Introduction
It has long been recognized that Lecidea hypnorum Lib. and its close associates form a distinctive group unrelated to Lecidea fuscoatra Ach., the type species of the genus. It appears more closely related to Clauzadea Hafellner & Bellem. (Hawksworth & Coppins Reference Hawksworth, Coppins, Purvis, Coppins, Hawksworth, James and Moore1992), although Meyer (Reference Meyer2002), in her study of Clauzadea, explicitly excluded L. hypnorum from that genus. The core species of the group were transferred to Mycobilimbia Rehm by Kalb & Hafellner in Wirth (Reference Wirth1987) although they are clearly not congeneric with M. obscurata (Sommerf.) Rehm, the type species of that genus. Recent molecular studies (Arup Reference Arup2004; Buschbom & Mueller Reference Buschbom and Mueller2004; Schmull et al. Reference Schmull, Miądlikowska, Pelzer, Stocker-Wörgötter, Hofstetter, Fraker, Hodkinson, Reeb, Kukwa and Lumbsch2011) have confirmed that Lecidea hypnorum cannot be accommodated in either Lecidea or Mycobilimbia, and that it is more closely related to Clauzadea, Farnoldia Hertel, Lecidoma Gotth. Schneid. & Hertel, and Romjularia Timdal.
Currently, some authors prefer to describe additional species in, or transfer them to, Mycobilimbia (e.g., Hafellner Reference Hafellner1989; Kalb & Hafellner Reference Kalb and Hafellner1992; Sarrión et al. Reference Sarrión, Aragón, Hafellner, Rico and Burgaz2003; Kantvilas et al. Reference Kantvilas, Messuti and Lumbsch2005), whereas others choose to retain the species in Lecidea (Hertel & Printzen Reference Hertel, Printzen, Nash, Ryan, Diederich, Gries and Bungartz2004; Coppins & Fryday Reference Coppins and Fryday2006; Fryday Reference Fryday2008; Aptroot et al. Reference Aptroot, Gilbert, Hawksworth, Coppins, Smith, Aptroot, Coppins, Fletcher, Gilbert, James and Wolseley2009). This situation, where two genera are available although the species are not congeneric with the type specimen of either, is clearly unsatisfactory and this study is an attempt to resolve the issue.
Materials & Methods
DNA extraction and PCR amplification
DNA was extracted from 2–3 apothecia per thallus using the QIAquick™ Plant Mini Kit (Qiagen) according to the manufacturer's instructions. Five gene loci were amplified with the following primers: ITS1F (Gardes & Bruns Reference Gardes and Bruns1993) and ITS4 (White et al. Reference White, Bruns, Lee, Taylor, Innis, Gelfand, Sninsky and White1990) for the internal transcribed spacer region of the ribosomal DNA (ITS), mrSSU1 (Zoller et al. Reference Zoller, Scheidegger and Sperisen1999) and MSU7 (Zhou & Stanosz Reference Zhou and Stanosz2001) for part of the small subunit of the mitochondrial ribosomal DNA (mtSSU), LR0R and LR7 (Vilgalys Laboratory, Duke University: http:/www.biology.duke.edu/fungi/mycolab/primers.htm) for the first part of the large subunit of the nuclear ribosomal DNA (nuLSU), RPB1-Af (Stiller & Hall Reference Stiller and Hall1997) and RPB1-Cr (Matheny et al. Reference Matheny, Liu, Ammirati and Hall2002) for part of the largest subunit of RNA polymerase II (RPB1), and RPB2-5f and RPB2-7Cr (Liu et al. Reference Liu, Whelen and Hall1999) for part of the second largest subunit of RNA polymerase II (RPB2). 