Introduction
Species identification is often emphasized as a basic prerequisite for the understanding of diversity, ecology and evolution of the living world (Hey et al., Reference Hey, Waples, Arnold, Butlin and Harrison2003; Olson and Tkach, Reference Olson and Tkach2005; Shaffer et al., Reference Shaffer, Davy and Bell2019). For the last three decades, DNA sequence data including DNA barcoding became an integral part of species characterization and identification. Nevertheless, the actual specimen associated with a particular DNA sequence must still be identified by the use of traditional morphology-based analysis to ensure that the sequence and identification are linked correctly (Blasco-Costa et al., Reference Blasco-Costa, Cutmore, Miller and Nolan2016; Schwelm et al., Reference Schwelm, Georgieva, Grabner, Kostadinova and Sures2021). Trematode species identification has been and continues to be based on morphological data collected from adult specimens since the larval stages often lack reliable distinguishing morphological characters. Thereafter, specimens of either adults or larval stages can be accurately identified if they match the sequence of the known species. DNA sequence databases rely on sequences that are derived from taxonomically correctly identified isolates. Although the DNA sequences became a primary source for assessment and measure of biodiversity, there is a growing problem of taxonomic misidentification in public DNA databases (Bridge et al., Reference Bridge, Roberts, Spooner and Panchal2003; Tautz et al., Reference Tautz, Arctander, Minelli, Thomas and Vogler2003; Vilgalys, Reference Vilgalys2003; Valkiūnas et al., Reference Valkiūnas, Atkinson, Bensch, Sehgal and Ricklefs2008; Locke et al., Reference Locke, Al-Nasiri, Caffara, Drago, Kalbe, Lapierre, McLaughlin, Nie, Overstreet, Souza, Takemoto and Marcogliese2015; Achatz et al., Reference Achatz, Martens, Kostadinova, Pulis, Orlofske, Bell, Fecchio, Oyarzún-Ruiz, Syrota and Tkach2021; Bensch et al., Reference Bensch, Inumaru, Sato, Lee Cruz, Cunningham, Goodman, Levin, Parker, Casanueva, Hernández, Moreno-Rueda and Rojo2021; Pantoja et al., Reference Pantoja, Faltýnková, O'Dwyer, Jouet, Skírnisson and Kudlai2021). The problems in each dataset are different. The sequences may be incorrectly labelled, of poor quality, incomplete for reliable comparison or without voucher specimens (Bridge et al., Reference Bridge, Roberts, Spooner and Panchal2003; Locke et al., Reference Locke, Al-Nasiri, Caffara, Drago, Kalbe, Lapierre, McLaughlin, Nie, Overstreet, Souza, Takemoto and Marcogliese2015; Blasco-Costa et al., Reference Blasco-Costa, Cutmore, Miller and Nolan2016).
One of the trematode groups that has largely benefited from application of the molecular genetic methods is the genus Diplostomum von Nordmann, 1832 – a species-rich genus with complex taxonomy distributed worldwide in freshwater ecosystems (Niewiadomska, Reference Niewiadomska, Gibson, Jones and Bray2002). The members of Diplostomum are important fish pathogens, with a three-host life-cycle encompassing lymnaeid snails, fish and fish-eating bird hosts; the most pathogenic stage are metacercariae infecting fish eyes or brain, which can impair vision and lead to cataract formation in wild and farmed fish (Shigin, Reference Shigin1986; Karvonen et al., Reference Karvonen, Seppälä and Valtonen2004; Karvonen and Marcogliese, Reference Karvonen, Marcogliese, Woo, Leong and Buchmann2020).
The development of suitable molecular markers, particularly the barcode region of the cytochrome c oxidase subunit (cox1), allowed a wealth of studies to prospect for Diplostomum with an aid of molecular genetic methods; thus, in North America, Europe, Africa and Asia an unexpectedly wide spectrum of lineages and complexes of cryptic species were revealed (Galazzo et al., Reference Galazzo, Dayanandan, Marcogliese and McLaughlin2002; Moszczynska et al., Reference Moszczynska, Locke, McLaughlin, Marcogliese and Crease2009; Locke et al., Reference Locke, McLaughlin and Marcogliese2010a, Reference Locke, Al-Nasiri, Caffara, Drago, Kalbe, Lapierre, McLaughlin, Nie, Overstreet, Souza, Takemoto and Marcogliese2015; Georgieva et al., Reference Georgieva, Soldánová, Pérez-del-Olmo, Dangel, Sitko, Sures and Kostadinova2013; Hoogendoorn et al., Reference Hoogendoorn, Smit and Kudlai2020).
The recent intensive studies resulted in generating sequence libraries, presenting a platform for further molecular delineation and linking larval stages with adults (Kudlai et al., Reference Kudlai, Oros, Kostadinova and Georgieva2017; Achatz et al., Reference Achatz, Martens, Kostadinova, Pulis, Orlofske, Bell, Fecchio, Oyarzún-Ruiz, Syrota and Tkach2021; Schwelm et al., Reference Schwelm, Georgieva, Grabner, Kostadinova and Sures2021). To date, libraries contain more than 40 species and lineages from Africa, Asia, Europe and North America (Hoogendoorn et al., Reference Hoogendoorn, Smit and Kudlai2020). However, still a small part of these sequences is based on adult isolates (18 species in total), of which 15 represent identified species, i.e. Diplostomum adamsi Lester & Huizinga, 1977, Diplostomum alarioides Dubois, Reference Dubois1937, Diplostomum alascense Dubois, 1969, Diplostomum ardeae Dubois, 1969, Diplostomum gavium (Guberlet, 1922), Diplostomum huronense (La Rue, 1927), Diplostomum indistinctum (Guberlet, 1923), Diplostomum marshalli Chandler, 1954 and Diplostomum scudderi Olivier, 1941 in North America (Galazzo et al., Reference Galazzo, Dayanandan, Marcogliese and McLaughlin2002; Locke et al., Reference Locke, Al-Nasiri, Caffara, Drago, Kalbe, Lapierre, McLaughlin, Nie, Overstreet, Souza, Takemoto and Marcogliese2015; Achatz et al., Reference Achatz, Martens, Kostadinova, Pulis, Orlofske, Bell, Fecchio, Oyarzún-Ruiz, Syrota and Tkach2021), Diplostomum lunaschiae Locke, Drago, Núñez, Rangel e Souza & Takemoto, 2020 in South America (Locke et al., Reference Locke, Drago, Núñez, Souza and Takemoto2020) and Diplostomum mergi Dubois, 1932, Diplostomum spathaceum (Rudolphi, 1819), Diplostomum pseudospathaceum Niewiadomska, 1984 and Diplostomum rauschi Shigin, 1993 in Europe (Pérez-del-Olmo et al., Reference Pérez-del-Olmo, Georgieva, Pula and Kostadinova2014; Selbach et al., Reference Selbach, Soldánová, Georgieva, Kostadinova and Sures2015; Achatz et al., Reference Achatz, Martens, Kostadinova, Pulis, Orlofske, Bell, Fecchio, Oyarzún-Ruiz, Syrota and Tkach2021; Schwelm et al., Reference Schwelm, Georgieva, Grabner, Kostadinova and Sures2021). However, this number is most likely to be changed as Achatz et al. (Reference Achatz, Martens, Kostadinova, Pulis, Orlofske, Bell, Fecchio, Oyarzún-Ruiz, Syrota and Tkach2021) questioned the identification of D. ardeae sensu Locke et al. (Reference Locke, Al-Nasiri, Caffara, Drago, Kalbe, Lapierre, McLaughlin, Nie, Overstreet, Souza, Takemoto and Marcogliese2015). Thanks to recent studies (particularly Achatz et al., Reference Achatz, Martens, Kostadinova, Pulis, Orlofske, Bell, Fecchio, Oyarzún-Ruiz, Syrota and Tkach2021) out of these 15 species, specimens of 13 species (D. alarioides, D. alascense, D. ardeae, D. gavium, D. huronense, D. indistinctum, D. lunaschiae, D. marshalli, D. mergi, D. pseudospathaceum, D. rauschi, D. scudderi and D. spathaceum) were obtained from naturally infected bird hosts and connected to the original description (Pérez-del-Olmo et al., Reference Pérez-del-Olmo, Georgieva, Pula and Kostadinova2014; Locke et al., Reference Locke, Al-Nasiri, Caffara, Drago, Kalbe, Lapierre, McLaughlin, Nie, Overstreet, Souza, Takemoto and Marcogliese2015; Heneberg et al., Reference Heneberg, Sitko and Těšínský2020; Achatz et al., Reference Achatz, Martens, Kostadinova, Pulis, Orlofske, Bell, Fecchio, Oyarzún-Ruiz, Syrota and Tkach2021) or described as new species (Locke et al., Reference Locke, Drago, Núñez, Souza and Takemoto2020). Still, most of the sequences available in GenBank are based on metacercariae, a stage with least distinguishing characters, and only a small portion of them is linked to voucher material, thus there is no unequivocal identification which would warrant assignment to valid species for many of isolates (Selbach et al., Reference Selbach, Soldánová, Georgieva, Kostadinova and Sures2015; Hoogendoorn et al., Reference Hoogendoorn, Smit and Kudlai2020). Another basic issue concerns the consistency and uniformity in naming unidentified species and lineages by different authors that often follow different nomenclature. This creates misidentifications and misinterpretations in later studies when authors solely rely on identifications provided for sequences in GenBank.
