INTRODUCTION
Schistosomiasis is a major poverty-related disease, affecting more than 200 million people in developing tropical countries. The genus Schistosoma comprises in total 26 species, all of which are infecting mammals and among which eight are infecting humans (Brant and Loker, Reference Brant and Loker2013). The major burden of human infection resides in sub-Saharan Africa where more than 90% of cases are registered; 112 million people are estimated to be infected with Schistosoma haematobium and 54 million with Schistosoma mansoni (Brindley and Hotez, Reference Brindley and Hotez2013). In addition, in Africa, three Schistosoma species are found with a significant impact on livestock: Schistosoma mattheei, Schistosoma bovis and Schistosoma curassoni (Leger and Webster, Reference Leger and Webster2016). A DNA barcoding approach was used to study the genetic variation and population structure of S. haematobium (Webster et al. Reference Webster, Emery, Webster, Gouvras, Garba, Diaw, Seye, Tchuente, Simoonga, Mwanga, Lange, Kariuki, Mohammed, Stothard and Rollinson2012) and S. mansoni (Morgan et al. Reference Morgan, Dejong, Adeoye, Ansa, Barbosa, Brémond, Cesari, Charbonnel, Corrêa, Coulibaly, D'Andrea, De Souza, Doenhoff, File, Idris, Incani, Jarne, Karanja, Kazibwe, Kpikpi, Lwambo, Mabaye, Magalhães, Makundi, Moné, Mouahid, Muchemi, Mungai, Séne and Southgate2005; Webster et al. Reference Webster, Webster, Gouvras, Garba, Lamine, Diaw, Seye, Tchuem Tchuenté, Simoonga, Mubila, Mwanga, Lwambo, Kabatereine, Lange, Kariuki, Mkoji, Rollinson and Stothard2013a) on a large geographical scale. Surprisingly low sequence variation was found within mitochondrial genes of S. haematobium (Webster et al. Reference Webster, Emery, Webster, Gouvras, Garba, Diaw, Seye, Tchuente, Simoonga, Mwanga, Lange, Kariuki, Mohammed, Stothard and Rollinson2012), which is in stark contrast with the high mtDNA diversity found in S. mansoni (Morgan et al. Reference Morgan, Dejong, Adeoye, Ansa, Barbosa, Brémond, Cesari, Charbonnel, Corrêa, Coulibaly, D'Andrea, De Souza, Doenhoff, File, Idris, Incani, Jarne, Karanja, Kazibwe, Kpikpi, Lwambo, Mabaye, Magalhães, Makundi, Moné, Mouahid, Muchemi, Mungai, Séne and Southgate2005; Webster et al. Reference Webster, Webster, Gouvras, Garba, Lamine, Diaw, Seye, Tchuem Tchuenté, Simoonga, Mubila, Mwanga, Lwambo, Kabatereine, Lange, Kariuki, Mkoji, Rollinson and Stothard2013a). Two main geographical groups were found in S. haematobium: one is found throughout the African mainland and the other is found exclusively at the Indian Ocean Islands and the neighbouring African coastal regions. A barcoding approach has also proven to be an ideal tool to study the diversity and interrelationships within the freshwater snails in the genus of Bulinus (Kane et al. Reference Kane, Stothard, Emery and Rollinson2008), of which some species act as an intermediate host for S. haematobium and S. bovis.
Schistosoma worms have a complex life cycle. Their transmission depends on both an intermediate snail host and a final vertebrate host. Schistosomes are the only genus of Trematoda with separate sexes; both males and females are required for sexual reproduction (Southgate et al. Reference Southgate, Jourdane and Tchuem Tchuenté1998; Loker and Brant, Reference Loker and Brant2006). Mating of adults takes place at the hepatic portal system and is required for migration towards the egg-laying site and maturation of female worms (Southgate et al. Reference Southgate, Jourdane and Tchuem Tchuenté1998). Although most worms show species-specific mating preferences, heterospecific mating occurs, even when a same species mate is available (Norton et al. Reference Norton, Webster, Kane and Rollinson2008). This may result in parthenogenesis or hybridization, depending on the schistosome species involved (Southgate et al. Reference Southgate, Jourdane and Tchuem Tchuenté1998). Upon mating, female worms release their eggs into the veins of the surrounding organs. Eggs migrate towards the bladder, liver and intestine, depending on the species and reach the environment through urine or feces.
The Senegal River Basin (SRB) represents a transmission hotspot for schistosomiasis. In 1988 the Diama Dam was constructed on the Senegal River and a year later the Mantali Dam was built on the Bafing River, an upstream tributary of the Senegal River in Mali. These dams provided fresh water for irrigation and new habitats for snails, triggering a huge outbreak of intestinal schistosomiasis in the late eighties. A few years following this outbreak the prevalence of S. haematobium, causing urinary schistosomiasis increased also (Picquet et al. Reference Picquet, Ernould, Vercruysse, Southgate, Mbaye, Sambou, Niang and Rollinson1996; Southgate et al. Reference Southgate, Tchuem Tchuenté, Sène, De Clercq, Théron, Jourdane, Webster, Rollinson, Gryseels and Vercruysse2001). Today, S. mansoni and S. haematobium are co-endemic in the SRB and along the shores of Lac de Guiers, resulting in mixed Schistosoma infections in humans (Meurs et al. Reference Meurs, Mbow, Vereecken, Menten, Mboup and Polman2012; Abdellahi et al. Reference Abdellahi, Ndir and Niang2016). The prevalence of S. haematobium still seems to increase while S. mansoni prevalence tends to decline (personal Observation: personal communication Elke Steinacher in 2014, poste de santé Catholique in Dagana). Schistosoma bovis and S. curassoni are also present in northern Senegal (Vercruysse et al. Reference Vercruysse, Southgate and Rollinson1984) but a recent study only found S. bovis in the SRB (Webster et al. Reference Webster, Diaw, Seye, Webster and Rollinson2013b). Both species cause intestinal schistosomiasis in ruminants.