25 µl PCR reactions were carried out using PCR-PuReTaq Ready-to-Go Beads™ (GE Healthcare). For ITS, mtSSU and nuLSU, reactions contained 5 µl of DNA extract, 1 µl of each forward- and reverse-primer (10 µM) and 18 µl of distilled water. Cycling conditions included initial denaturation at 94°C for 5 min; 5 cycles of 94°C for 30 s, 54°C for 30 s, 72°C for 1 min; 33 cycles of 94°C for 30 s, 48°C for 30 s, 72°C for 1 min; and a final extension step at 72°C for 10 min. For RPB1 and RPB2, PCR reactions contained 10 µl of DNA extract, 3·5 µl of each forward- and reverse-primer (10 µM) and 8 µl of distilled water. Cycling conditions were as follows: for RPB1 initial denaturation at 95°C for 5 min, 8 cycles of 95°C for 1 min, 58°C for 1 min, 72°C for 1 min 45 s, 34 cycles of 95°C for 1 min, 50°C for 1 min, 72°C for 1 min 45 s, and a final extension step at 72°C for 10 min; for RPB2 initial denaturation at 92°C for 2 min, 8 cycles of 94°C for 1 min, 59°C for 1 min, 72°C for 2 min, 33 cycles of 95°C for 30 s, 50°C for 30 s, 72°C for 2 min, and a final extension step at 72°C for 10 min. PCR products were run on agarose gels, bands cut out and purified using the QIAquick Gel Extraction Kit (Qiagen). Purified DNA was labelled with the BigDye™ Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) and cycle sequenced at 94°C for 30 s, with 29 cycles of 95°C for 15 s, 45°C for 15 s, and 60°C for 4 min using the PCR primers. Sequences were determined on an ABI PRISM® 3730 DNA Analyzer (Applied Biosystems), and assembled and edited using Geneious Pro, version 5.6.5 (Biomatters Inc.).
BLAST searches in GenBank were performed to ascertain that all sequences used in the phylogenetic analyses originated from the lichens and not from contaminating organisms such as parasymbiotic fungi. Single gene datasets containing the sequences listed in Table 1 were compiled and aligned in Geneious Pro, version 5.6.5, using the Muscle algorithm with default settings. Regions of uncertain alignment were removed using GBlocks version 0.91b (Castresana Reference Castresana2000), applying default settings but allowing gap positions in half of the sequences. After excluding uncertain alignment, the RPB1 and RPB2 sequences of Lecidea auriculata consisted only of missing data. Accordingly, the species was removed from these datasets. Final alignments comprised 25 sequences, 373 bp (ITS), 26 seq., 620 bp (mtSSU), 27 seq., 902 bp (nuLSU), 12 seq., 663 bp (RPB1) and, 13 seq., 702 bp (RPB2).
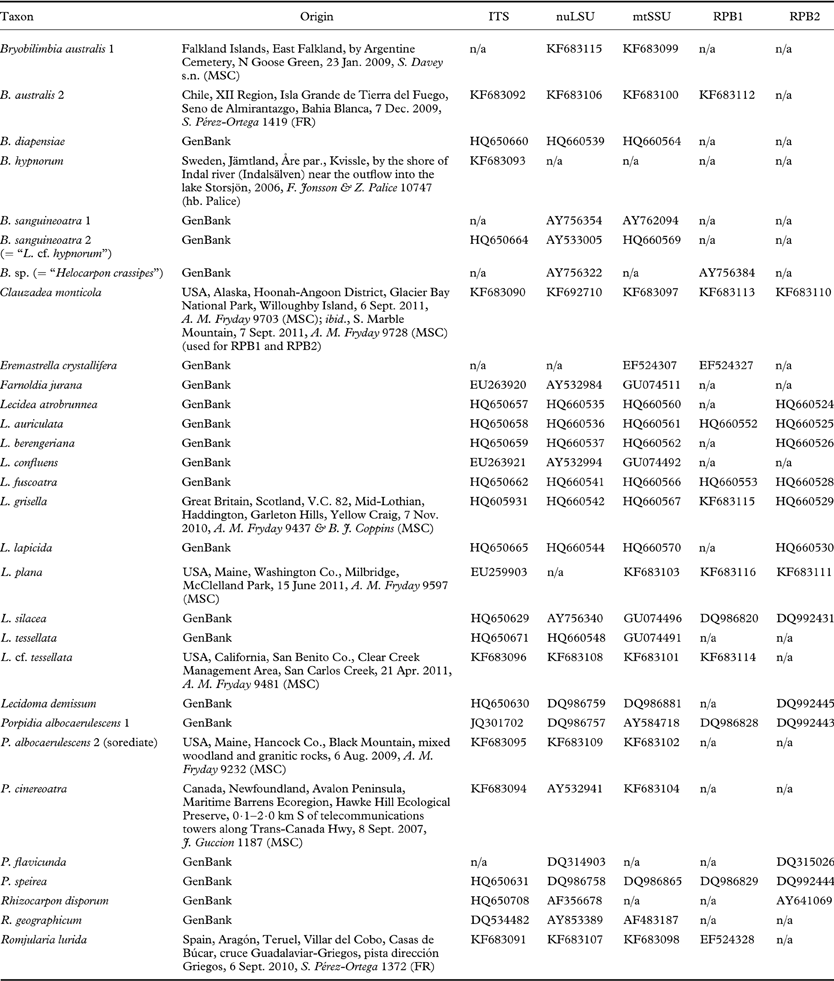
Table 1. Taxa and sequences used in this study. Collection data are given for samples that were used to generate new sequences.