In this study, we provide species identification for the ‘questionable’ lineage of Diplostomum of Georgieva et al. (Reference Georgieva, Soldánová, Pérez-del-Olmo, Dangel, Sitko, Sures and Kostadinova2013) in Europe and we extend the recommendation of Blasco-Costa et al. (Reference Blasco-Costa, Cutmore, Miller and Nolan2016) for the ‘best practice’ in molecular approaches to trematode systematics. We characterize morphologically and molecularly (cox1, ITS-5.8S-ITS2 and 28S) adults of Diplostomum baeri based on new material collected from naturally infected Larus canus Linnaeus in Karelia, Russia; thus, we link the original description by Dubois (Reference Dubois1937, Reference Dubois1938, Reference Dubois1970) with our morphological and DNA sequence data. Furthermore, via molecular tools we reveal that sequences of our isolates of D. baeri are identical with those of the lineage Diplostomum sp. clade Q of Georgieva et al. (Reference Georgieva, Soldánová, Pérez-del-Olmo, Dangel, Sitko, Sures and Kostadinova2013). Also, we review the morphology of cercariae reported in the literature as D. baeri or Diplostomum volvens Nordmann, 1832 (based on views of Shigin, Reference Shigin1993 and Niewiadomska, Reference Niewiadomska2010), and we accompany our data with new material of cercariae from Ireland, with DNA sequences and morphology corresponding to Diplostomum sp. clade Q of Georgieva et al. (Reference Georgieva, Soldánová, Pérez-del-Olmo, Dangel, Sitko, Sures and Kostadinova2013) and Selbach et al. (Reference Selbach, Soldánová, Georgieva, Kostadinova and Sures2015), respectively. The current study is another step in elucidating the species spectrum of Diplostomum based on integrative taxonomy with a well-described morphology of adults, linked to sequences.
Materials and methods
Sample collection
Samples of adults were collected from a single specimen of the common gull L. canus found dead on the shore of the Kostomukshskoye Lake (64°39′34″N, 30°48′10″E), Karelia, northwest Russia, in June 2010. The bird was transported on ice to the laboratory and immediately dissected following the protocol of Dubinina (Reference Dubinina1971). A total of 154 worms of the genus Diplostomum were found in the duodenum and small intestine. Collected digeneans were preserved in 96% ethanol for both morphological investigation (without additional pressure) and DNA extraction; a total of five adult worms were used for molecular and morphological analyses. Samples of cercariae were obtained from snails Radix balthica (Linnaeus) [Ampullaceana balthica (Linnaeus) being considered senior synonym by Aksenova et al., Reference Aksenova, Bolotov, Gofarov, Kondakov, Vinarski, Bespalaya, Kolosova, Palatov, Sokolova, Spitsyn, Tomilova, Travina and Vikhrev2018] (two snails infected out of a total of 573) collected in the Lake Lough Corrib (53°20′24.3″N, 9°05′28.6″W), Ireland in July 2019. In the laboratory, snails were placed individually in plastic cups filled with dechlorinated tap water and left for 24 h to detect emergence of cercariae. Emerged cercariae were studied alive, fixed in 96% ethanol for DNA isolation and in 4% formalin for measurements.
Morphological examination
Specimens recovered from the bird were identified as members of the genus Diplostomum, based on the generic diagnosis provided by Dubois (Reference Dubois1970), Shigin (Reference Shigin1993) and Niewiadomska (Reference Niewiadomska, Gibson, Jones and Bray2002). Specimens of adults (n = 5) selected for molecular analysis were vouchered following the concept of Pleijel et al. (Reference Pleijel, Jondelius, Norlinder, Nygren, Oxelman, Schander, Sundberg and Thollesson2008) and the recommendation of Blasco-Costa et al. (Reference Blasco-Costa, Cutmore, Miller and Nolan2016) and series of photomicrographs of vouchers were taken with a digital camera of an Olympus СX41 and BX51 microscope. Thereafter, a small piece of worm body was excised and used for DNA extraction. The remaining voucher (hologenophore) was stained in iron acetocarmine, dehydrated in ethanol, cleared in clove oil, mounted in Canada balsam and used for detailed morphological analyses. Measurements were taken from the digital photomicrographs and total mounts in Canada balsam with the use of the Levenhuk C1400 NG, Levenhuk ToupView image analysis software, V.3.5 and the QuickPHOTO CAMERA 2.3 image analysis software. All measurements in the descriptions and tables are in μm. For the description of morphological characters, we followed the terminology of Niewiadomska (Reference Niewiadomska, Gibson, Jones and Bray2002); for anterior and posterior parts of body, we used the terms ‘prosoma’ and ‘opisthosoma’ proposed by Achatz et al. (Reference Achatz, Martens, Kostadinova, Pulis, Orlofske, Bell, Fecchio, Oyarzún-Ruiz, Syrota and Tkach2021). Morphometric variables were used as in Dubois (Reference Dubois1970) and Shigin (Reference Shigin1993). The voucher specimens were deposited in the Helminthological Collection of Karelian Research Centre RAS, Petrozavodsk, Russia (nos. DB1LC26 and DB2LC26) and in the Helminthological Collection of the Institute of Parasitology (IPCAS, D-829, three hologenophores; D-845, two vials with cercariae), Biology Centre of the Czech Academy of Sciences, České Budějovice, Czech Republic. The morphology of cercariae was studied on live specimens under a light microscope and series of photomicrographs (of three specimens) were taken with a digital camera on Olympus BX51 to obtain measurements with the aid of QuickPHOTO CAMERA 2.3 image analysis software. Cercariae were identified following description and DNA sequences of Selbach et al. (Reference Selbach, Soldánová, Georgieva, Kostadinova and Sures2015). For abbreviations and explanations of characters measured, see Table 2.
DNA amplification and sequencing
Genomic DNA was isolated from an excised part of adult worms and 20–25 ethanol-fixed cercariae following the protocol described by Antar et al. (Reference Antar, Georgieva, Gargouri and Kostadinova2015) or using DNA-Extran kits (Synthol, Moscow). As suggested in Blasco-Costa et al. (Reference Blasco-Costa, Cutmore, Miller and Nolan2016), we used ribosomal and mitochondrial molecular genetic markers in the current study.
The cox1 region of the mtDNA was amplified using the primers Dice1F (forward: 5′-ATTAACCCTCACTAAATTWCNTTRGATCATAAG-3′) and Dice14R (reverse: 5′-TAATACGACTCACTATACCHACMRTAAACATATGATG-3′) (van Steenkiste et al., Reference van Steenkiste, Locke, Castelin, Marcogliese and Abbott2015), or Plat-diploCOX1F (5′-CGTTTRAATTATACGGATCC-3′) and Plat-diploCOX1R (5′-GCATAGTAATMGCAGCAGC-3′) (Moszczynska et al., Reference Moszczynska, Locke, McLaughlin, Marcogliese and Crease2009).
The 28S region of the rDNA was amplified using the primers digl2 (5′-AAGCATATCACTAAGCGG-3′) and 1500R (5′-GCTATCCTGAGGGAAACTTCG-3′) (Snyder and Tkach, Reference Snyder and Tkach2001); additional internal primers ECD2 (5ʹ-CCTTGGTCCGTGTTTCAAGACGGG-3ʹ) (Littlewood et al., Reference Littlewood, Rohde and Clough1997) and 300F (5ʹ-CAAGTACCGTGAGGGAAAGTTG-3ʹ) (Littlewood et al., Reference Littlewood, Curini-Galletti and Herniou2000) were used for sequencing. The ITS1-5.8S-ITS2 region of the rDNA was amplified using the primers D1 (F) (5′-AGGAATTCCTGGTAAGTGCAAG-3′) and D2 (R) (5′-CGTTACTGAGGGAATCCTGGT-3′) (Galazzo et al., Reference Galazzo, Dayanandan, Marcogliese and McLaughlin2002).
Polymerase chain reactions (PCRs) (25 μL) included 12.5 μL of MyFi™ mix, 1.25 μL of each oligonucleotide primer (10 mm), 8 μL of H2O and 1.5 μL of genomic DNA. Cycling parameters of PCR amplification were the same as in van Steenkiste et al. (Reference van Steenkiste, Locke, Castelin, Marcogliese and Abbott2015) and Moszczynska et al. (Reference Moszczynska, Locke, McLaughlin, Marcogliese and Crease2009) for cox1, Tkach et al. (Reference Tkach, Littlewood, Olson, Kinsella and Swiderski2003) for 28S and Galazzo et al. (Reference Galazzo, Dayanandan, Marcogliese and McLaughlin2002) for ITS1-5.8S-ITS2. The amplified products were purified with the Exo-SAP-IT Kit™ Express Reagent (Thermo Fisher Scientific Baltics UAB, Vilnius, Lithuania) following the manufacturer's instructions, sequenced using the same primers of PCRs and the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems-Perkin Elmer, Waltham, Massachusetts) in a MegaBACE sequencer (GE Healthcare Life Sciences). Contiguous sequences were assembled using Geneious v. 11 (Biomatters, Auckland, New Zealand) and deposited in GenBank.
Phylogenetic analyses
Identity of newly generated sequences was checked with the Basic Local Alignment Search Tool (BLAST) (www.ncbi.nih.gov/BLAST/). The novel sequences (Table 1) (cox1, 473 and 836 nucleotides (nt); ITS1-5.8S-ITS1, 1250 nt; 28S, 1300 nt) were aligned with the representative sequences of Diplostomum spp. (n = 38 species/lineages) previously reported from Europe (Supplementary Table S1) with MUSCLE (Edgar, Reference Edgar2004) implemented in Geneious v.11. The ITS1-5.8S-ITS2 sequence of a single species, D. adamsi (syn. D. baeri; AY123042; Galazzo et al., Reference Galazzo, Dayanandan, Marcogliese and McLaughlin2002) reported from North America was included in analyses due to its relevance to the current study. Two datasets (cox1 and ITS-5.8S-ITS2) were prepared. The cox1 alignment (356 nt) comprised of seven novel sequences and 35 sequences of the representatives of Diplostomum from GenBank. The ITS-5.8S-ITS alignment (960 nt) included four novel sequences and 24 sequences of Diplostomum spp. from GenBank. Sequences of Tylodelphys clavata (von Nordmann, 1832) (JX986908, cox1; JQ665459, ITS1-5.8S-ITS2) (Digenea: Diplostomidae) were used as the outgroup based on the results of the phylogenetic analyses of Georgieva et al. (Reference Georgieva, Soldánová, Pérez-del-Olmo, Dangel, Sitko, Sures and Kostadinova2013).