The SRB (Fig. 1) was among the first places where hybridization between the human host species S. haematobium and the cattle host species S. bovis was observed (Huyse et al. Reference Huyse, Webster, Geldof, Stothard, Diaw, Polman and Rollinson2009). This hybrid was first described by Brémond et al. (Reference Brémond, Sellin, Sellin, Nameoua, Labbo, Theron and Combes1993) in Niger. Schistosoma haematobium and S. bovis belong both to the S. haematobium species group. Several species pairs within this group are known to hybridize experimentally or in the wild (Tchuem Tchuenté et al. Reference Tchuem Tchuenté, Southgate, Combes and Jourdane1996; Webster et al. Reference Webster, Southgate and Littlewood2006; Leger and Webster, Reference Leger and Webster2016). Laboratory experiments have shown that hybrids may have an enhanced transmission potential. For example, F1 and F2 hybrids between S. haematobium and S. guineensis exhibited a greater infectivity for snail intermediate hosts than the parental schistosome species (Webster and Southgate, Reference Webster and Southgate2003). In the SRB, several Bulinus species act as intermediate snail host for S. haematobium, the main intermediate snail host is Bulinus globosus. Bulinus truncatus, the most common bulinid snail in the SRB, is host of S. bovis (Diaw et al. Reference Diaw, Vassiliades, Thiongane, Seye, Sanrr and Diouf1998; Sène et al. Reference Sène, Southgate and Vercruysse2004). If the S. haematobium × S. bovis hybrids show infectivity to both B. globosus and B. truncatus, this could induce an increased transmission of these hybrids and therefore lead to higher prevalences of urinary schistosomiasis. However, the exact intermediate snail host range of the hybrid species is unknown at present. The S. haematobium × S. bovis hybrid seems widespread and has also been found elsewhere in Senegal (Webster et al. Reference Webster, Diaw, Seye, Webster and Rollinson2013b), Corsica and Benin (Moné et al. Reference Moné, Holtfreter, Allienne, Mintsa-Nguéma, Ibikounlé, Boissier, Berry, Mitta, Richter and Mouahid2015) and Mali (Soentjens et al. Reference Soentjens, Cnops, Huyse, Yansouni, De Vos, Bottieau, Clerinx and Van Esbroeck2016). Also, hybridization between the two phylogenetically remotely related human species S. haematobium and S. mansoni has been found in the SRB (Huyse et al. Reference Huyse, Van den Broeck, Hellemans, Volckaert and Polman2013). Despite the common observation of hybrids in the SRB (Huyse et al. Reference Huyse, Webster, Geldof, Stothard, Diaw, Polman and Rollinson2009; Webster et al. Reference Webster, Diaw, Seye, Webster and Rollinson2013b), little is known about their spatiotemporal distribution.

Fig. 1. Map with the sample locations in the Senegal River Basin including in the Middle Valley the villages of Diatar, Dioundiou, and Kodit; towards the coast, in the Senegal River Delta the village of Rhonne; in the vicinity of Lac de Guiers on the eastern shore the village of Mbane, on the western shore the villages of Ndieumeul and Diokhor Tack, and north of the lake the village of Pakh; in the Lampsar River, a tributary of the Senegal River, the villages Mbodienne and Pont Gendarme. For each region, the percentage of hybrids (i.e. parasites with a S. bovis cox1 mtDNA profile) is indicated by a pie chart.
Our first aim was to determine and explain the spatiotemporal distribution of hybridization in the SRB. Hybrids were detected through barcoding, based on the genetic profile of a mitochondrial marker (partial cytochrome c oxidase subunit I (cox1)). We compared the frequency of S. haematobium × S. bovis hybrids in the SRB in ten villages, sampled between 2006 and 2010, along four regions in the SRB; the Delta and the Middle Valley of the Senegal River, the Lampsar River and Lac de Guiers (Fig. 1). Secondly, we aimed to understand the potential impact of S. haematobium × S. bovis hybrids on the local epidemiology of schistosomiasis. The recent increase in S. haematobium prevalence across the SRB might be linked with the presence of hybrids. Therefore the frequency of hybrids, per village, was related to the prevalence of urinary and intestinal schistosomiasis.
MATERIAL AND METHODS
Study area and data collection
We sampled parasite stadia (eggs and miracidia) for genetic analysis and parasitological information on the prevalence and infection intensity of schistosomiasis in ten villages located in four regions in Northern Senegal (Fig. 1). Data were collected during parasitological surveys in 2006, 2007, 2009 and 2010 (Table 1). From the village of Diatar (1993) material from adult worms was obtained from the Schistosomiasis Collection at the Natural History Museum (London, UK), where field-collected cercariae were passaged through mice (SCAN; Emery et al. Reference Emery, Allan, Rabone and Rollinson2012).
Table 1. Overview of parasite barcoding and prevalence of schistomiasis in Northern Senegal per geographical region, village and year of sampling: the ITS rDNA genotype and cox1 haplotype of Schistosoma haematobium miracidia collected from urines; the prevalence of urinary and intestinal schistosomiasis.

Ind, individuals; NA, not applicable, S.h, S. haematobium; S.b, S. bovis; S.m, S. mansoni.
a Genotype: The ITS rDNA genotype is only reported when it displayed a hybrid or a S. mansoni profile. In all other cases, the ITS rDNA profile showed a S. haematobium genotype. The abbreviations S.h /S.b and S.h /S.m indicate that at the diagnostic sites two chromatogram peaks were visible after Sanger sequencing.
b Pool: In 2006, Schistosoma eggs were isolated by sedimentation after pooling positive urine samples from several human individuals. For Mbane (2006) the exact number of human urines that was pooled is not known.
c These studies cover the whole population of a village, including school-age children and adults.
