INTRODUCTION
Bartonella species are gram-negative bacteria that infect erythrocytes of their respective hosts and most are believed to be transmitted by arthropod vectors (Billeter et al. Reference Billeter, Levy, Chomel and Breitschwerdt2008). There are currently more than 20 recognized Bartonella species and many of those are known or suspected zoonotic pathogens (Breitschwerdt et al. Reference Breitschwerdt, Maggi, Chomel and Lappin2010). Bartonella DNA has also been detected in a wide variety of biting arthropods including ticks, fleas, lice, mites, flies, and keds (Billeter et al. Reference Billeter, Levy, Chomel and Breitschwerdt2008). The detection of DNA, however, does not demonstrate viability of the organism or vector competency of the arthropod. To date, only 6 Bartonella species have been confirmed to be transmitted by an arthropod experimentally; B. bacilliformis is transmitted by Lutzomyia verrucarum sandflies, B. quintana is transmitted by Pediculus humanus body lice, B. birtlesii is transmitted by Ixodes ricinus ticks, B. henselae is transmitted by Ctenocephalides felis fleas, and B. grahamii and B. taylorii are transmitted by Ctenophthalmus nobilis fleas (Swift, Reference Swift1920; Battistini, Reference Battistini1931; Chomel et al. Reference Chomel, Kasten, Floyd-Hawkins, Chi, Yamamoto, Roberts-Wilson, Gurfield, Abbott, Pederson and Koehler1996; Bown et al. Reference Bown, Bennett and Begon2004; Billeter et al. Reference Billeter, Levy, Chomel and Breitschwerdt2008; Reis et al. Reference Reis, Cote, Le Rhun, Lecuelle, Levin, Vayssier-Taussat and Bonnet2011).
Eidolon helvum, the straw-coloured fruit bat, is distributed throughout much of sub-Saharan Africa and feeds on flowers, leaves, and fruits. These bats tend to roost in colonies of over 100 000, often in urban areas, and are used as a human food source in Central and West Africa. Bats are known reservoirs of zoonotic viruses including rabies, Nipah virus, coronaviruses, and others, but their role in the transmission of human pathogenic bacteria has not been explored in detail (Wong et al. Reference Wong, Lau, Woo and Yuen2007). Results from our laboratory have shown that some bats, including E. helvum, are carriers for several unique Bartonella species but their ability to transmit these organisms to humans and other animals is unknown (Kosoy et al. Reference Kosoy, Bai, Lynch, Kuzmin, Niezgoda, Franka, Agwanda, Breiman and Rupprecht2010). It is also unknown whether any arthropod vectors are responsible for the transmission of Bartonella species between bats.
Bat flies (Diptera) are obligate, blood-sucking ectoparasites often found living in the fur and on wing membranes of the bat host (Dick and Patterson, Reference Dick, Patterson, Morand, Krasnov and Poulin2006). Bat flies are most closely related to tse-tse flies (Glossinidae) and Hippoboscid flies and all belong to the same super family, Hippoboscoidea (Dittmar et al. Reference Dittmar, Porter, Murry and Whiting2006). Bat flies are divided into 2 families: Streblidae, most commonly found in the Western Hemisphere, and Nycteribiidae, predominantly in the Eastern Hemisphere (Dick and Patterson, Reference Dick, Patterson, Morand, Krasnov and Poulin2006).
Although Bartonella DNA was detected in a bat fly, Trichobius major (Diptera: Streblidae) collected from a Myotis austroriparius in Florida (Reeves et al. Reference Reeves, Loftis, Gore and Dasch2005), no surveys have been performed to screen Nycteribiidae flies for the presence of Bartonella or demonstrate viability of the organism in these flies. Because of this, a study was performed to examine bat flies from western Africa for the presence of Bartonella using culture and PCR analysis.
MATERIALS AND METHODS
Bat fly collection and identification
Bat flies were collected from E. helvum in Ghana and on 2 islands in the Gulf of Guinea, Annobón and Bioko; in total 137 flies were examined (Fig. 1). From the study site in Accra, Ghana, 46 adult flies were collected, placed into pots, and transferred to −80°C shortly thereafter. From the island of Annobón, a total of 32 flies was collected: 16 were suspended in 70% ethanol and 16 were placed at −80°C. A total of 59 bat flies was collected from E. helvum on the island of Bioko and all samples were placed in tubes containing 70% ethanol. Samples were subsequently shipped overnight on dry ice (those samples frozen at −80°C) or ice packs (samples suspended in ethanol) to the Ft. Collins CDC, Bartonella Laboratory.