Table 1. Summary data for the sequences of Diplostomum baeri generated in the current study and used for morphological analyses
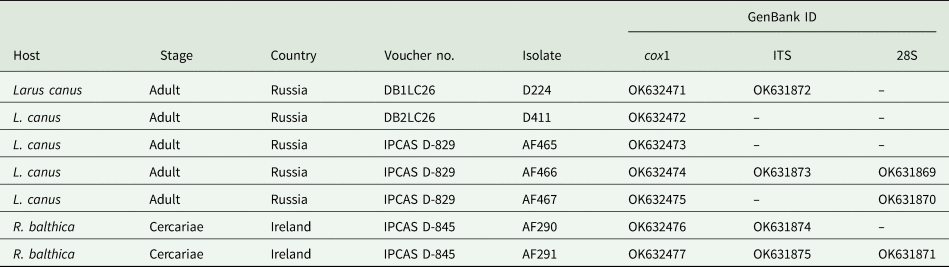
To assess the phylogenetic relationships of Diplostomum spp., we used Bayesian inference (BI) and maximum likelihood (ML) analyses for both datasets. Prior to analyses, the best-fitting model was estimated with jModelTest 2.1.2 (Darriba et al., Reference Darriba, Taboada, Doallo and Posada2012). This was the general time-reversible model incorporating invariant sites and gamma distributed among-site rate variations (GTR + I + G) for both alignments. BI analyses were conducted using MrBayes software (ver. 3.2.3) (Ronquist et al., Reference Ronquist, Teslenko, Van Der Mark, Ayres, Darling, Höhna and Huelsenbeck2012). Markov chain Monte Carlo chains were run for 3 000 000 generations, log-likelihood scores were plotted and only the final 75% of trees were used to produce the consensus tree. ML analyses were conducted using PhyML version 3.0 (Guindon et al., Reference Guindon, Dufayard, Lefort, Anisimova, Hordijk and Gascuel2010) run on the ATGC bioinformatics platform (http://www.atgc-montpellier.fr/). Nodal support was estimated by performing 100 bootstrap pseudoreplicates. FigTree ver. 1.4 software (Rambaut, Reference Rambaut2012) was used to visualize the trees. Genetic distances (uncorrected P-distance) were calculated in MEGA ver. 6. The unique cox1 haplotypes collected in Ireland and Russia in the current study and in Germany and Spain in the previous studies were identified with DnaSP (Rozas et al., Reference Rozas, Sánchez-DelBarrio, Messeguer and Rozas2003). A haplotype network was reconstructed using the median-joining method in PopART software (Population Analysis with Reticulate Trees, http://popart.otago.ac.nz).
Results
Description of the molecular voucher material
Diplostomidae Poirier, 1886
Diplostomum Nordmann, 1832
Diplostomum baeri Dubois, Reference Dubois1937
Synonym: Diplostomum sp. Clade Q of Georgieva et al. (Reference Georgieva, Soldánová, Pérez-del-Olmo, Dangel, Sitko, Sures and Kostadinova2013).
Host: Larus canus canus Linnaeus.
First intermediate host: Radix balthica (Linnaeus).
Locality: Kostomukshskoye Lake, Karelia, northwest Russia (64°39′34″N, 30°48′10″E) (adults); Lough Corrib, Ireland (53°20′24.3″N, 9°05′28.6″W) (cercariae).
Site in host: small intestine and duodenum in bird (adult stage); hepatopancreas in snail (larval stage).
Infection rates: prevalence, 1 of 1 bird; 2 of 573 snails (0.35%); intensity, 5 specimens per bird.
Material: two voucher specimens (DB1LC26, DB2LC26), three hologenophores (IPCAS D-829).
Representative DNA sequences: 28S, three sequences (OK631869–OK631871); ITS1-5.8S-ITS2, four sequences (OK631872–OK631875), cox1, seven sequences (OK632471–OK632477).
Adult (Fig. 1, Table 2)
[Description based on five specimens.] Body distinctly bipartite, partly retroflexed, i.e. prosoma and opisthosoma usually forming dorsally a sharp angle (Fig. 1B). Prosoma elongate-oval, dorso-ventrally flattened, anterior extremity tapered and trilobed, with maximum width at the level of holdfast organ, longer than opisthosoma, posterior rim of prosoma elevated ventrally, slightly forming a cup. Opisthosoma cylindrical, with stout, rounded posterior extremity; slightly narrower anteriorly, maximum width at its mid-level. Tegument smooth.

Fig. 1. Adult Diplostomum baeri ex Larus canus (IPCAS D-829): (A) ventral view and (B) partly retroflexed specimen, ventral view of prosoma, lateral view of opisthosoma.
Table 2. Metrical data of adults of Diplostomum spp.
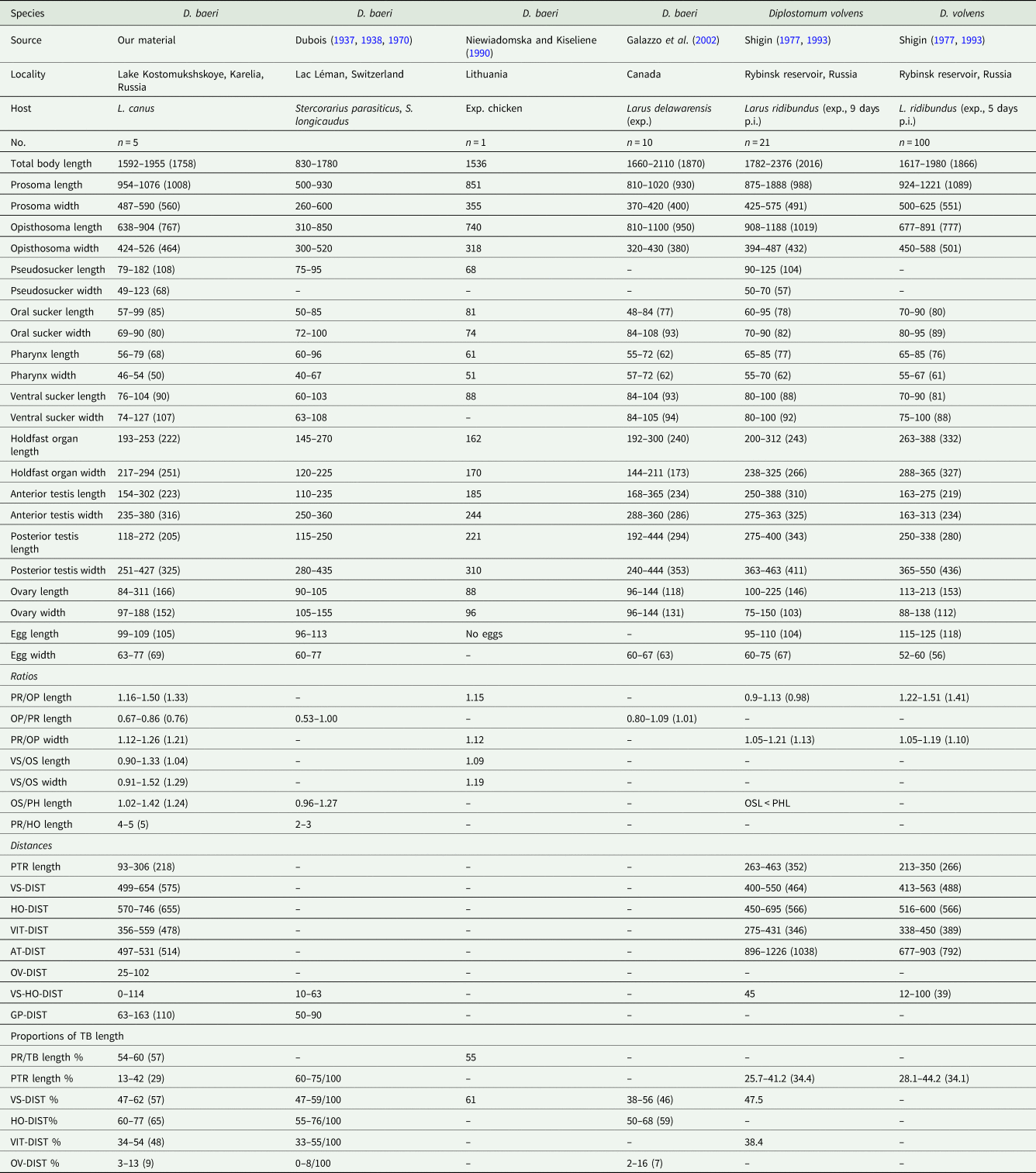
PR, prosoma; OP, opisthosoma; VS, ventral sucker; OS, oral sucker; PH, pharynx; HO, holdfast organ; TB, total body. PTR, post-testicular region; VS-DIST, distance of centre of ventral sucker from anterior margin of prosoma; HO-DIST, distance of anterior margin of holdfast organ from anterior margin of prosoma; VIT-DIST, distance between front level of vitelline follicles and anterior margin of prosoma; AT-DIST, distance of anterior margin of anterior testis from posterior margin of opisthosoma; OV-DIST, distance of ovary from anterior margin of opisthosoma, VS-HO-DIST, distance between ventral sucker and holdfast organ; GP-DIST, distance of genital pore from posterior margin of opisthosoma.
Oral sucker small, weakly muscular, ventro-subterminal, subspherical. Pseudosuckers well developed, posterolateral to oral sucker, reaching back to the level of pharynx. Prepharynx very short. Pharynx well-developed, small. Oesophagus shorter than pharynx. Intestinal bifurcation in first quarter of prosoma. Caeca long, narrow, terminate blindly close to posterior extremity of opisthosoma. Ventral sucker weakly muscular, transversely oval to oval, positioned in its third quarter (Fig. 1A) or at the mid-level of prosoma (Fig. 1B); slightly larger than oral sucker (Table 2). Holdfast organ sub-globular, with median slit, posterior to ventral sucker and contiguous or separated (no further than diameter of ventral sucker).