++Adult worms.
The prevalence of S. haematobium and S. mansoni
In March 2006 urine and stool samples were collected from 20 school-age children in each of the villages of Dioundiou, Kodit, Mbane, Ndieumeul, Mbodienne and Pont Gendarme (Table 1). In 2007 parasitological data were collected from 75 school-age children in Diatar, Dioundiou, Rhonne, Mbane, Ndieumeul and Mbodienne. In 2009 and 2010 parasitological data were collected from both children (older than 4) and adults at the villages of Pakh, Diokhor and Ndieumeul (Table 1). Schistosoma haematobium was detected by filtering 10 mL of urine on a Nucleopore filter (Isopore, USA). Schistosoma mansoni infection was diagnosed by a duplicate (two slides of 25 mg) Kato Katz (Katz et al. Reference Katz, Chaves and Pellegrino1972) per stool sample. In the studies of 2006 and 2007, one urine and one stool sample were collected, in the studies of 2009 and 2010 two urine and two stool samples per person were collected. The prevalence of Schistosoma species in urine and stool are reported per village and per sampling year (Table 1). In the studies of 2006 and 2007 S. haematobium and S. mansoni prevalence is calculated as the proportion of infected school-aged children. In the studies of 2009 and 2010 prevalence is calculated as the proportion of infected individuals across all ages.
Molecular analysis of S. haematobium stadia
Schistosoma eggs from positive urine samples were isolated by sedimentation. Isolated eggs hatched through exposure to sunlight in Petri dishes with clean bottled water to hatch. Individual miracidia were pipetted onto FTA® Classic cards (Whatman) in a volume of 3 µL. Eggs that did not hatch were stored on 100% ethanol. DNA extraction of eggs and miracidia was performed as described by Van den Broeck et al. (Reference Van den Broeck, Geldof, Polman, Volckaert and Huyse2011). DNA from adult worms was extracted using the Nucleospin® Tissue kit following manufacturer's instructions (Macherey-Nagel).
mtDNA genotyping. A rapid diagnostic ‘multiplex’ one-step polymerase chain reaction (RD-PCR) was performed to discriminate between S. haematobium, S. bovis and S. mansoni (Webster et al. Reference Webster, Rollinson, Stothard and Huyse2010; Van den Broeck et al. Reference Van den Broeck, Geldof, Polman, Volckaert and Huyse2011). A partial cytochrome c oxidase subunit I (cox1) mtDNA fragment of parasite DNA was amplified in a total reaction volume of 25 µL consisting of 1 × PCR buffer, 1.5 mm MgCl2, 200 µ m of each dNTP, 0.4 µ m Asmit 1 (Eurogentec, forward primer TTTTTTGGTCA-TCCTGAGGTGTAT, Bowles et al. Reference Bowles, Blair and McManus1992), 0.2 µ m of each reverse primer [Eurogentec; S. haematobium (Sh.R.) TGATAATCAATGACCCTGCAATAA 543 bp (Webster et al. Reference Webster, Rollinson, Stothard and Huyse2010), S. bovis (Sb.R) CACAGGATCAGACAAACGAGTACC 306 bp (Webster et al. Reference Webster, Rollinson, Stothard and Huyse2010), S. mansoni (Sm.R) TGCAGATAAAGCCACCCCTGTG 375 bp (Van den Broeck et al. Reference Van den Broeck, Geldof, Polman, Volckaert and Huyse2011)], 1 unit Taq polymerase (Eurogentec), milli-Q water and 2 µL of DNA. PCR parameters during thermal cycling were 3 min at 95 °C, followed by 40 cycles of 30 s at 94 °C, 1 min 30 s at 58 °C and 1 min 30 s at 72 °C followed by a final cycle at 72 °C for 7 min. PCR products were visualized on a 1% Midori Green agarose gel.
ITS rDNA genotyping. Samples were subjected to PCR amplification of the complete rDNA region (981 bp; primers ITS4: TCCTCCGCTTATTGATATGC and ITS5: GGAAGTAAAAGT-CGTAACAAG (Barber et al. Reference Barber, Mkoji and Loker2000)). PCR amplification was performed in a volume of 25 µL containing 1 × PCR buffer, 2 mm MgCl2, 200 µ m of each dNTP, 2 mm of each primer, 1 unit Taq polymerase (Eurogentec), milli-Q water and 2 µL of DNA. Thermal cycling started with 3 min at 95 °C, followed by 40 cycles of 30 s at 94 °C, 45 s at 54 °C and 1 min 30 s at 72 °C followed by a final cycle at 72 °C for 7 min. We brought 4 µL of the PCR product on a 1% Midori Green agarose gel to visualize whether the PCR reaction was successful. PCR products of successful PCR reactions were further analysed by restriction fragment length polymorphism (RFLP) and visualized on a 1% Midori Green agarose gel (See Figures S1 and S2, supplementary information). Digestion of the ITS fragment with MboI (Fermentas) distinguishes S. haematobium from S. bovis and S. mansoni. Schistosoma haematobium PCR products were digested in fragment sizes of (in bp) 280, 174, 130, 100, 85 and 36 bp; S. bovis in fragment sizes of 380, 174, 167, 130, 83 and 36 bp; for S. mansoni in sizes of 380, 176, 174, 130, 81, 16 and 9 bp. Samples showing a S. bovis or S. mansoni MboI pattern were subsequently digested with AluI (Fermentas), which differentiates between S. bovis (no cut; 981 bp) and S. mansoni (fragments of 545 and 421 bp). We used for each RFLP 8.8 µL of the same PCR product in a reaction of 10 µL (1 µL buffer, 0.2 µL enzyme) for digestion with the MboI and the AluI enzyme, respectively. To confirm the genotype based on restriction enzymes, the ITS rDNA region was sequenced for 19 randomly chosen parasite larvae by Sanger sequencing. The PCR products were purified using the E.Z.N.A Cycle Pure kit (Omega Bio-tek, Norcross, USA). Sanger sequencing was done using a Big Dye Chemistry Cycle Sequencing kit v3.1 in a 3130 Genetic Analyzer (Applied Biosystems). Sequences were assembled and manually edited using Geneious® v6.1.6. (http://geneious.com).