Upon arrival, all samples were placed at −80°C until further analysis. After DNA extraction, exoskeletons were kept as DNA vouchers and were subsequently slide mounted in PVA (BioQuip Products, Rancho Dominguez, CA, USA). Bat flies were sexed and identified using the species keys of Theodor (Reference Theodor1967). All specimens keyed to Cyclopodia greefi greefi, based on the characteristic setation of the fore-tibiae, the presence of a quadratic patch of long dorsal abdominal setae on the female, characteristic head morphology, a broad, open haltere groove, and a single dorsal thoracic seta. Specimens collected from Annobón were slightly heavier sclerotized, but otherwise adhered to the key.
Fig. 1. Map of western Africa, including the sites of bat fly collection. A total of 137 adult Cyclopodia greefi greefi flies were collected from Eidolon helvum: 46 flies from Accra, Ghana, 32 flies from the island of Annobón, and 59 flies from the island of Bioko.
Culturing of Bartonella spp. from flies
All frozen bat fly specimens were thawed, subjected to surface sterilization using a Wescodyne wash (20 min) followed by a 70% ethanol wash (5 min), and were then rinsed in sterile 1x PBS (0·15 M, pH: 7·50, Atlanta, CDC). Flies were cut in half longitudinally using a sterile scalpel blade so that half could be used for DNA extraction and half for culture analysis. For culture, flies were suspended in 500 μl of Brain Heart Infusion (BHI) broth (CDC) containing 20% fungizone; 300 μl was aliquoted onto a BHI agar plate containing 5 or 10% defibrinated rabbit blood (CDC). Plates were placed at 35°C, 5% CO2 and examined daily for up to 4 weeks. Any Bartonella-like colonies were subcultured by streaking onto a fresh rabbit blood agar plate.
DNA extraction and PCR amplification
DNA extraction of 137 adult bat flies and second passage colonies was performed using a Qiagen QIAamp tissue kit (QIAGEN, Valencia, CA, USA) according to the manufacturer's instructions. Prior to DNA extraction, the bat flies in ethanol were triturated using a sterile scalpel blade. For the cultured Bartonella, 6 individual colonies from 1 isolate were picked using a sterile loop and suspended in 200 μl of BHI broth. Bat flies were examined for the presence of Bartonella DNA by conventional PCR. Primers 443f (5′ GCT ATG TCT GCA TTC TAT CA) (Birtles and Raoult, Reference Birtles and Raoult1996) and 1210r (5′ GAT CYT CAA TCA TTT CTT TCC A) were designed to amplify a portion of the citrate synthase gene, gltA. Each 50 μl PCR reaction contained 28·75 μl of nuclease-free water, 5x PCR buffer containing MgCl2, 0·2 mM dNTPs, 15 pmol of each primer, and 1·25 U of GoTaq DNA polymerase (Promega, Madison, WI, USA). PCR was performed using a Bio-Rad Laboratories iCycler (Hercules, CA, USA) with the following cycling conditions: 94°C for 2 min followed by 45 cycles of 94°C for 30 sec, 48°C for 1 min, 72°C for 1 min, and 1 cycle of 72°C for 7 min.
PCR products were separated using electrophoresis in a 1·5% agarose gel containing ethidium bromide and were visualized under UV light. The positive control consisted of B. doshiae DNA and nuclease-free water was used as a negative control.
Gene sequencing and analysis
Amplicons were purified using the QIAquick PCR purification kit (QIAGEN) and sequenced using an Applied Biosystems Model 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). DNA sequences were analysed using the Lasergene v8 sequence analysis software (DNASTAR, Madison, WI, USA) to determine a consensus sequence for the PCR product from each bat fly. All sequences from this study were subsequently cropped, ∼307 bp, for further phylogenetic analysis. Sequences obtained in this study were considered similar to validated Bartonella species if similarity over the gltA fragment was >96·0% (La Scola et al. Reference La Scola, Zeaiter, Khamis and Raoult2003). The Clustal W program in Megalign (Lasergene) was used to compare sequences obtained from this study to Bartonella sequences available in GenBank. GltA sequences from 5 novel Bartonella species, isolated from E. helvum in Kenya, were also included in the phylogenetic analyses. Five species, identified as ‘E1’, ‘E2’, ‘E3’, ‘Ew’ (GenBank number: HM363765 – HM363768) and ‘E-124’ (GenBank number: JN190887), represent bat specific Bartonella.