Testes 2, large, entire, tandem, contiguous or overlapping, in mid-part of opisthosoma. Anterior testis asymmetrical, with one developed lappet. Posterior testis symmetrical, with two lappets turned ventrally. Seminal vesicle coiled, posttesticular, median, contiguous with posterior testis. Ovary subspherical to transversely oval, entire, sub-median, pretesticular, contiguous with anterior testis; close to anterior extremity of opisthosoma or in its first quarter. Vitellarium follicular, vitelline follicles numerous, small; in prosoma most dense in its posterior part at the level of holdfast organ, anteriorly protruding in three or four branches on each side of ventral sucker and extending in front of it. In opisthosoma, vitelline follicles most dense in its anterior and posterior extremity; confluent in front of testes, forming a ventral field at the level of both testes and being also confluent in posttesticular region. Uterus short, with few (1–7), large eggs. Copulatory bursa small, hermaphroditic duct short, opening dorso-subterminally (Fig. 1B). Excretory vesicle not observed.
Remarks
The present material agrees well with the diagnosis of the genus Diplostomum of Niewiadomska (Reference Niewiadomska, Gibson, Jones and Bray2002) in the presence of a distinctly bipartite body, a trilobate anterior extremity with pseudosuckers, vitelline follicles distributed in prosoma and opisthosoma, tandem testes with the anterior one being asymmetrical, a non-protrusible copulatory bursa and ovary being pretesticular. Diplostomum baeri was originally described by Dubois (Reference Dubois1937) ex Stercorarius longicaudus Vieillot and Stercorarius parasiticus (Linnaeus) from Lac Léman in Switzerland; the description of Dubois (Reference Dubois1937) was very brief, thus the species was redescribed by Dubois (Reference Dubois1938, Reference Dubois1970) and provided with drawings. The morphology of the present material of adults agrees well with the description of D. baeri of Dubois (Reference Dubois1938, Reference Dubois1970) in the ratio of the opisthosoma to the prosoma length (OPL/PRL = 0.67–0.86 vs 0.53–1.00), i.e. the prosoma is always longer than the opisthosoma as stated in the description by Dubois (Reference Dubois1970), although the range of the ratio given by him indicates that the body segments can be up to the same length. We infer that the typical character for this species is that the prosoma is longer than the opisthosoma or can be nearly equal in length. Furthermore, our material agrees in the vitelline follicles extending in front of the ventral sucker, in the elongate-oval shape of the prosoma being trilobed anteriorly and exhibiting the maximum width at the level of holdfast organ, in the position of the pseudosuckers (posterolateral to oral sucker) and in the ovary being close to the anterior extremity of the opisthosoma. The body dimensions in our material and in D. baeri Dubois, 1937 are very similar and overlap, including the large, not too numerous eggs; only the minima for total body length and length and width of both prosoma and opisthosoma in our material exhibit higher values, while the prosoma in our material is longer (954–1076 vs 500–930 μm) than that in D. baeri of Dubois (Reference Dubois1970) (Table 2). Because of the correspondence in morphology, dimensions of body and internal organs and ratios of dimensions we consider our material of adults identical with D. baeri Dubois, 1937.
In Canada, adults under the name D. baeri were obtained experimentally ex Larus delawarensis Ord by Galazzo et al. (Reference Galazzo, Dayanandan, Marcogliese and McLaughlin2002). However, Schwelm et al. (Reference Schwelm, Georgieva, Grabner, Kostadinova and Sures2021) re-classified this material as D. adamsi based on morphology of adults and the microhabitat of metacercariae (located in the peripheral retina). We agree with this concept, because the sequences (ITS1-5.8S-ITS2) of the adult worms obtained by Galazzo et al. (Reference Galazzo, Dayanandan, Marcogliese and McLaughlin2002) do not match ours (see below). Our material of D. baeri resembles that of Galazzo et al. (Reference Galazzo, Dayanandan, Marcogliese and McLaughlin2002) in dimensions, of which almost all overlap (Table 2). However, our adult worms differ in the OPL/PRL ratio [0.67–0.86 (0.76) vs 0.80–1.09 (1.01)], and although there is a slight overlap, still it indicates that the worms of Galazzo et al. (Reference Galazzo, Dayanandan, Marcogliese and McLaughlin2002) have a prosoma shorter than opisthosoma, which is never the case in our material, neither it is in the American subspecies, D. baeri bucculentum Dubois & Rausch, 1948 described from Michigan, USA (Dubois, Reference Dubois1970). Moreover, the worms of D. adamsi from Canada differ in their biology, as the adults were obtained from metacercariae recovered from the vitreous humour of eyes of Perca flavescens (Mitchill) by Galazzo et al. (Reference Galazzo, Dayanandan, Marcogliese and McLaughlin2002) or retina (see Schwelm et al., Reference Schwelm, Georgieva, Grabner, Kostadinova and Sures2021), while the European D. baeri occurs in the eye lens of cyprinid fishes (see below).
The other most similar species to our material, and also to D. baeri of Dubois (Reference Dubois1970), is D. volvens. This species was characterized by Shigin (Reference Shigin1977), who at first recognized it under the name D. yogenum (Cort and Brackett, Reference Cort and Brackett1937), based on the description by Cort and Brackett (Reference Cort and Brackett1937) of Cercaria yogena ex Stagnicola emarginata (Say) and Stagnicola palustris elodes (Say) from Michigan, USA. Later Shigin (Reference Shigin1993) considered D. yogenum a synonym of D. volvens. The present material resembles D. volvens of Shigin (Reference Shigin1977, Reference Shigin1993) in the shape of the whole body and prosoma, and in dimensions which are very similar (Table 2). However, the main difference is in the prosoma being longer than opisthosoma in our material vs prosoma being shorter or of similar length as opisthosoma in D. volvens [PRL/OPL ratios: 1.16–1.50 (1.33) vs 0.9–1.13 (0.98)/1.22–1.51 (1.41)]. Another similar worm is the one presented as D. baeri by Niewiadomska and Kiseliene (Reference Niewiadomska and Kiseliene1990), who obtained one adult experimentally from chicken, the material originating from cercariae in Lithuania. Our material and D. volvens of Shigin (Reference Shigin1977, Reference Shigin1993) resemble to the specimen of Niewiadomska and Kiseliene (Reference Niewiadomska and Kiseliene1990) in shape of prosoma and opisthosoma, in the vitelline follicles reaching in front of ventral sucker, in the prosoma being longer than the opisthosoma, and in similar dimensions (Table 2). However, the cercariae and metacercariae (found outside the eye lens) described and linked to that adult by Niewiadomska and Kiseliene (Reference Niewiadomska and Kiseliene1990) are clearly different from our material of cercariae and from metacercariae characterized by Pérez-del-Olmo et al. (Reference Pérez-del-Olmo, Georgieva, Pula and Kostadinova2014) as Diplostomum sp. clade Q (see below for details). Another material collected in Ireland and identified as D. volvens is that of McKeown and Irwin (Reference McKeown and Irwin1995), who did not provide comparable measurements, however, from their figure it is clear that the prosoma is shorter than the opisthosoma.
From D. mergi Dubois, 1932 which is similar in body shape, our material differs in the prosoma to opisthosoma length ratio (PRL/OPL), which is lower than in D. mergi [1.16–1.50 vs 1.06–2.43 of Dubois (Reference Dubois1970), 1.83–2.52 of Shigin (Reference Shigin1993)] and in showing higher minima for body size and internal organs. From Diplostomum nordmanni Shigin & Shapirov, 1986, which has a similar PRL/OPL ratio (1.09–1.46) and was found in Karelia and is typically occurring in larids (Shigin, Reference Shigin1993), our material differs in body shape (stout vs slender) and in being smaller (mean: 1758 vs 2443 μm); moreover, D. nordmanni has a smaller holdfast organ (193–253 vs 120–175).
Cercariae (Fig. 2)
Remarks
Cercariae of our material from Ireland are genetically identical (see below) and agree well with the morphology of Diplostomum sp. clade Q described by Selbach et al. (Reference Selbach, Soldánová, Georgieva, Kostadinova and Sures2015), in the presence of the same pattern of body spination, i.e. number of pre-oral spines (Fig. 2C), shape and number of rows of post-oral spines, number of transverse rows on body, non-converging lateral spined fields posterior to ventral sucker, the number of rows and spines on ventral sucker, the spination on tail stem and furca and the fish-fin like fin-fold on furca (Fig. 2D). We newly add the information on the resting position of the cercariae, which is characteristic with a bent tail stem and widespread furcae (Table 3, Fig. 2A). As stated in Selbach et al. (Reference Selbach, Soldánová, Georgieva, Kostadinova and Sures2015), their cercariae of Diplostomum sp. clade Q agree in part with the description of D. spathaceum of Niewiadomska and Kiseliene (Reference Niewiadomska and Kiseliene1994), however, they differ in tail stem and furca spination (spined vs devoid of spines) and in presence of a fin-fold on furcae, with which we agree.
