching rate was verified, indicating a likely role of RmGRP in embryo development. On the other hand, since Ixodidae ticks present several GRPs, which may have different or overlapping roles (Maruyama et al. Reference Maruyama, Anatriello, Anderson, Ribeiro, Brandão, Valenzuela, Ferreira, Garcia, Szabó, Patel, Bishop and de Miranda-Santos2010; Kim et al. Reference Kim, Tirloni, Pinto, Moresco, Yates, da Silva Vaz and Mulenga2016), the low hatching rate of dsRmGRP treated females could also be at least partly derived from a possible cross-reaction with other GRPs, which would share similar mRNA sequences. Indeed, GRPs were already detected in R. microplus frustrated larvae transcriptome (Lew-Tabor et al. Reference Lew-Tabor, Moolhuijzen, Vance, Kurscheid, Rodriguez Valle, Jarrett, Minchin, Jackson, Jonsson, Bellgard and Guerrero2010) and in unfed larvae proteome (Untalan et al. Reference Untalan, Guerrero, Haines and Pearson2005), while Rodriguez-Valle et al. (Reference Rodriguez-Valle, Lew-Tabor, Gondro, Moolhuijzen, Vance, Guerrero, Bellgard and Jorgensen2010) detected transcripts of GGY domain proteins in frustrated, but not in unfed larvae, suggesting that these proteins are associated to the cement cone and the transcription in early stages indicates a role in tick–host attachment process. It should be emphasized that R. microplus is a one-host tick, attaching to and feeding on only one host throughout all three life stages. Engorged ticks spontaneously detach from the host for oviposition, after which it is not possible to attach the ticks on bovines again. Consequently, it is necessary to apply a different strategy to conduct gene silencing by RNAi in adult females, as compared with three-host tick species, which has been successfully achieved employing the capillary artificial feeding system (Reck et al. Reference Reck, Klafke, Webster, Dall'Agnol, Scheffer, Souza, Corassini, Vargas, dos Santos and Martins2014; Antunes et al. Reference Antunes, Merino, Lérias, Domingues, Mosqueda, de la Fuente and Domingos2015; Lara et al. Reference Lara, Pohl, Gandara, Ferreira, Nascimento-Silva, Bechara, Sorgine, Almeida, da Silva Vaz and Oliveira2015).
The evaluation of rmgrp expression by qRT–PCR in eggs and adult tissues corroborates the antigenicity analysis with the rabbit-raised antisera, including the lack of rRmGRP recognition by the anti-gut serum. The transcriptome analysis, on the other hand, shows the presence of rmgrp in the guts of partially and fully engorged females, but in very low levels, indicating the presence of at least a basal expression in all adult tissues evaluated. The lack of recognition of rRmGRP by the anti-gut serum indicates that this tissue does not present RmGRP (as well as other possible cross-reacting GRPs) in quantity and/or form capable of generating an IgG-based response as intense as the other tissues tested presented. Concerning adult males, the mRNA transcription of rmgrp and the presence of molecules recognized by the anti-rRmGRP serum compatible with RmGRP are in accordance with transcriptomes and proteomes analyses that found GRPs in saliva and salivary glands of R. appendiculatus (de Castro et al. Reference de Castro, de Klerk, Pienaar, Latif, Rees and Mans2016), R. pulchellus (Tan et al. Reference Tan, Francischetti, Slovak, Kini and Ribeiro2015) and Ornithodoros moubata (Díaz-Martín et al. Reference Díaz-Martín, Manzano-Román, Valero, Oleaga, Encinas-Grandes and Pérez-Sánchez2013) males. Furthermore, in R. appendiculatus and R. pulchellus, these proteins presented higher expression levels in salivary glands from males than females (Tan et al. Reference Tan, Francischetti, Slovak, Kini and Ribeiro2015; de Castro et al. Reference de Castro, de Klerk, Pienaar, Latif, Rees and Mans2016). This high expression was attributed to feeding and mating behavior of males, which attach and detach where the females are feeding and, therefore, need constantly to secrete salivary proteins, as GRPs, to fix to the host and/or to ‘help’ the coupled female to modulate the immune and hemostatic systems (Wang et al. Reference Wang, Paesen, Nuttall and Barbour1998).