The neighbor-joining (N-J) method based on the Kimura 2-parameter model and the bootstrap calculations were carried out with 1000 re-samplings.
Nucleotide sequence Accession numbers
Sequences, representative of each genotype obtained from this study, were deposited in GenBank under Accession numbers JN172035 – JN172074.
RESULTS
Prevalence of Bartonella DNA in bat flies
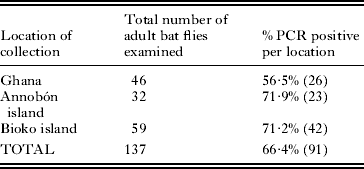
Of the 137 adult bat flies examined, 66·4% were PCR positive for the presence of Bartonella DNA using gltA specific primers (Table 1). Positivity from each location ranged from 56·5% in Ghana to 71·9% on the island of Annobón.
Table 1. Prevalence of Bartonella DNA in adult bat flies from western Africa
Genetic heterogeneity
Of 82 sequences examined, a total of 39 genotypes (genotype consisting of 1 or more nucleotide differences) were found (Table 2). Eight genotypes were represented by 2–30 identical sequences. The sequence similarity among Bartonella sequences detected in bat flies ranged between 71·4 and 100%, while similarity among Bartonella sequences obtained from bat flies and those identified previously from E. helvum ranged from 71·9 to 100%. Fifty-five of the 82 sequences were ⩾96% similar to Bartonella previously isolated from E. helvum or other Bartonella species, suggesting that the remaining 27 sequences may represent novel species (Fig. 2).
Table 2. The total number of 39 Bartonella genotypes, from 82 sequences examined, detected in bat flies from western Africa

Isolation of a Bartonella sp. from a bat fly
Although attempts to culture Bartonella spp. from bat flies were largely unsuccessful due to overgrowth of contaminants, a single isolate from a bat fly was obtained. A Bartonella sp. isolate, 98·4% identical to Bartonella sp. ‘E1-105’ isolated from bats in Kenya based on gltA sequence analysis (GenBank Accession: HM363765), was cultured from 1 bat fly collected in Ghana (fly Cg 23 Q22-1). Further molecular characterization was performed by sequence analysis of the 16S rRNA gene, ftsZ, and rpoB using methods described previously (Marchesi et al. Reference Marchesi, Sato, Weightman, Martin, Fry, Hiom, Dymock and Wade1998; Renesto et al. Reference Renesto, Gouvernet, Drancourt, Roux and Raoult2001; Zeaiter et al. Reference Zeaiter, Liang and Raoult2002). Comparison of sequence data from Bartonella ‘E1-105’ demonstrated sequence similarity of 100%, 100%, and 99·8% within the 16S rRNA gene, ftsZ, and rpoB genes, respectively (GenBank Accession numbers for Bartonella ‘E1-105’: HM363785, HM363770 and HM363775).
DISCUSSION
Within the current study, we demonstrated (1) the presence of Bartonella DNA in bat flies from western Africa and (2) the successful culture of Bartonella from a bat fly collected in Ghana. Bartonella DNA has previously been detected in bat ectoparasites, specifically in a T. major bat fly, Cimex adjunctus (Hemiptera: Cimicidae) bat bug, Carios kelleyi (Acari: Argasidae) tick, Sternopsylla texanus (Siphonaptera: Ischnopsyllidae) flea, and a Steatonyssus sp. mite (Acari: Mesostigmata) (Loftis et al. Reference Loftis, Gill, Schriefer, Levin, Eremeeva, Gilchrist and Dasch2005; Reeves et al. Reference Reeves, Loftis, Gore and Dasch2005, Reference Reeves, Dowling and Dasch2006a, Reference Reeves, Rogers, Durden and Dasch2007). Our data, unlike previous surveys, reveals that a large percentage (up to 66·4%) of C. greefi greefi harboured Bartonella DNA and, in at least 1 case, viability of the bacteria could be confirmed. More than half (65·9% of 82) of the sequences obtained were identical or similar to Bartonella species previously isolated from E. helvum (Kosoy et al. Reference Kosoy, Bai, Lynch, Kuzmin, Niezgoda, Franka, Agwanda, Breiman and Rupprecht2010), suggesting a potential role of bat flies in the transmission of Bartonella between bats. When compared with Bartonella species isolated from other bat species in Kenya, the bat fly Bartonella genotypes detected in this study appear to be very host specific (data not shown). Interestingly, 27 (32·9%) Bartonella sequences did appear to be unique to bat flies. It is unclear, at this point, whether Bartonella species may circulate among bat flies outside of the host or whether these 27 sequences represent potential symbionts.