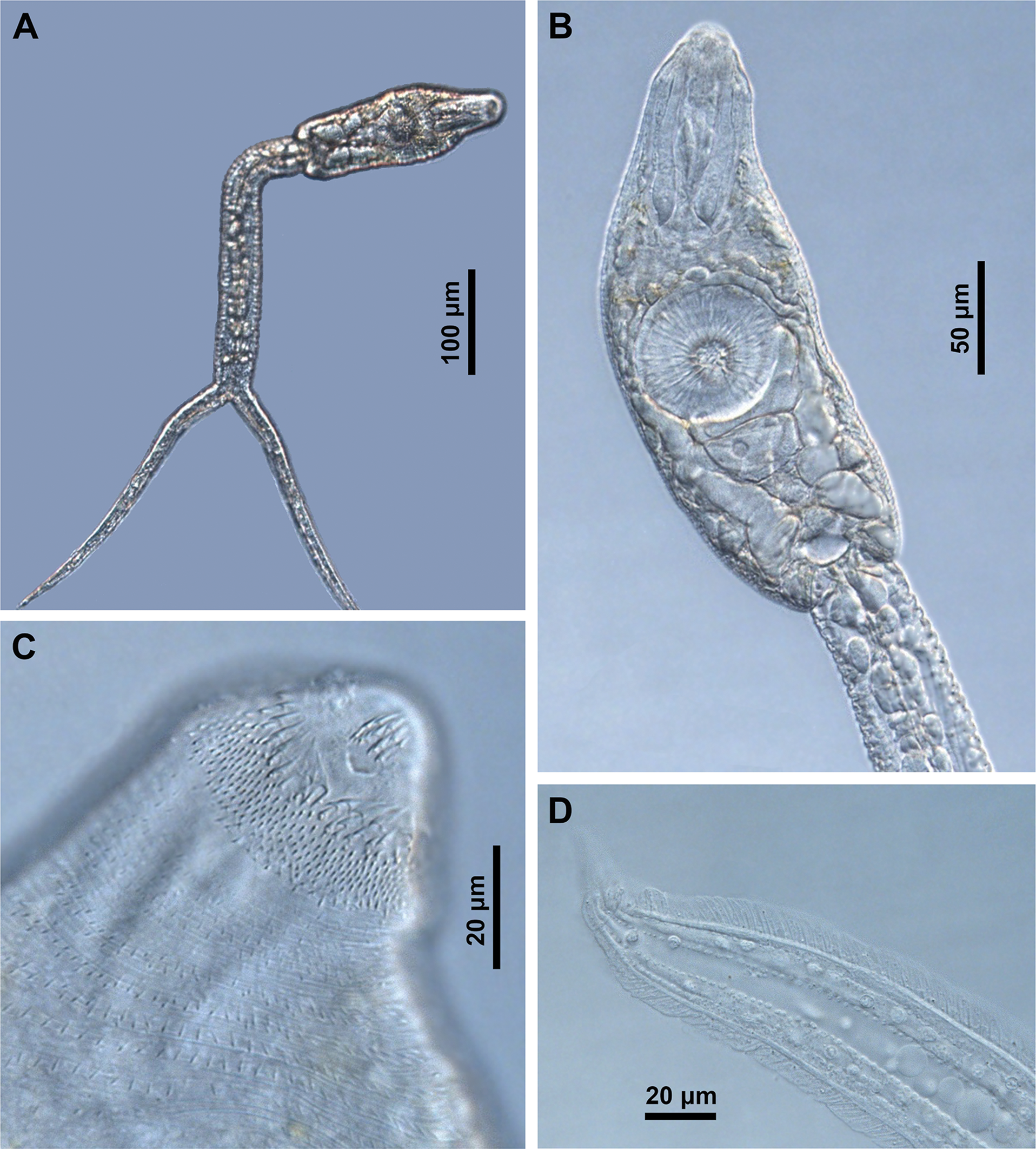
Fig. 2. Cercaria of Diplostomum baeri ex Radix balthica (IPCAS D-845): (A) total view with resting position; (B) ventral view of body; (C) anterior extremity with pre-oral and post-oral tegumental spines, ventral view and (D) detail of fish-fin like fin-fold on furca.
Table 3. Distinctive characters of cercariae of D. baeri and cercariae originally associated with it
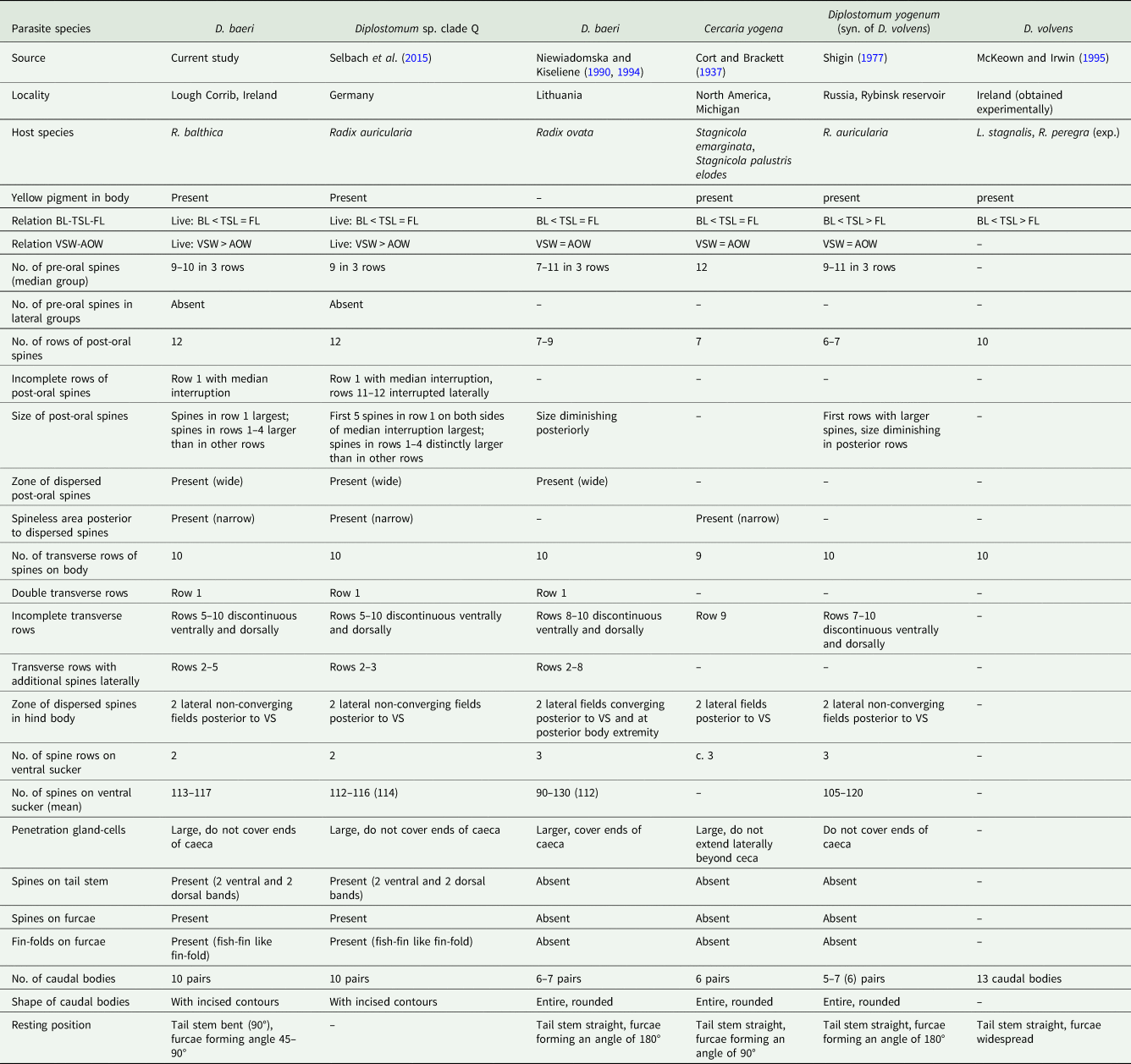
BL, body length; BW, maximum body width; AOW, anterior organ width; VS, ventral sucker; VSW, ventral sucker width; TSL, tail stem length; FL, furca length.
The current cercariae clearly differ in morphology from those assigned to D. volvens, i.e. C. yogena of Cort and Brackett (Reference Cort and Brackett1937), D. yogenum of Shigin (Reference Shigin1977) and D. volvens of McKeown and Irwin (Reference McKeown and Irwin1995), and to D. baeri as presented by Niewiadomska and Kiseliene (Reference Niewiadomska and Kiseliene1990); the most striking difference is the resting position (Fig. 2A, tail stem bent in our material vs tail stem straight in all four descriptions), furcae with a clearly visible fin-fold (Fig. 2D) vs no fin-fold, a tail stem with caudal bodies with incised contours vs tail stem with smooth caudal bodies; also, the arrangement of tegumental spines on body differs (Table 3).
Phylogenetic results
Fourteen novel sequences including seven cox1 (473 and 836 nt), four ITS1-5.8S-ITS2 (1250 nt) and three 28S (1300 nt) were obtained from seven isolates (Table 1). The 28S sequences were identical.
A phylogram resulted from BI and ML analyses based on the cox1 sequences generated in the current study (Table 1) and 35 sequences of Diplostomum spp. retrieved from GenBank (Supplementary Table S1) is presented in Fig. 3. Seven novel sequences clustered in a strongly supported clade with the sequences of the isolates previously identified as Diplostomum sp. clade Q collected from their first intermediate hosts, Radix auricularia (Linnaeus) and R. cf. peregra in Germany and second intermediate hosts, Rutilus rutilus (Linnaeus) and Cyprinus carpio Linnaeus in Germany and Spain, respectively. The sequence divergence within this clade was 0–1.4% (0–5 nt) which corresponds to the intraspecific level for members of Diplostomum. Sequences of the isolates that were identified to belong to the ‘D. baeri’ complex sensu Georgieva et al. (Reference Georgieva, Soldánová, Pérez-del-Olmo, Dangel, Sitko, Sures and Kostadinova2013), including Diplostomum sp. lineage 3 of Blasco-Costa et al. (Reference Blasco-Costa, Faltýnková, Georgieva, Skírnisson, Scholz and Kostadinova2014) (‘D. baeri Lineage 1’ of Georgieva et al., Reference Georgieva, Soldánová, Pérez-del-Olmo, Dangel, Sitko, Sures and Kostadinova2013) and Diplostomum sp. lineage 4 of Blasco-Costa et al. (Reference Blasco-Costa, Faltýnková, Georgieva, Skírnisson, Scholz and Kostadinova2014) (‘D. baeri Lineage 2’ Georgieva et al., Reference Georgieva, Soldánová, Pérez-del-Olmo, Dangel, Sitko, Sures and Kostadinova2013) and recently published complete mitochondrial genome sequences of metacercarial isolates from Salmo trutta Linnaeus identified as D. baeri (see Landeryou et al., Reference Landeryou, Ropiquet, Kett, Wildeboer and Lawton2020) clustered in a distant clade. Importantly, the metacercarial isolates used for generation of mitochondrial genome were identified based solely on DNA sequence data. Within this clade, sequences of D. baeri of Landeryou et al. (Reference Landeryou, Ropiquet, Kett, Wildeboer and Lawton2020) clustered with sequences of Diplostomum sp. lineage 3 of Blasco-Costa et al. (Reference Blasco-Costa, Faltýnková, Georgieva, Skírnisson, Scholz and Kostadinova2014) (‘D. baeri Lineage 1’ Georgieva et al., Reference Georgieva, Soldánová, Pérez-del-Olmo, Dangel, Sitko, Sures and Kostadinova2013). The sequence divergence between these species was 1.3–2% (6–7 nt) suggesting that they belong to the same species.