Tick GRPs have been shown to be expressed with considerable variability in tissues/developmental stages. While rmgrp was detected in all female adult tissues tested, Zhou et al. (Reference Zhou, Gong, Zhou, Xuan and Fujisaki2006) showed expression of a GRP from R. hemaphysaloides (RH50) only in salivary glands after blood feeding, but not in ovaries, guts and fat bodies of unfed and fed females. On the other hand, Jiang et al. (Reference Jiang, Gao, Wang, Xu, Li, Qi, Yang, Ji, Zhang, Luo and Yin2014) detected Hq15 (an alanine-, proline-, glycine-, threonine- and serine-rich protein of H. qinghaiensis) in larvae, nymphs, adult ticks, eggs, salivary glands and carcasses, but not in midguts. Furthermore, Shahein et al. (Reference Shahein, Abouelella, Hussein, Hamed, El-Hakim, Abdel-Shafy and Tork2013) analysed the GRPs from A. variegatum RaSaI1, RaSaI2 and RaSaI3, showing that all genes were expressed in 12- and 18-day-old eggs, but only RaSaI2 showed a very low expression level in 6-day-old eggs. RaSaI and RmGRP seem to be expressed in a very similar embryogenesis period, although these GRPs do not present similar sequences. Lew-Tabor et al. (Reference Lew-Tabor, Moolhuijzen, Vance, Kurscheid, Rodriguez Valle, Jarrett, Minchin, Jackson, Jonsson, Bellgard and Guerrero2010) reported the expression of a R. microplus GRP in adult males and female salivary glands, presenting a significant higher expression in nymphs, but not in adult female guts and ovaries. Indeed, Perner et al. (Reference Perner, Provazník, Schrenková, Urbanová, Ribeiro and Kopáček2016) detected two transcripts encoding GRPs in midgut from females only during the slow feeding phase of I. ricinus. Due to the large description of tick transcriptomes and proteomes, GRPs have been identified in tissues and development stages from a variety of other tick species, including A. americanum (Radulović et al. Reference Radulović, Kim, Porter, Sze, Lewis and Mulenga2014), H. flava (Xu et al. Reference Xu, Cheng, Yang and Liao2016) and Dermacentor variabilis (Anderson et al. Reference Anderson, Sonenshine and Valenzuela2008).
Potential candidate antigens/molecules for anti-tick vaccines can be classified as exposed (usually secreted in saliva and capable to interact with the host immune system during a natural infestation) or concealed (hidden from the host immune system in a natural infestation) (Willadsen and Kemp, Reference Willadsen and Kemp1988; Mulenga et al. Reference Mulenga, Sugimoto and Onuma2000). In addition to its secretion in saliva, RmGRP was recognized by sera from infested bovines, which makes it an exposed antigen. The rRmGRP recognition by almost all tested sera from naturally infested B. indicus or experimentally infested B. taurus indicates its high antigenicity, what have been reported for other tick GRPs (Bishop et al. Reference Bishop, Lambson, Wells, Pandit, Osaso, Nikonge, Morzaria, Musoke and Nene2002; Trimnell et al. Reference Trimnell, Hails and Nuttall2002; Zhou et al. Reference Zhou, Gong, Zhou, Xuan and Fujisaki2006). The experimentally infested B. taurus sera also showed a heterogeneous pattern of recognition between and within bovines, under light and heavy infestations. Such heterogeneity has already been reported when analysing tick tissues/developmental stages protein extracts (Cruz et al. Reference Cruz, Silva, Mattos, da Silva Vaz, Masuda and Ferreira2008) or recombinant R. microplus paramyosin (Leal et al. Reference Leal, Seixas, Mattos, Coutinho, Masuda, da Silva Vaz and Ferreira2013). In contrast, all three bovines presented a clear recognition following the first infestation. Indeed, as bovines one and three showed strong recognitions among sera from light infestations and one heavy infested serum bovine did not recognize rRmGRP, it can be inferred that anti-RmGRP IgG levels are naturally modulated during consecutive infestations and are influenced greatly by immune characters other than the amount of antigen disposal. The recognition of rRmGRP by