Hybrid definition
The hybrid definition used in this study builds on barcoding; hybrids are defined based on the genetic profile of a mitochondrial marker (partial cytochrome c oxidase subunit I (cox1)) and a nuclear marker (ITS rDNA). If discordance between both profiles was found (mito-nuclear discordance) the parasite was defined as ‘hybrid’. If both markers showed a S. haematobium profile, the parasite was defined as S. haematobium, although introgression of other nuclear S. bovis genes cannot be ruled out (Boon et al. Reference Boon, Fannes, Rombouts, Polman, Volckaert and Huyse2017). Parasites with a S. bovis cox1 profile that lacked an ITS rDNA profile were also defined as S. haematobium × S. bovis hybrids, assuming that human infection by pure S. bovis parasites is absent or at most rare (Boissier et al. Reference Boissier, Grech-Angelini, Webster, Allienne, Huyse, Mas-Coma, Toulza, Barré-Cardi, Rollinson, Kincaid-Smith, Oleaga, Galinier, Foata, Rognon, Berry, Mouahid, Henneron, Moné, Noel and Mitta2016). Hence, in this study, the frequency of hybrids is a synonym for the frequency of eggs or miracidia with a S. bovis cox1 profile.
Data analyses
R v3.3.1-based packages (R Development Core Team, 2016) were used for all statistical analyses. The genetic profile of the mitochondrial cox1 fragment and the nuclear ITS rRNA was reported for each parasite stadium (adult worms, eggs and miracidia). To test whether the frequency of hybrids differed among the four regions, a Kruskal–Wallis test (Kruskal and Wallis, Reference Kruskal and Wallis1952) was performed with the frequency of hybrids per village as dependent variable and region as grouping factor. Afterwards, pairwise comparisons among the four regions were performed using Dunn's test (Dunn, Reference Dunn1964) with a Bonferroni correction for multiple comparisons. Both tests were performed with the R-package dunn.test (Dinno, Reference Dinno2016).
We explored with a principal component analysis (PCA) with R-package ade4 (Dray and Dufour, Reference Dray and Dufour2007) the correlation between the frequency of hybrids per village, S. haematobium prevalence and S. mansoni prevalence. The prevalence of S. haematobium and S. mansoni was based on microscopy-based egg counts, and the frequency of hybrids was based on the molecular identification of eggs or miracidia. The statistical significance of the association between the frequency of hybrids per village and S. mansoni prevalence was investigated with a linear regression based on the percentage of hybrids per village (separated per year of sampling) as the dependent variable and S. mansoni prevalence as the independent variable.
RESULTS
Species identification of the mitochondrial cox1 and nuclear ITS marker
The number and percentage of parasites with a Schistosoma bovis, S. haematobium or S. mansoni profile for each genetic marker is presented in Table 2. The cox1 fragment was successfully genotyped in 1236 parasite stadia; the ITS rDNA was successfully genotyped by RFLP in 268 parasite stadia. The lower success rate of the latter may be due to the deteriorating DNA quality since the ITS PCR was performed 5 years after the cox1 PCR. The ITS rDNA profile was successfully confirmed with Sanger sequencing in a test panel of 12 miracidia. Despite the relatively low number of ITS profiles (n = 268), we achieved a good geographical coverage for both markers (Table 1). Table 1 shows the frequency of parasite stadia with a S. bovis mitochondrial cox1 profile for each geographic region and village, per year of sampling.
Table 2. The cox1 haplotype (A) and ITS rDNA genotype (B) of all schistosome adult worms, eggs and miracidia sampled in Northern Senegal between 2006 and 2010. S.h /S.b and S.h /S.m indicate that two species bands were observed during the RD-PCR or fragment analysis, or that two peaks were visible after Sanger sequencing.
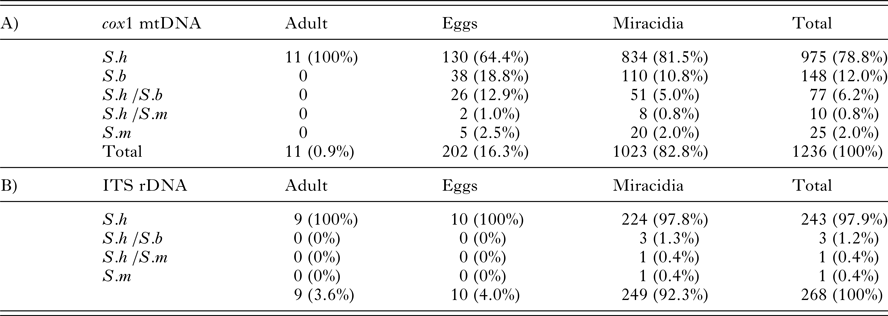
S.h, S. haematobium; S.b, S. bovis ;S.m, S. mansoni.