Datasets were concatenated to yield a final alignment of 30 sequences and 3260 bp length. The optimal partitioning scheme and substitution models for each data partition, inferred with the help of PartitionFinder version 1.0.1 (Lanfear et al. Reference Lanfear, Calcott, Ho and Guindon2012) using default settings and suggesting eleven data blocks (ITS1, 5.8S rDNA, ITS2, mtSSU, nuLSU, and three independent codon positions for RPB1 and RPB2), are listed in Table 2. We used the Markov Chain Monte Carlo (MCMC) approach implemented in MrBayes, version 3.2 (Ronquist et al. Reference Ronquist, Teslenko, van der Mark, Ayres, Darling, Höhna, Larget, Liu, Suchard and Huelsenbeck2012) to infer phylogenetic trees for the single gene datasets, applying the substitution models and partitioning schemes inferred with PartitionFinder and default settings of MrBayes with three exceptions. We used a proportional model on partition-specific rates, gamma distributed rates across sites modelled as six discrete categories and with a mean 1 exponential prior, and an empirical Bayes approach to select the mean of the exponential branch length prior. The mean of the branch length prior was inferred by calculating ML trees for all single gene datasets and the concatenated dataset using raxmlGUI version 0.9 beta 2 (Stamatakis Reference Stamatakis2006; Silvestro & Michalak Reference Silvestro and Michalak2010) and applying either an unpartitioned GTRGAMMAI model (mtSSU, nuLSU) or a partitioned model with separate, proportional GTRGAMMAI models (the rest) and 20 runs. The mean branch lengths of the ML trees were then used as means of the exponential distributions.
Table 2. Optimal partitioning scheme and substitution models for each data partition inferred by PartitionFinder, version 1.0.1, and used in the phylogenetic analyses.

The single gene MCMC trees were compared to identify conflicting phylogenetic signals between datasets. Three supported conflicts were detected (data not shown), which concerned the positions of Lecidea lapicida and L. silacea. In mtSSU, L. lapicida appeared as the sister taxon of L. silacea, in nuLSU its position was outside a well-supported clade of L. silacea, L. confluens and Porpidia speirea. In RPB2, it formed a well-supported clade with L. fuscoatra, L. grisella and P. speirea that excluded L. silacea. The different positions of L. silacea in nuLSU and RPB2, either inside or outside a clade with P. speirea, were also well supported. Because none of these conflicts concerned taxa connected with the L. hypnorum group, we decided to infer phylogenetic relationships based on a concatenated dataset containing all five gene regions. The inferred branch length prior for the MCMC analysis of this dataset followed an exponential distribution with mean 1/19. MrBayes was set to sample every 500th tree from three independent runs, each with four chains with the temperature increment parameter set to 0·15. The average standard deviation of bipartition frequencies among runs was calculated every 1 M generations to infer convergence of the Markov Chains, discarding the first 50% of the trees sampled as burn-in and including only those bipartitions with a frequency of at least 10%. The analysis was stopped after 2 M generations when the standard deviation had dropped below 0·01. Finally, we also calculated a ML bootstrap tree for the concatenated dataset using raxmlGUI and unlinked GTRGAMMAI models for the five partitions inferred by PartitionFinder.