Fig. 3. BI tree for Diplostomum spp. based on the partial cox1 mtDNA sequences. Nodal support from BI and ML analyses is indicated as BI/ML; values <0.90 (BI) and <70 (ML) are not shown. The scale bar indicates the expected number of substitutions per site. The newly generated sequences are highlighted in bold. Yellow rectangle indicates the clade with published and novel sequences of D. baeri. Sequence names followed by abbreviations: AP, Pérez-del-Olmo et al. (Reference Pérez-del-Olmo, Georgieva, Pula and Kostadinova2014); BG, Behrmann-Godel (Reference Behrmann-Godel2013); CS, Selbach et al. (Reference Selbach, Soldánová, Georgieva, Kostadinova and Sures2015); SL, Locke et al. (Reference Locke, Al-Nasiri, Caffara, Drago, Kalbe, Lapierre, McLaughlin, Nie, Overstreet, Souza, Takemoto and Marcogliese2015). Haplotype network for D. baeri based on published and novel cox1 sequences. Unsampled intermediate haplotype is represented by short intersecting line; each branch corresponds to a single mutational difference and connective lines represent one mutational step. Circle size is proportional to the number of isolates sharing a haplotype; haplotype frequency is indicated by colourless circles.
Diplostomum sp. clade Q was delineated by Georgieva et al. (Reference Georgieva, Soldánová, Pérez-del-Olmo, Dangel, Sitko, Sures and Kostadinova2013) in Europe as consisting of eight sequences, five identical ITS1 sequences: two of cercarial isolates identified originally as D. spathaceum (AF419275 and AF419276) and one identified as D. parviventosum (AF419278) ex Radix ovata in Poland by Niewiadomska and Laskowski (Reference Niewiadomska and Laskowski2002); another cercarial isolate submitted to GenBank under the name D. mergi (JQ665458) but designated as D. spathaceum ex R. auricularia in Germany by Behrmann-Godel (Reference Behrmann-Godel2013); one of a metacercarial isolate submitted to GenBank as D. cf. parviventosum/spathaceum (JF775727) ex R. rutilus in Finland by Rellstab et al. (Reference Rellstab, Louhi, Karvonen and Jokela2011). And the three cox1 sequences were from R. auricularia (JQ639179) and from R. rutilus (JQ639177 and JQ639178) added by Behrmann-Godel (Reference Behrmann-Godel2013). Pérez-del-Olmo et al. (Reference Pérez-del-Olmo, Georgieva, Pula and Kostadinova2014) obtained two more cox1 and ITS1-5.8S-ITS2 sequences (KP025770 and KP025788) for a metacercaria from the eye lens ex C. carpio in Spain, which also clustered with Diplostomum sp. clade Q; they were the first to combine their genetic data with a morphological description of the metacercaria, as the previously obtained sequences were not linked to any voucher material. Pérez-del-Olmo et al. (Reference Pérez-del-Olmo, Georgieva, Pula and Kostadinova2014) assumed the questionable clade, which was most close to the ‘D. mergi’ complex, could represent D. parviventosum. This assumption was disproved by Selbach et al. (Reference Selbach, Soldánová, Georgieva, Kostadinova and Sures2015) who by integrative taxonomy characterized cercariae of both Diplostomum sp. clade Q and D. parviventosum and proved that they differed genetically and morphologically. While cercariae of D. parviventosum corresponded to those described by Niewiadomska and Kiseliene (Reference Niewiadomska and Kiseliene1994), cercariae of Diplostomum sp. clade Q did not correspond to any of their descriptions.
The 14 cox1 sequences (359 nt) of D. baeri generated in the present (n = 7) and previous (n = 7; identified as Diplostomum sp. clade Q) studies were collapsed into eight haplotypes (Fig. 3) including seven unique haplotypes and one haplotype shared by seven isolates collected from L. canus in Russia (AF465 and AF467), R. cf. peregra (KR271470 and KR271471; Locke et al., Reference Locke, Al-Nasiri, Caffara, Drago, Kalbe, Lapierre, McLaughlin, Nie, Overstreet, Souza, Takemoto and Marcogliese2015), R. auricularia (KR149554; Selbach et al., Reference Selbach, Soldánová, Georgieva, Kostadinova and Sures2015) and R. rutilus (JQ639178; Behrmann-Godel, Reference Behrmann-Godel2013) in Germany, and from C. carpio in Spain (KP025770; Pérez-del-Olmo et al., Reference Pérez-del-Olmo, Georgieva, Pula and Kostadinova2014).
Figure 4 presents the phylogram resulting from BI and ML analyses based on the ITS1-5.8S-ITS2 dataset. Four novel sequences obtained from L. canus in Russia and R. balthica in Ireland clustered with a sequence of the isolate collected from R. auricularia in Germany (Supplementary Table S1). Sequences in this clade were identical. Importantly, a sequence of the adult isolate identified as D. adamsi and reported from North America (Galazzo et al., Reference Galazzo, Dayanandan, Marcogliese and McLaughlin2002) clustered in a distant clade with representatives previously considered to belong to the ‘D. baeri’ complex and recently published sequences of D. baeri obtained from the metacercarial isolates by Landeryou et al. (Reference Landeryou, Ropiquet, Kett, Wildeboer and Lawton2020). Our sequences of D. baeri differed from the sequence of Galazzo et al. (Reference Galazzo, Dayanandan, Marcogliese and McLaughlin2002) by 2.7% (26 nt) suggesting that these species are not conspecific. Similar to the cox1 analyses, sequences of D. baeri by Landeryou et al. (Reference Landeryou, Ropiquet, Kett, Wildeboer and Lawton2020) clustered with sequences of Diplostomum sp. lineage 3 (D. baeri lineage 1) and Diplostomum sp. lineage 4 (D. baeri lineage 2). The comparative analysis of our sequences and sequences originally assigned to the clade of Diplostomum sp. clade Q (Fig. 5 in Georgieva et al., Reference Georgieva, Soldánová, Pérez-del-Olmo, Dangel, Sitko, Sures and Kostadinova2013) restricted to the ITS1 region showed no nucleotide difference.
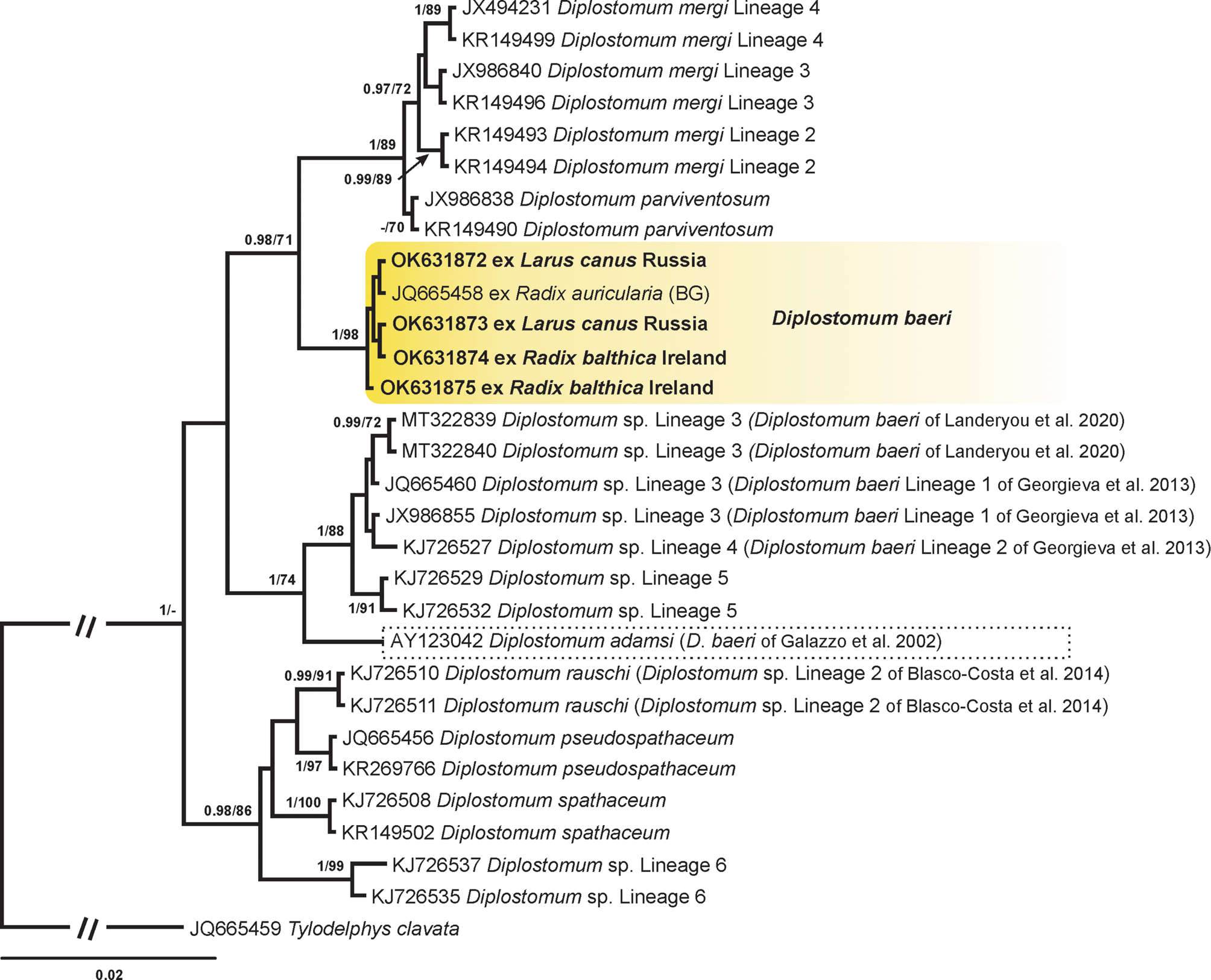
Fig. 4. BI tree for Diplostomum spp. based on ITS1-5.8S-ITS2 sequences. Nodal support from BI and ML analyses is indicated as BI/ML; values <0.90 (BI) and <70 (ML) are not shown. The scale bar indicates the expected number of substitutions per site. The newly generated sequences are highlighted in bold. Yellow rectangle indicates the clade with published and novel sequences of D. baeri. Dotted rectangle indicates the sequence of Diplostomum adamsi identified as D. baeri by Galazzo et al. (Reference Galazzo, Dayanandan, Marcogliese and McLaughlin2002) in North America. Sequence name followed by abbreviation: BG, Behrmann-Godel (Reference Behrmann-Godel2013).