Averaged over all villages, 79% of the schistosome specimens had a S. haematobium (S.h) cox1 profile and 12% had a S. bovis (S.b) profile (Table 2). The percentage of the S.b profile was higher in eggs (19%) than in miracidia (11%). Six percent of the samples displayed a double-banded Sh/Sb cox1 profile; the majority in eggs (13% compared with 5% in miracidia).
Of the 268 parasites successfully genotyped with ITS rDNA, 243 showed a S. haematobium profile (97.9%, Table 2). Three miracidia (1%) presented a hybrid S.h /S.b ITS genotype with RFLP, two of them in the village of Pakh and one in Kodit. The hybrid ITS genotype was confirmed in each case by Sanger sequencing; double chromatogram peaks were clearly visible at the diagnostic point mutations between S. haematobium (Acc. no FJ588857) and S. bovis (Acc. no FJ588862). Two of those miracidia had a S.h cox1 profile, while the third was not successfully amplified for cox1. Not a single S.b ITS profile was found. For 213 parasites we successfully genotyped both cox1 and ITS rDNA. Twenty of these (9%) showed a S.b or a S.h /S.b cox1 profile with a S.h ITS profile and were thus confirmed to be hybrids. This percentage is slightly lower than the total number of hybrids found when adopting the barcoding approach by only taking the cox1 data into account (12%). The latter statistic is based on a total of 1236 parasite stages (Table 2).
Five eggs and 20 miracidia showed a S. mansoni cox1 haplotype; their ITS rDNA was not successfully genotyped. We assume that they are pure S. mansoni eggs excreted via urine. Two eggs (from Ndieumeul) and eight miracidia (one from Mbodienne, five from Rhonne and two from Ndieumeul) had both an S.h and S.m band at cox1. From five of these the ITS locus was successfully genotyped as S.h. In one miracidium the ITS genotype was confirmed to be S.m with Sanger sequencing (one insertion in comparison with Acc. no JQ289742) while a double S.h and a S.b band was found at cox1.
Prevalence of Schistosoma haematobium and S. mansoni infection in humans
The prevalence of S. haematobium was high in all regions (Table 2) and ranged between 57.1% in Diatar and 100% in Kodit. The prevalence of S. mansoni in stools was highly variable between regions. In the Middle Valley of the Senegal River S. mansoni prevalence was lowest with values between 8% (Diatar) and 33% (Dioundiou in 2006). The highest prevalence was found in Rhonne (81%) and in the villages bordering Lac de Guiers, except for Pakh (17%) (Table 1). The same pattern was observed for co-infection intensities with S. haematobium and S. mansoni (data not shown).
Geographical distribution of hybrids
The frequency of hybrids, defined as Schistosoma stadia with a S. bovis cox1 profile, was not homogeneously distributed among the four regions in Northern Senegal (Kruskal–Wallis test, P = 0.01). Villages bordering Lac de Guiers harboured significantly more schistosomes with a S. bovis cox1 profile (on average 27.5% in 2006 and 2007 and 25.7% in 2009 and 2010) than villages in the Middle Valley of the Senegal River (on average 2.4%, P value Dunn's = 0.01) and the Lampsar River (on average 1.8%, P value Dunn's = 0.046). In Rhonne, in the Delta of the Senegal River, the hybrid frequency was 17% (see Table 2, Fig. 1).
Association between the frequency of hybrids and the prevalence of Schistosoma species
The frequency of the Schistosoma bovis cox1 profile was very high in Diokhor Tack; 74% of the parasite larvae were identified as hybrids. We excluded this outlying observation in the statistical analysis because it is based on parasite eggs from a single host. The PCA biplot (Fig. 2) shows the clustering of villages per geographical region. All villages from the Middle Valley of the Senegal River and the Lampsar River were found at the left side of the first principal component. They had low hybrid frequencies and a relatively low S. mansoni prevalence. However, these villages varied in the direction of the second principal component, which corresponds with S. haematobium prevalence. Rhonne and the villages bordering Lac de Guiers were found at the right site of the PCA biplot, corresponding to a high frequency of hybrids and a high prevalence of S. mansoni. Pakh and Ndieumeul (sampling year 2009/2010) were found at the bottom of the figure. They had the lowest S. haematobium prevalence. The incorporation of adults instead of only school-age children (which is the case in sampling year 2009/2010) is expected to lead to a lower S. haematobium prevalence. Schistosoma haematobium and S. mansoni prevalences show a typical age-related pattern with a rapid increase in children, a peak in adolescents and a decline in adults (Meurs et al. Reference Meurs, Mbow, Vereecken, Menten, Mboup and Polman2012). The percentage of hybrids was significantly positively associated with S. mansoni prevalence (linear regression: P = 0.004, R 2 = 46.7%). Both variables were highest in Rhonne and the villages bordering Lac de Guiers.
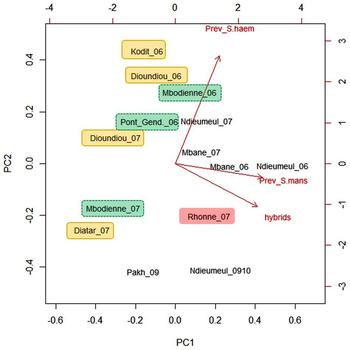
Fig. 2. Biplot of a principal component analysis on the percentage of larval schistosomes with S. bovis cox1 mtDNA (hybrids), the prevalence of S. haematobium (Prev_S. haem) and the prevalence of S. mansoni (Prev_S.mans) in nine villages in Northern Senegal by year of sampling. The cumulative proportion is 0.91 for the first two principal components (PC1: 0.61; PC2: 0.30). Observations were coloured per geographical region (Middle Valley Senegal River: yellow; Delta Senegal River: red; Lampsar River: green; Lac de Guiers: colourless). The prevalence of S. haematobium and S. mansoni was based on egg counts (microscopy), the frequency of hybrids was based on the molecular identification of eggs or miracidia.