Results
The results of the molecular analysis (Fig. 1) indicate that L. hypnorum belongs to a well-supported group of closely related species that also includes Mycobilimbia australis Kantvilas & Messuti, Lecidea diapensiae Th. Fr. and L. sanguineoatra auct. Because these species are also morphologically and anatomically similar, we describe below the new genus Bryobilimbia to accommodate them. Lecidea berengeriana (A. Massal.) Nyl., which is superficially similar to these species but can be distinguished by a number of characters (see below), is most closely related to Romjularia lurida (Ach.) Timdal. Clauzadea monticola (Ach.) Hafellner & Bellem. appears basal to this clade. These groupings receive high support from ML bootstrap values and posterior probabilities. The position of Lecidoma demissum (Rutstr.) Gotth. Schneid. & Hertel and Farnoldia jurana (Schaer.) Hertel is less clear. In the phylogenetic tree they form a sister group to the Clauzadea-Romjularia clade but the inter-relationships between these two clades and Bryobilimbia are not supported. The family Lecideaceae s. str., including Lecidea and Porpidia but also Eremastrella, appears monophyletic but the genera Lecidea and Porpidia are non-monophyletic.

Fig. 1. Maximum likelihood phylogenetic tree of Bryobilimbia (shaded) and related genera and species. Two species of Rhizocarpon were used as the outgroup. Numbers on and beside branches denote ML bootstrap values and MCMC posterior probabilities. Bold branches have BP≥70% and PP≥0·95. Grey branches were only supported in one of the analyses. Total length was 3·076 for the ML tree and 2·319 for the MCMC tree.
Our phylogenetic analysis resulted in an anomalous position for two of the specimens for which sequences were obtained from GenBank; the sequences for Helocarpon crassipes Th. Fr. (AY756322, AY756384) placed this specimen firmly within our new genus, whereas the sequences for Lecidea hypnorum (HQ650664, AY533005, HQ660569) resulted in this species appearing paraphyletic with our newly obtained sequences of this species. However, examination of the specimens from which the GenBank sequences were obtained showed that they had been incorrectly identified: the H. crassipes specimen was a species of Bryobilimbia, although its identity is currently unclear, whereas the L. hypnorum collections were referable to B. sanguineoatra, which is consistent with their position in our analysis.
Taxonomic Innovations
Bryobilimbia Fryday, Printzen & S. Ekman gen. nov.
MycoBank No.: MB805035
Distinguished from Lecidea and Mycobilimbia by having a Porpidia-type ascus, from Clauzadea and Porpidia by the much thinner gelatinous coat in mature ascospores, and from Lecidoma and Romjularia by the inconspicuous thallus and darker hypothecium. Its distinctness is also supported by molecular phylogenetic analyses based on ITS, nuclear LSU, mitochondrial SSU, RPB1 and RPB2 sequences.
Type species: Bryobilimbia hypnorum (Lib.) Fryday, Printzen & S. Ekman.
Bryobilimbia ahlesii (Körb.) Fryday, Printzen & S. Ekman comb. nov.
MycoBank No.: MB805040
Biatora ahlesii Körb., Parerga lichenol. 161 (1865).—Lecidea ahlesii (Körb.) Nyl., Flora 55: 356 (1872); type: [Germany, Baden-Württemberg], in sylvis “montanus” [montanis] p[rope]. Heidelberg, 1852; [W.] Zwackh (H-Nyl. 20416—neotype, designated by Meyer Reference Meyer2002).
Lecidea delincta Nyl., Flora 55: 356 (1872); type: [Finland, Tavastia Australis], Kuhmois, 1866, J. P. Norrlin 362 (H-Nyl. 20423—lectotype, designated by Meyer Reference Meyer2002).
Lecidea valentior Nyl., Flora 60: 229 (1877); type: Hibernia [Ireland, Galway], Bois du Lough Inagh, 1876, C. Larbalestier (H-Nyl. 20822—lectotype, designated by Meyer Reference Meyer2002).
Bryobilimbia ahlesii var. nemoralis (J. Lowe) Fryday, Printzen & S. Ekman comb. nov.
MycoBank No.: MB805047
Lecidea nemoralis J. Lowe, Lloydia 2: 264 (1939).—Lecidea ahlesii var. nemoralis (J. Lowe) Fryday & Coppins, Bryologist 109: 12 (Reference Coppins and Fryday2006); type: USA, New York, Adirondack Region, The Huntingdon Forest at Newcomb, on rock in brook bed, 1934, J. L. Lowe 5016 (MICH—holotype!).