all tested sera from naturally infested B. indicus suggests that these putative naturally acquired resistant hosts maintain an IgG response under field conditions, although levels of IgG do not necessaryly correlate with naturally acquired resistance following tick infestations (Cruz et al. Reference Cruz, Silva, Mattos, da Silva Vaz, Masuda and Ferreira2008; Piper et al. Reference Piper, Jonsson, Gondro, Vance, Lew-Tabor and Jackson2016), even when dealing with essential molecules, as the functionally important portions may not be targeted or immunodominant (Havlíková et al. Reference Havlíková, Roller, Koči, Trimnell, Kazimírová, Klempa and Nuttall2009). Additionally, the presence of the protein in non-salivary tissues, such as ovaries and fat bodies, as well as in eggs and larvae, suggest that this protein could be targeted in a ‘dual action strategy’, presenting properties of exposed and concealed antigens. ‘Dual action’ vaccines target both exposed and concealed antigenic epitopes, thus, host antibodies against the salivary RmGRP could cross-react with hidden epitopes, becoming more efficient (Nuttall et al. Reference Nuttall, Trimnell, Kazimirova and Labuda2006), as previously described for 64P (Trimnell et al. Reference Trimnell, Hails and Nuttall2002). However, the suggested reinforce of the immune response by natural infestations (Nuttall et al. Reference Nuttall, Trimnell, Kazimirova and Labuda2006) should be evaluated on a ‘case- by-case’ basis, as infestation density and host genetic heterogeneity seem to influence immune responses greatly (Ogden et al. Reference Ogden, Casey, French, Adams and Woldehiwet2002; Cruz et al. Reference Cruz, Silva, Mattos, da Silva Vaz, Masuda and Ferreira2008), as could be seen for RmGRP. In this sense, a different R. microplus GRP was used as immunogen in a four-antigen vaccine evaluation, which resulted in a reduction of female numbers and engorgement weight, but the tick infestation did not stimulate the anti-GRP IgG response (Maruyama et al. Reference Maruyama, Garcia, Teixeira, Brandão, Anderson, Ribeiro, Valenzuela, Horackova, Veríssimo, Katiki, Banin, Zangirolamo, Gardinasse, Ferreira and de Miranda-Santos2017). Other vaccination trials using GRPs as antigen were already described, such as RH50 of R. haemaphisaloides, which resulted in a low rate of attachment and high mortality of nymphs (Zhou et al. Reference Zhou, Gong, Zhou, Xuan and Fujisaki2006), and p29 of H. longicornis, which showed a decrease of weight of fed females and an increase of larvae and nymphs mortality (Mulenga et al. Reference Mulenga, Sugimoto, Ingram, Ohashi and Onuma1999).
In the present study, we described the cDNA and amino-acid sequences encoding to RmGRP, which presented glycine content and repeats characteristic from the superfamily of GRPs. The presence of the rmgrp mRNA in various tissues and developmental stages indicates its probable importance in different periods of the R. microplus life cycle. The rRmGRP recognition by naturally and experimentally infested bovines sera, the low larvae hatching from females silenced with dsRmGRP, the probable RmGRP presence in most adult tissues/developmental stages, in addition to important effects in ticks by previous vaccination trials with other GRPs, turns rRmGRP a potential antigen that deserves a further investigation in order to compose an anti-tick vaccine. We can also infer that RmGRP is possibly essential in functions other than the host–parasite interface, such as embryonic development, thereby opening new challenges for unravelling the importance of GRPs in tick biology.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182017001998
Acknowledgements
We thank Luís Fernando Parizi, Maria Claudia Rosa Garcia, Joanna Ines Ayres Reis, Betânia Souza de Freitas and Gaby Renard for their technical help and Sérgio Silva da Silva by providing of infested bovine sera.
Financial support
This is study was supported by INCT-Entomologia Molecular, CNPq, CAPES and FAPERGS.