Villages that were sampled more than once clustered together, which indicates that over time the frequency of hybrids and the prevalence of Schistosoma species remained constant. Only the two observations of the village Mbodienne did not cluster together: among the 28 genotyped parasites two were hybrids (7%) in 2006, while no hybrids were found among 104 parasites in 2007. The prevalence of S. haematobium and S. mansoni was also lower in 2007 in Mbodienne.
DISCUSSION
This study supports previous research and confirms that, in Senegal, the majority of hybrid parasites typically present a Schistosoma haematobium nuclear (ITS rDNA) and a S. bovis mitochondrial (cox1) profile (Huyse et al. Reference Huyse, Webster, Geldof, Stothard, Diaw, Polman and Rollinson2009 and Webster et al. Reference Webster, Webster, Gouvras, Garba, Lamine, Diaw, Seye, Tchuem Tchuenté, Simoonga, Mubila, Mwanga, Lwambo, Kabatereine, Lange, Kariuki, Mkoji, Rollinson and Stothard2013a, Reference Webster, Diaw, Seye, Webster and Rollinsonb). No pure S. bovis ITS profiles were found; only 1% of the ITS profiles displayed both parental profiles. This stands however in contrast with the observation by Webster et al. (Reference Webster, Webster, Gouvras, Garba, Lamine, Diaw, Seye, Tchuem Tchuenté, Simoonga, Mubila, Mwanga, Lwambo, Kabatereine, Lange, Kariuki, Mkoji, Rollinson and Stothard2013a, Reference Webster, Diaw, Seye, Webster and Rollinsonb) in the village Nder at Lac de Guière where 18% (n = 59) of the genotyped miracidia from children's urine (n = 372) presented a hybrid profile. Of those hybrids, 19% had a S. haematobium ITS and S. bovis cox1 profile while 80% had a S. bovis or a double-banded ITS rDNA profile. The low quality of the ITS rDNA PCR in this study might have resulted in an underestimation of S. bovis rDNA ITS fragments. However, the difference with the study in Nder may also be due to genuine geographical variation in the distribution of the different types of hybrids.
Barcoding and hybrids
Before discussing the spatiotemporal distribution of hybrids we have to consider the tools used to detect hybrids. Hybrid schistosomes are typically defined based on the discordance between the genetic profile of the mitochondrial cox1 marker and the nuclear ITS rDNA marker (Sang et al. Reference Sang, Crawford and Stuessy1995; Steinauer et al. Reference Steinauer, Hanelt, Mwangi, Maina, Agola, Kinuthia, Mutuku, Mungai, Wilson, Mkoji and Loker2008; Boon et al. Reference Boon, Fannes, Rombouts, Polman, Volckaert and Huyse2017). The observed discordance may be the result from historical hybridization followed by introgression or from contemporary hybridization (Detwiler and Criscione, Reference Detwiler and Criscione2010; Hedrick, Reference Hedrick2013). In case of the latter, a mixed nuclear profile, indicating a F1 hybrid, would be expected. In case of the former, the ITS profile will have become identical to one of the parental species due to concerted evolution in the ribosomal RNA cistron (Sang et al. Reference Sang, Crawford and Stuessy1995). In that case, the hybridization event would be masked and the number of hybrids underestimated. For example, hybrids resulting from a cross between a male S. bovis with a female S. haematobium may, after a few generations of backcrossing with S. haematobium, no longer be detectable with the ITS marker. In the reverse cross, hybridization would still be detectable by the S. bovis mitochondrial profile.
When detecting a S. bovis mitochondrial profile in schistosome larvae collected from human samples, one can be absolutely sure of past or ongoing hybridization. Therefore, we argue that the rapid diagnostic PCR assay based on the barcoding fragment is a useful tool to screen large number of human samples, even if it will underestimate the number of hybrids, as it cannot detect the reverse crosses that display a S. haematobium cox1 profile and a S. bovis ITS profile. In our study this cross was much less prevalent than the former cross; hybrid parasites with a S. bovis ITS fragment must be rare or very focal, as they have been mostly found in one village (Nder) by Webster et al. (Reference Webster, Diaw, Seye, Webster and Rollinson2013b). When screening cercariae, both the nuclear and the mitochondrial marker should be used, as pure S. bovis and pure S. haematobium species can sometimes use the same snail species.
Two factors may contribute to the more commonly observed mitochondrial introgression (18%) compared with nuclear introgression (1%): drift and selection (see also below). Because mtDNA is maternally inherited, it is more prone to drift (and indirectly to introgression) in comparison with bi-parentally inherited nuclear DNA (Ballard and Whitlock, Reference Ballard and Whitlock2004; Currat et al. Reference Currat, Ruedi, Petit and Excoffier2008). For example, hybrids have been detected with a complete replacement of mtDNA of one species by another (mitochondrial capture), but without evidence for introgression in the nuclear genome. This pattern was for instance found for two chipmunk species (Good et al. Reference Good, Vanderpool, Keeble and Bi2015). Looking at mitochondrial DNA only may, in this way, provide a biased view of the overall contribution of introgression to the genomes of hybridizing species (Good et al. Reference Good, Vanderpool, Keeble and Bi2015).