Although we did not include B. ahlesii in our phylogenetic analysis, it has all the features characteristic of the genus and so we have no hesitation in including it in Bryobilimbia. It is unusual within the genus in that both varieties occur only on damp siliceous rock.
Bryobilimbia australis (Kantvilas & Messuti) Fryday, Printzen & S. Ekman comb. nov.
MycoBank No.: MB805049
Mycobilimbia australis Kantvilas & Messuti, Lichenologist 37: 252 (2004); type: Australia, Tasmania, summit plateau of Projection Bluff, 41°43′S, 146°42′E, 1260 m, on peaty soil in alpine heathland, 1994, G. Kantvilas (3/94) & J. Jarman (HO—holotype; BCRU, BM, CHR—isotypes).
Bryobilimbia diapensiae (Th. Fr.) Fryday, Printzen & S. Ekman comb. nov.
MycoBank No.: MB805052
Lecidea diapensiae Th. Fr., Lichenes Arctoi: 209 (1860).—Biatora diapensiae (Th. Fr.) Hellb., Öfvers. kongl. Svenska Vetensk.-Akad. Förhandl. 1875: 68 (1875); type: [Norway, Finnmark], Østfinnmark, Syd-Varanger, Elvenes, 15 July 1857, Th. M. Fries (UPS—lectotype, designated by Printzen in Biblioth. Lich. 60: 171, 1995).
The lectotype has an olivaceous subhymenium and this character is shared by many collections from Diapensia. However, other collections from Diapensia lack this pigment and are otherwise similar to B. sanguineoatra. The taxonomic status of these collections is currently unclear and warrants further study.
Bryobilimbia hypnorum (Lib.) Fryday, Printzen & S. Ekman comb. nov.
MycoBank No.: MB805054
Lecidea hypnorum Lib., Plantae cryptogamicae quas in Arduenna collegit M. A. Libert, fasc. 1, #12. (1830).—Mycobilimbia hypnorum (Lib.) Kalb & Hafellner, in Wirth, Die Flechten Baden-Württembergs. Verbreitungsatlas: 511 (Reference Wirth1987); type: [?Belgium, ‘Ardennes’], ad rupes supra muscos, Libert [PRA—lectotype, designated by Vězda in Lichenes Selecti Exs., Fasc. L (No. 1233), 1974, FR—isolectotype!].
Lecidea templetonii Taylor, in Mackay, Flora Hibernica 2: 123 (Reference Mackay1836); type: Ireland, near Belfast, Templeton (FH—lectotype!, designated here; BM—probable isolectotype!).
Lecidea hypnorum was first described by Libert in 1830 (not 1853 as given by Zahlbruckner) and therefore has priority over L. templetonii Taylor.
Taylor (in Mackay Reference Mackay1836) mentions two collections of his new taxon: “on moss, near Belfast, Mr. Templeton; on turf, near Bantry, Miss Hutchins”. Both collections are present in Taylor's herbarium in FH, with a further specimen of Templeton's collection in BM (ex hb. Crombie). The Hutchins collection (near Bantry) is Micarea inquinans (Tul.) Coppins, but the Templeton collection (near Belfast) is clearly conspecific with Lecidea hypnorum. There are two specimens in the packet in FH and the upper one (Bar Code: 00377084) is designated here as the lectotype. There is a space on the sheet beside this specimen where another specimen has been removed and it is probable that this is the specimen that is now in BM.
Bryobilimbia sanguineoatra (Wulfen) Fryday, Printzen & S. Ekman comb. nov.
MycoBank No.: MB805055
Lichen sanguineoater Wulfen, in Jacquin, Coll. Botan. 3: 117 (1789)—Lecidea sanguineoatra (Wulfen) Ach., Method. Lich.: 50 (1803).—Mycobilimbia sanguineoatra (Wulfen) Kalb & Hafellner nom. inval., Herzogia 9: 75 (1992)); type: Ueber Mossen [sic, not “Moosen”], besonders Hypnum cupressif. auf Fichtenwurzeln im Walde hinter Schernfeld bei Eichstätt, Sommer 1862, [Arnold, Lichenes exsiccati (Lichenes Jurae) #229: UPS—neotype!; FR —isoneotype!, designated here].