Based on the results of the phylogenetic analyses, we conclude that (i) isolates previously reported as Diplostomum sp. clade Q belong to the species of D. baeri, (ii) species of D. baeri lineage 1 and D. baeri lineage 2 do not belong to the ‘D. baeri’ complex and should be referred to as Diplostomum sp. lineage 3 and Diplostomum sp. lineage 4 as proposed by Blasco-Costa et al. (Reference Blasco-Costa, Faltýnková, Georgieva, Skírnisson, Scholz and Kostadinova2014), (iii) sequence of complete mitochondrial genome and the rest of sequences obtained from isolates identified as D. baeri in the study of Landeryou et al. (Reference Landeryou, Ropiquet, Kett, Wildeboer and Lawton2020) belong to Diplostomum sp. lineage 3 of Blasco-Costa et al. (Reference Blasco-Costa, Faltýnková, Georgieva, Skírnisson, Scholz and Kostadinova2014) and (iv) the isolate of D. adamsi [originally identified as D. baeri by Galazzo et al. (Reference Galazzo, Dayanandan, Marcogliese and McLaughlin2002)] represent a distinct species different from our material.
Discussion
The present new material of adult D. baeri corresponds well with the description of D. baeri Dubois, Reference Dubois1937 by Dubois (Reference Dubois1937, Reference Dubois1938, Reference Dubois1970). Therefore, we consider D. baeri as a species occurring in Europe in birds L. canus, S. longicaudus and S. parasiticus with metacercariae using cyprinid fishes with location in the eye lens and using snails R. auricularia and R. balthica (syn. R. ovata) as first intermediate hosts. With molecular genetic analyses we proved that this species is identical to Diplostomum sp. clade Q. Also, our new material of cercariae is identical in morphology and in DNA sequences with Diplostomum sp. clade Q characterized by Selbach et al. (Reference Selbach, Soldánová, Georgieva, Kostadinova and Sures2015). As the most typical characters in adults of D. baeri we view the ratio of prosoma to opisthosoma length, which means that the prosoma is always longer than the opisthosoma or almost of the same size, but never shorter than opisthosoma; the other features are the extent of vitelline follicles not far in front of the ventral sucker and a relatively stout body. For morphological investigations always a set of more specimens is needed, as the PRL/OPL ratio can be dependent on age of the worms.
We reveal that our material of D. baeri cannot be assigned to D. volvens recognized by Shigin (Reference Shigin1977, Reference Shigin1993), although both species are highly similar and their PRL/OPL ratio is overlapping, i.e. also in D. volvens the prosoma can be of almost the same length as the opisthosoma, however, it can be shorter, which is never true for D. baeri. Neither can our material be assigned to D. baeri of Niewiadomska and Kiseliene (Reference Niewiadomska and Kiseliene1990, Reference Niewiadomska and Kiseliene1994), which has a similar PRL/OPL ratio, because the corresponding cercariae are different in morphology (resting position, presence/absence of fin-folds on furca, spination of body and tail). It is possible that both authors (Niewiadomska and Kiseliene, Reference Niewiadomska and Kiseliene1990 and Shigin, Reference Shigin1977, Reference Shigin1993) might have had the same species, or they eventually had a mixture of species, because for experimental infections, they used whole fish eyes of Ctenopharyngodon idella (Valenciennes), Oncorhynchus mykiss (Walbaum) and Perca fluviatilis Linnaeus (potentially there could have been simultaneous infections with specimens from different locations in eyes). This overlap in characters (PRL/OPL) signals that there is no feature reliable enough to distinguish unambiguously between D. baeri of Dubois (Reference Dubois1937, Reference Dubois1970) and D. baeri of Niewiadomska and Kiseliene (Reference Niewiadomska and Kiseliene1990) and D. volvens of Shigin (Reference Shigin1977, Reference Shigin1993), and the high morphological similarity documents the difficulties in identification and proves that it is possible to reliably distinguish the species only with both molecular analyses and detailed morphological examination.
Another typical feature of D. baeri is the host specificity of metacercariae, which were so far found in the eye lens in cyprinid fishes (C. carpio, R. rutilus) by Rellstab et al. (Reference Rellstab, Louhi, Karvonen and Jokela2011), Behrmann-Godel (Reference Behrmann-Godel2013) and Pérez-del-Olmo et al. (Reference Pérez-del-Olmo, Georgieva, Pula and Kostadinova2014). However, this contradicts the previous data on the life-cycle of D. volvens (syn. D. baeri), because metacercariae of D. baeri of Shigin (Reference Shigin1968), D. volvens and D. yogenum were consistently reported from the Percidae or Lottidae and never from eye lens, i.e. from retina or between sclera (see Shigin, Reference Shigin1977, Reference Shigin1993; McKeown and Irwin, Reference McKeown and Irwin1995). Metacercariae of D. baeri of Niewiadomska and Kiseliene (Reference Niewiadomska and Kiseliene1990) were reported from C. idella, however they were obtained experimentally, and they were located outside the lens. The metacercariae differ also morphologically, those of Shigin (Reference Shigin1968) and Niewiadomska and Kiseliene (Reference Niewiadomska and Kiseliene1990) are much larger than Diplostomum sp. clade Q of Pérez-del-Olmo et al. (Reference Pérez-del-Olmo, Georgieva, Pula and Kostadinova2014) (body size: 405 × 205 μm and 518 × 244 μm, respectively vs 229 × 180 μm). In summary, the present results contradict those of the previous studies reporting that percids (and P. fluviatilis in particular) serve as the main fish hosts for D. baeri [D. volvens in view of Shigin (Reference Shigin1986, Reference Shigin1993) and McKeown and Irwin (Reference McKeown and Irwin1995)] and that the metacercariae are located between the sclera and retina or even deeper in the eye, under the retina (Shigin, Reference Shigin1993), it indicates that the authors (Shigin, Reference Shigin1977, Reference Shigin1993; Niewiadomska and Kiseliene, Reference Niewiadomska and Kiseliene1990, Niewiadomska and Laskowski, Reference Niewiadomska and Laskowski2002) were dealing with a species different from D. baeri.
The first material characterized molecularly under the name D. baeri was a metacercaria recovered from P. fluviatilis by Niewiadomska and Laskowski (Reference Niewiadomska and Laskowski2002); the morphological identification was based on the concept of Niewiadomska and Kiseliene (Reference Niewiadomska and Kiseliene1990). This identification was followed by authors subsequently providing the corresponding sequences of metacercariae from outside lens of percid fishes (e.g. Rellstab et al., Reference Rellstab, Louhi, Karvonen and Jokela2011; Behrmann-Godel, Reference Behrmann-Godel2013; Georgieva et al., Reference Georgieva, Soldánová, Pérez-del-Olmo, Dangel, Sitko, Sures and Kostadinova2013; Landeryou et al., Reference Landeryou, Ropiquet, Kett, Wildeboer and Lawton2020), and Schwelm et al. (Reference Schwelm, Georgieva, Grabner, Kostadinova and Sures2021) claimed that metacercariae of all species/lineages of the ‘D. baeri’ complex represent non-lens-dwelling forms. However, these metacercariae identified as belonging to the ‘D. baeri’ complex are genetically distant from Diplostomum sp. clade Q. Therefore, the sequences labelled as the ‘D. baeri’ complex sensu Georgieva et al. (Reference Georgieva, Soldánová, Pérez-del-Olmo, Dangel, Sitko, Sures and Kostadinova2013) (‘D. baeri Lineage 1’ and ‘D. baeri Lineage 2’); sensu Blasco-Costa et al. (Reference Blasco-Costa, Faltýnková, Georgieva, Skírnisson, Scholz and Kostadinova2014) [Diplostomum sp. lineages 3–5 of Blasco-Costa et al. (Reference Blasco-Costa, Faltýnková, Georgieva, Skírnisson, Scholz and Kostadinova2014)], D. adamsi and Diplostomum sp. lineages 2, 5–7 of Locke et al. (Reference Locke, McLaughlin and Marcogliese2010a, Reference Locke, McLaughlin, Dayanandan and Marcogliese2010b); and sensu Schwelm et al. (Reference Schwelm, Georgieva, Grabner, Kostadinova and Sures2021) (D. adamsi, D. phoxini, Diplostomum sp. lineages 3–5 of Blasco-Costa et al. (Reference Blasco-Costa, Faltýnková, Georgieva, Skírnisson, Scholz and Kostadinova2014), Diplostomum spp. lineages 5–7 of Locke et al. (Reference Locke, Al-Nasiri, Caffara, Drago, Kalbe, Lapierre, McLaughlin, Nie, Overstreet, Souza, Takemoto and Marcogliese2015), Diplostomum sp. of Lebedeva et al. (Reference Lebedeva, Chrisanfova, Ieshko, Guliaev, Yakovleva, Mendsaikhan and Semyenova2021) do not represent lineages of D. baeri.
Another important outcome of the molecular genetic analyses is that the recently published complete mitochondrial genome of D. baeri and the rest of sequences obtained from isolates identified as D. baeri in the study of Landeryou et al. (Reference Landeryou, Ropiquet, Kett, Wildeboer and Lawton2020) do not correspond to our material of D. baeri and in fact belong to an unknown species of Diplostomum, Diplostomum sp. lineage 3 of Blasco-Costa et al. (Reference Blasco-Costa, Faltýnková, Georgieva, Skírnisson, Scholz and Kostadinova2014) (see also Schwelm et al., Reference Schwelm, Georgieva, Grabner, Kostadinova and Sures2021). A metacercarial isolate used for construction of the mitochondrial genome was identified as D. baeri based on a comparison with the published cox1 and ITS1-5.8S-ITS2 sequences of the ‘D. baeri’ complex obtained in Germany and Iceland (Georgieva et al., Reference Georgieva, Soldánová, Pérez-del-Olmo, Dangel, Sitko, Sures and Kostadinova2013; Blasco-Costa et al., Reference Blasco-Costa, Faltýnková, Georgieva, Skírnisson, Scholz and Kostadinova2014; Unger and Palm, Reference Unger and Palm2017), however without specifying to which of the lineages it belongs.