Spatiotemporal distribution of hybrids in the SRB
The frequency of hybrid schistosomes remained stable in almost all villages that were sampled twice or more (e.g. for twice in Dioundiou, Mbane; four times in Ndieumeul). Although this study spans only a short time period, it suggests that hybridization between S. haematobium and S. bovis may already have happened in the past and has led to the spread of, and a stable frequency of S. bovis mtDNA among the human population. Hybrid frequency differed between regions, varying between <5% in the Middle Valley of the Senegal River and >30% in villages bordering Lac de Guiers. To explore the potential influence of hybridization in the SRB on schistosomiasis epidemiology, we associated the frequency of hybrids with the prevalence of schistosomiasis. In the discussion below, to find drivers for this heterogeneous pattern, we integrated also data from the literature (e.g. distribution of cattle and, intermediate snail host) with the occurrence of hybrids across the SRB. We start from two hypotheses to explain the geographical heterogeneity. First, in regions with a high hybrid frequency, local conditions may facilitate hybridization between S. haematobium and S. bovis (presently or in the past) more than in other regions. Secondly, in regions with a higher frequency of hybrids, the introgression of S. bovis mtDNA (or nuclear DNA) may be more adaptive and under positive selection (Irwin, Reference Irwin2002; Toews and Brelsford, Reference Toews and Brelsford2012).
Link between hybrid distribution and the prevalence of schistosomiasis
The prevalence of Schistosoma haematobium was high in all the villages while the prevalence of S. mansoni was more patchy with a high prevalence in villages along Lac de Guiers and Rhonne, and a low prevalence in the east of the SRB. The frequency of hybrids was not linked with the prevalence of urinary schistosomiasis. This could advocate against the hypothesis that hybrids would alter schistosomiasis epidemiology, by increasing the transmission of urinary schistosomiasis in the SRB. However, S. haematobium prevalence showed little variation across the villages, which makes it less plausible to find a link.
A remarkable observation was the positive association between the frequency of S. haematobium × S. bovis hybrids and the prevalence of S. mansoni parasites. This suggests that in villages with a high S. mansoni prevalence there is also a higher risk of infection with hybrids than in villages with mainly urinary schistosomiasis. We slightly violated the conditions for this linear regression: the errors of this model are not fully independent. Some of the villages were sampled twice and there may be some geographical auto-correlation. We did not have sufficient observations to the model dependence of the residuals. However, also within geographical regions and even within villages that were sampled twice we always found the same trend: a positive association between the percentage of hybrids and S. mansoni prevalence (Fig. 3).

Fig. 3. Scatter plot of the association between the percentage of hybrids and Schistosoma mansoni prevalence. The linear regression of the percentage of hybrids per village and per year, for all regions combined, is indicated by the dotted regression line (P = 0.004, R 2 = 46.7%, regression coefficient = 0.32). The coloured regression lines show the associations for each geographical region separately.
To explain this association we extrapolate our results (at the village level) to the individual level and look at parasite transmission between individual hosts. Parasites are known to have a different transmission potential in individual hosts with mixed infections in comparison with hosts with single infections (Alizon et al. Reference Alizon, de Roode and Michalakis2013). These interactions can be direct (e.g. competition for host resources, heterologous mating (Tchuem Tchuenté et al. Reference Tchuem Tchuenté, Southgate, Combes and Jourdane1996)) or indirect, by evoking cross-reactive immune responses (Alizon et al. Reference Alizon, de Roode and Michalakis2013). One possible explanation is that once an individual is infected with a schistosome species (e.g. S. mansoni), (s)he will become more susceptible to infection by schistosome species that are usually unable to infect humans (e.g. S. bovis or hybrid species). For example, S. mansoni actively suppresses immune responses (McSorley and Maizels, Reference McSorley and Maizels2012). In this way, S. mansoni infections, or mixed infections with S. mansoni and S. haematobium, may first facilitate infections with S. bovis or first generation hybrids, followed by hybridization with S. haematobium and ultimately drive the transmission of hybrid schistosomes. In order to test this hypothesis, future studies should focus on the individual level, and link the presence of hybrids within a single host with Schistosoma infection intensity. Also, other host-related factors (e.g. host-immunology, ethnicity, water contact behavior) may be associated with the frequency of hybrids. In mycobacteria for example, such variable host–parasite interactions were found, where specific mycobacterial lineages were adapted to particular human populations (Gagneux et al. Reference Gagneux, DeRiemer, Van, Kato-Maeda, de Jong, Narayanan, Nicol, Niemann, Kremer, Gutierrez, Hilty, Hopewell and Small2006).
Other explanations of the geographically heterogeneous hybrid distribution
Many other factors may explain or add to the heterogeneous distribution of S. haematobium × S. bovis hybrids. The occurrence of hybrids may be associated with the (historical) prevalence of S. bovis. Schistosoma bovis inhabits the intestinal veins of cattle, sheep and goats in parts of West Africa including the SRB. A study by Diaw et al. (Reference Diaw, Vassiliades, Thiongane, Seye, Sanrr and Diouf1998) mentions that along the shores of Lac de Guiers, where most hybrids are found, the breeding of livestock is much more developed and more intensive than in the delta of the Senegal River. As for human schistosomiasis, Diaw et al. (Reference Diaw, Vassiliades, Thiongane, Seye, Sanrr and Diouf1998) showed also a sharp increase of bovine schistosomiasis and the appearance of new infection foci at the Senegal River Delta and Lac de Guiers after the construction of the dams. Diaw et al. (Reference Diaw, Vassiliades, Thiongane, Seye, Sanrr and Diouf1998) found that S. bovis was more prevalent than S. curassoni in Northern Senegal. They related this to the wide distribution of the snail Bulinus truncatus, intermediate host of S. bovis in Senegal. In a more recent study, no S. curassoni was found in the SRB (Webster et al. Reference Webster, Diaw, Seye, Webster and Rollinson2013b); nor were worms with a pure or a hybrid S. haematobium profile found in cattle. Hence it remains unknown whether they are a reservoir for hybrid schistosomes. Webster et al. (Reference Webster, Webster, Gouvras, Garba, Lamine, Diaw, Seye, Tchuem Tchuenté, Simoonga, Mubila, Mwanga, Lwambo, Kabatereine, Lange, Kariuki, Mkoji, Rollinson and Stothard2013a, Reference Webster, Diaw, Seye, Webster and Rollinsonb) observed hybrid crosses between S. haematobium and S. curassoni in children living in Tambacounda and the Vallée du Ferlo but not in the SRB. In the same regions where S. haematobium × S. curassoni hybrids were found in children, S. curassoni was found in domestic animals. These observations support the hypothesis that the occurrence of schistosome hybrids in humans may be associated with the occurrence of the paternal bovine species in livestock in the same region. More animal studies are needed to confirm this for the SRB.