Previous investigations have also pointed to the likely potential of Bartonella transmission by other hippoboscid flies. Lipoptena cervi, or the deer ked, is the suspected vector of B. schoenbuchensis in deer. Dehio et al. (Reference Dehio, Sauder and Hiestand2004) successfully isolated B. schoenbuchensis from deer ked collected in Germany and demonstrated the presence of the bacteria in the mid-gut of the flies. Bartonella DNA has also been detected in L. cervi from Massachusetts and L. mazamae collected in South Carolina (Reeves et al. Reference Reeves, Nelder, Cobb and Dasch2006b; Matsumoto et al. Reference Matsumoto, Berrada, Klinger, Goethert and Telford III2008). Halos et al. (Reference Halos, Jamal, Maillard, Girard, Guillot, Chomel, Vayssier-Taussat and Boulouis2004) detected Bartonella DNA in a large percentage of L. cervi (94% of 48 adults), Hippobosca equina (71% of 17 adults), and Melophagus ovinus (100% of 20 adults) collected from 4 different ruminant species. Bartonella DNA was also found in 100% (10 total) of the M. ovinus pupae screened suggesting the bacteria may be vertically transmitted in this species.
Further investigation is warranted to decipher what role, if any, bat flies or other bat ectoparasites might play in the transmission of Bartonella spp. between bats. Future studies will be performed to screen bat flies from other bat species and locations for the presence of Bartonella. Attempts will also be made to isolate viable Bartonella from pupae to ascertain whether vertical transmission of the bacteria may occur, as suggested by Halos et al. (Reference Halos, Jamal, Maillard, Girard, Guillot, Chomel, Vayssier-Taussat and Boulouis2004). Bat flies undergo adenotrophic vivaparity (i.e. the complete development of 3 larval stages inside the adult female) and deposition of a single pre-pupae. This unique reproductive ability may facilitate vertical transmission of parasites, including Bartonella spp. (Dittmar et al. Reference Dittmar, Porter, Murry and Whiting2006). Importantly, efforts also are on-going to determine whether these agents are responsible for human illnesses.

Fig. 2. Tree topology displaying similarity of Bartonella DNA detected in Cyclopodia greefi greefi with known Bartonella sequences based upon partial citrate synthase gene, gltA. The topology was constructed by the neighbor-joining method based on the Kimura-2 parameter model of nucleotide substitution. Bootstrap values are based on 1000 replicates. The tree was rooted by the use of Brucella melitensis 16MT as the out-group. Bartonella sequences obtained from 5 Eidolon helvum, collected in Kenya (Kosoy et al. Reference Kosoy, Bai, Lynch, Kuzmin, Niezgoda, Franka, Agwanda, Breiman and Rupprecht2010), are included in the tree (GenBank Accession numbers: HM363765 – HM363768 and JN190887). Unique Bartonella sequences identified in bat flies from Annobón are represented by GenBank Accession numbers JN172049 – JN172057, Bartonella sequences obtained from bat flies from Bioko are represented by GenBank Accession numbers JN172058 – JN172074, and Bartonella sequences obtained from Ghana bat flies are represented by GenBank Accession numbers JN172035 – JN172048.
ACKNOWLEDGEMENTS
S. A. Billeter is supported through the American Society of Microbiology/Centers for Disease Control and Prevention Post-Doctoral Associates Program in Infectious Diseases and Public Health Microbiology. D. T. S. Hayman is supported by a Wellcome Trust Research Training Fellowship, J. L. N. Wood is supported by the Alborado Trust, and both are supported by the RAPIDD program of the Science & Technology, Directorate, Department of Homeland Security. K. Baker is supported by a Wellcome Trust Research Training Fellowship. A. A. Cunningham is supported by a Royal Society Wolfson Research Merit award.