Based on the results of ITS1-5.8S-ITS2 sequence analyses we showed that the isolate identified as D. baeri in Canada by Galazzo et al. (Reference Galazzo, Dayanandan, Marcogliese and McLaughlin2002) does not cluster with the ‘true’ D. baeri (Fig. 4) and represents a different species, which is in accordance with Achatz et al. (Reference Achatz, Martens, Kostadinova, Pulis, Orlofske, Bell, Fecchio, Oyarzún-Ruiz, Syrota and Tkach2021) and Schwelm et al. (Reference Schwelm, Georgieva, Grabner, Kostadinova and Sures2021), who re-classified it as D. adamsi; the metacercariae were obtained from the vitreous humour of eyes of P. flavescens and the experimentally obtained adults differ morphologically from our material in the PRL/OPL ratio. Consequently, sequences from the North American isolates identified to belong to the ‘D. baeri’ complex [Diplostomum sp. lineages 2, 5–7 of Locke et al. (Reference Locke, McLaughlin and Marcogliese2010a, Reference Locke, McLaughlin, Dayanandan and Marcogliese2010b)] by Blasco-Costa et al. (Reference Blasco-Costa, Faltýnková, Georgieva, Skírnisson, Scholz and Kostadinova2014) represent an unknown species.
Diplostomum baeri is widely distributed in Europe, as the known range is from north/east Europe (Finland and Northwest Russia) via Poland, Germany and Switzerland up to most west Europe (Ireland and Spain). This was also supported by the results of the haplotype analysis. Out of eight haplotypes of D. baeri identified, one was shared between isolates from all three hosts of species life-cycle distributed in Germany, Russia and Spain (Fig. 3). However, despite numerous previous studies on metacercariae of Diplostomum from various fish species, including cyprinids, these parasites were found sporadically (Rellstab et al., Reference Rellstab, Louhi, Karvonen and Jokela2011; Behrman-Godel, Reference Behrmann-Godel2013; Pérez-del-Olmo et al., Reference Pérez-del-Olmo, Georgieva, Pula and Kostadinova2014). Potential previous records of metacercariae from eye lens of fishes could be masked by putting all findings under the collective name D. spathaceum or Diplostomum sp.
DNA sequence data for Diplostomum spp. have accumulated from around the world (Locke et al., Reference Locke, Al-Nasiri, Caffara, Drago, Kalbe, Lapierre, McLaughlin, Nie, Overstreet, Souza, Takemoto and Marcogliese2015), however, the majority of these data is represented by unidentified species of larval isolates, thus the identification and interpretation are not satisfactory yet and keeping vouchers is still nearly non-existent. This indicates that there is still a long way to comprehend the importance of vouchering samples including host species, and yet again, the holistic approach (which considers the parasite morphology, biology and ecology) in interpreting and evaluating data should be advocated as done by Blasco-Costa et al. (Reference Blasco-Costa, Cutmore, Miller and Nolan2016). Consequently, the taxonomy of Diplostomum will remain unclear until molecular data from adults are available for accurate species identification. However, at present it is crucial to collect standardized data that can be later integrated into analyses to answer questions related to systematics, biology, ecology and evolution of Diplostomum spp. Therefore, we extend the recommendations by Blasco-Costa et al. (Reference Blasco-Costa, Cutmore, Miller and Nolan2016) and provide a checklist including key aspects that need to be considered when studying Diplostomum spp. at any life-cycle stage in a molecular-taxonomic work.
(i) What is the host species of Diplostomum spp.? The accurate identification of the host organism is crucial and reliable information will aid identification of Diplostomum spp. Moreover, recording host species spectrum data will aid studies of ecology, host–pathogen coevolution and epidemiological aspects of Diplostomum spp. (see Thompson et al., Reference Thompson, Phelps, Allard, Cook, Dunnum, Ferguson, Gelang, Khan, Paul, Reeder, Simmons, Vanhove, Webala, Weksler and Kilpatrick2021). Patterns of host–parasite associations revealed by recent molecular genetic studies indicated higher host specificity in metacercariae than reported by previous morphology or experiment-based studies (Blasco-Costa and Locke, Reference Blasco-Costa, Locke, Rollinson and Stothard2017).
(ii) What is the microhabitat of Diplostomum spp. in the host? Cercariae and adults of Diplostomum spp. occur in the same discrete microhabitats in their snail and bird hosts – hepatopancreas and digestive tract, respectively. Metacercariae of Diplostomum spp. occur in either eyes or brain of their fish host. Recent molecular genetic surveys demonstrated that metacercariae of different species in fish eye may restrict their distributions to precise locations such as retina, vitreous humour or lens (Locke et al., Reference Locke, McLaughlin, Dayanandan and Marcogliese2010b; Blasco-Costa et al., Reference Blasco-Costa, Faltýnková, Georgieva, Skírnisson, Scholz and Kostadinova2014; Schwelm et al., Reference Schwelm, Georgieva, Grabner, Kostadinova and Sures2021).
(iii) What type of voucher is required for the sample? Morphological vouchers are crucial for taxonomic identification of genetic lineages especially when multiple infections occur. The best practice in vouchering of material is described by Pleijel et al. (Reference Pleijel, Jondelius, Norlinder, Nygren, Oxelman, Schander, Sundberg and Thollesson2008). For adults we suggest excising a small piece of tissue from the lateral side of forebody or hindbody without removing informative morphological structures. For larval stages which are substantially smaller than adults, electronic vouchers (E-vouchers), i.e. photomicrographs of each individual showing details of morphology should be applied. E-voucher individuals are used for generation of DNA sequences. Live cercariae and metacercariae are preferable for E-vouchers as some details are not visible on fixed individuals. Morphological vouchers need to be deposited in recognized parasitological collections and E-vouchers are available in Supplementary materials.
(iv) Which morphological traits should be considered and of which life-cycle stage? Morphology of any of the life-cycle stages of Diplostomum spp. is valuable and will contribute to completing the picture of the particular species. Characters in adults and cercariae are the most informative. In adults (which are most decisive for species description), the body shape and proportions (prosoma to opisthosoma ratio and sucker ratio), the extent and development of vitelline follicles, and the position of gonads are important. In cercariae, particularly the arrangement and numbers of tegumental spines are important and should be best studied with aid of scanning electron microscopy to obtain reliable counts; furthermore, proportions of body to tail stem and furca, sucker ratios and prominence of penetration gland cells should be considered as well. The metacercariae are the stages with least characters (size of body, suckers, and holdfast organ, number of excretory granules), however, together with other traits (host species, microhabitat and geographical distribution), they are an important counterpart to genetic data. Photos of live material (see above for E-vouchers) can be used for descriptions and measurements. When possible, a wider lot of specimens of one species should be examined to catch the variability.
(v) Which molecular markers can aid identification? The results of molecular genetic studies of Diplostomum depend on the use of standardized molecular markers and obtaining DNA sequences that are compatible with those available in molecular library. Thus far, the mitochondrial cox1 gene and the ribosomal gene cluster ITS1-5.8S-ITS2 are the most employed markers and therefore, their corresponding sequences are available for the majority of Diplostomum spp. in GenBank for analysis. DNA-based identification of Diplostomum spp. relies on phylogenetic analyses and estimation of genetic divergence (P-distance).
(vi) Has the species been reported previously? Linking morphological descriptions of Diplostomum spp. to the existing descriptions as well as linking novel sequences to those available in molecular library helps to identify material. The name used for the new material should correspond to the names (or provisional names) used in previous studies. If an unidentified isolate of a larval stage is novel it requires a unique name, and we recommend the numbering system as suggested by Locke et al. (Reference Locke, Al-Nasiri, Caffara, Drago, Kalbe, Lapierre, McLaughlin, Nie, Overstreet, Souza, Takemoto and Marcogliese2015).
(vii) Does the species require morphological description? Morphological descriptions with illustrations need to be provided for adults representing a new species of Diplostomum and unidentified isolates of larval stages of Diplostomum reported for the first time. Also, sequences of Diplostomum spp. published for the first time should be supplemented with a description or illustration of morphological vouchers.
(viii) Do the names of the species and metadata in GenBank correspond to data in publication? Publication is the main source for the identification. Currently, there are discrepancies between data in the publications and GenBank annotations (Locke et al., Reference Locke, Al-Nasiri, Caffara, Drago, Kalbe, Lapierre, McLaughlin, Nie, Overstreet, Souza, Takemoto and Marcogliese2015) and therefore, identification should not be based solely on the comparison of DNA sequences in GenBank but should always be checked with the original publication and recently published papers reporting on any changes in taxonomy. The most updated revision of classification and nomenclature of the species and species-level lineages of Diplostomum until 2021 are presented in Schwelm et al. (Reference Schwelm, Georgieva, Grabner, Kostadinova and Sures2021).
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182021002092.
Data
Data are available on request from the authors and in the Supplementary material. Newly generated sequences were deposited in GenBank with the following accession numbers: OK631869–OK631875 and OK632471–OK632477.
Acknowledgements
The authors are grateful to Dr Alexandr Artem'ev (Laboratory of Zoology, IB, KRC RAS) for help with bird identification. We thank Katie O'Dwyer (Marine and Freshwater Research Centre, Galway-Mayo Institute of Technology) for help with sampling and processing trematode material in Ireland.
Author contributions
Concept and design of the study: AF, DL, OK. Writing: AF, DL, OK. Review and editing: CP, GY. Morphological analysis: AF. Molecular analyses: CP, OK, DL. Fieldwork and data processing: AF, CP, GY, DL. All authors approved the final version of the manuscript before submission.
Financial support
The current study was funded by the Russian Ministry of Science and Education (state order 0218-2019-0075) and the Czech Grant Agency (project No. 18-18597S).
Conflict of interest
None.