Another explanation may be that the introgressed mtDNA is under selection. Mitochondrial DNA has been shown to be highly adaptive (Ballard and Whitlock, Reference Ballard and Whitlock2004; Ballard and Melvin, Reference Ballard and Melvin2010; Hill, Reference Hill2015) and important in determining aspects of life-history traits (Ballard and Melvin, Reference Ballard and Melvin2010). Mitochondria are responsible for the majority of ATP production; 13 genes encode for subunits of the electron transport chain where carbohydrates and fats are oxidized to generate carbon dioxide, water and ATP. The additional genes encode for the translational machinery of the mtDNA itself (Ballard and Whitlock, Reference Ballard and Whitlock2004). It is predicted that important ecological differences are mediated by mitochondrial changes. For example, evolutionary changes in metabolic rates under stress could be effected by changes in the mtDNA sequence (Ballard and Whitlock, Reference Ballard and Whitlock2004). It was found that negative frequency-dependent selection may act on mtDNA and maintain mitochondrial genetic variation (Kazancioǧlu and Arnqvist, Reference Kazancioǧlu and Arnqvist2014). If different mitochondrial haplotypes are favored in different locations, phylogeographic breaks (the occurrence of sharp breaks in mtDNA genealogy that often distinguish regional sets of populations across a species’ range) can persist, even in the face of gene flow (Irwin, Reference Irwin2002).
In our setting, one of the two mtDNA cox1 variants (the S. bovis or the S. haematobium profile) might be adaptive in certain water bodies or villages, leading to the fixation of the S. bovis variant in those areas, and thereby resulting in the observed heterogeneous distribution of hybrids.
Also nuclear S. bovis genes may have introgressed that are adaptive or under selection. Several studies have demonstrated evidence for positive selection on introgressed alleles in animals (Harrison and Larson, Reference Harrison and Larson2014). For example, a mutation that causes melanism in North American wolves was derived from past hybridization with domestic dogs. The nuclear locus seemed under positive selection in specific habitats: in forested habitats it had risen to high frequencies (Anderson et al. Reference Anderson, Bridgett, Candille, Musiani, Stahler, Smith, Padhukasahasram, Randi, Leonard, Bustamante, Ostrander, Tang, Robert and Barsh2009). The susceptibility of a snail host depends at least partly on the geographic origin of the snail and the parasite strain (Theron et al. Reference Theron, Rognon, Gourbal and Mitta2014). For example, in the Middle Valley (where we found a much lower hybrid frequency), S. haematobium is compatible with B. truncatus while in the other regions of the SRB B. globosus is the main intermediate host (Southgate et al. Reference Southgate, de Clercq, Séne, Rollinson, LY and Vercruysse2000; Sène et al. Reference Sène, Southgate and Vercruysse2004). In light of this, one may hypothesize that hybrids may have a higher compatibility with the snail strains present around Lac de Guiers compared with other regions in the SRB. Also, the near-total absence of hybrids in the Middle Valley might be linked with the (near-total) absence of B. globosus in that area (Huyse, personal observations), if indeed B. globosus is a better host for hybrids.
In conclusion, in order to improve insight into potential drivers of hybridization in the SRB, more information on the snail host distribution and the occurrence of S. bovis is needed. It is also necessary to screen potential animal reservoirs for hybrid infection. Snail infectivity experiments on the other hand can shed more light on hybrid-snail compatibility.
SUPPLEMENTARY MATERIAL
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182018000525
ACKNOWLEDGEMENTS
We thank the field team of Richard Toll, A. Yague, M. Diop, M. Wade and N. Sy for assisting with the sample collection and microscopic analysis. We thank B. Hellemans and S. Geldof for assisting with the molecular work. We also thank Moustapha Mbow for useful suggestions on an earlier version of this manuscript. A special word of thanks goes to the people of the Senegalese villages for their kind participation in the study.
FINANCIAL SUPPORT
N.B. benefited from a Flemish Interuniversity Council (VLIR) VLADOC fellowship and travel grants from the Research Foundation – Flanders and VLIR. Part of the research was covered by a Research Grant of the Research Foundation—Flanders (contract G.0552.10) and a Krediet to navorsers (contract 1516913N) to TH.
ETHICAL STATEMENT
Scientific material was collected in Senegal between 2006 and 2010 in the frame of the SCHISTOINIR project (www.york.ac.uk/res/schistoinir). For this project, approval was obtained from the review board of the Institute of Tropical Medicine (Antwerp, Belgium), the Ethical Committee of Antwerp University Hospital (Antwerp, Belgium) and ‘Le Comité National d'Ethique de la Recherche en Santé’ (Dakar, Senegal). For the sample from Diatar (1993) approval was obtained from the NHS-LREC of Imperial College (London, UK). Informed consent was obtained from all participants prior to incorporation in the study. At the end of the study inhabitants of participating villages were treated with praziquantel at a dose of 40 mg kg−1 according to World Health Organization (WHO) (WHO, 2006).
CONFLICTS OF INTEREST
None.