Based on Wulfen's description, Arnold (Reference Arnold1882) referred the taxon to Bilimbia sabuletorum (Schreb.) Arnold, the basionym of which (Lichen sabuletorum Schreb.) was described 18 years earlier than Lichen sanguineoater. Because no original material of Wulfen's taxon is found in either M or W to confirm or reject Arnold's determination, subsequent authors referred to the species as L. sanguineoatra auct. (e.g., Aptroot et al. Reference Aptroot, Gilbert, Hawksworth, Coppins, Smith, Aptroot, Coppins, Fletcher, Gilbert, James and Wolseley2009), or L. sanguineoatra sensu A. L. Smith (e.g., Index Fungorum 2012). In order to avoid further confusion, we here designate a neotype for Wulfen's name that matches current usage.
Discussion
Systematic position
In the study by Buschbom & Mueller (Reference Buschbom and Mueller2004), Bryobilimbia sanguineoatra [as Mycobilimbia hypnorum (Lib.) Kalb & Hafellner] grouped with Clauzadea, which in turn formed a sister group, although with little support, to a group containing Farnoldia, Melanolecia Hertel, Notolecidea Hertel and Pachyphysis R. C. Harris & Ladd, in a group outside the core Lecideaceae. However, for want of a better alternative, all of these genera were included in the Lecideaceae by Lumbsch & Huhndorf (Reference Lumbsch and Huhndorf2010).
Subsequent molecular work by Schmull et al. (Reference Schmull, Miądlikowska, Pelzer, Stocker-Wörgötter, Hofstetter, Fraker, Hodkinson, Reeb, Kukwa and Lumbsch2011) has shown that the L. hypnorum-group is not congeneric with the ‘Lecidea’ berengeriana group but is more closely related to Lecidoma, and also indicated an anomalous position of these two groups outside the major groups in Lecanoromycetidae. Our study confirms these relationships but does little to resolve them. It does, however, indicate that Bryobilimbia and L. berengeriana are more closely related to each other than suggested by Schmull et al. (Reference Schmull, Miądlikowska, Pelzer, Stocker-Wörgötter, Hofstetter, Fraker, Hodkinson, Reeb, Kukwa and Lumbsch2011). Further work, including other species of Bryobilimbia and the L. berengeriana group (see excluded species), along with other taxa with a Lecidea or Porpidia-type ascus that were shown by Buschbom & Mueller (Reference Buschbom and Mueller2004) to be related to Farnoldia (e.g., Melanolecia, Pachyphysis, Poeltiaria, Poeltidea, etc.), is required to clarify the higher systematic position of these two groups of species.
Conidiomata
Pycnidia are very rare in Bryobilimbia. To our knowledge, they have been reported only from B. australis by Kantvilas et al. (Reference Kantvilas, Messuti and Lumbsch2005), who described them as “uncommon, black, rather glossy, superficial, 0·10–0·15 mm wide, resembling apothecial initials; ostiole gaping at maturity, becoming excavate; wall in section dark brown to purple-brown with blue-black, K+ aeruginose pigment. Conidia bacilliform, 4–5×1 µm”. Unfortunately, Kantvilas et al. (Reference Kantvilas, Messuti and Lumbsch2005) did not record details of the conidiophores or the attachment of the conidia. However, inspection of a collection of B. australis housed in MSC (Imshaug 42125) confirmed the description given by Kantvilas et al. (Reference Kantvilas, Messuti and Lumbsch2005) and revealed that the conidia were acrogenous, borne singly on conidiophores with an unbranched terminal cell measuring 12–15×3 µm.
Comparison of Bryobilimbia with similar genera
A number of other genera, all with Porpidia-type asci, have characters that are similar to those of our new genus. The differences between Bryobilimbia and similar genera are shown in detail in Table 3.
Table 3. Comparison of characters of Bryobilimbia and similar genera (significant characters in bold)

Clauzadea. The species of this genus differ in having branched, partly moniliform paraphyses, the ascospores having a poorly- to well-developed perispore and/or gelatinous coat, and in the extremely rare occurrence of blue-green granules in the hymenium, hypothecium, and excipulum. Clauzadea is also a genus of calcareous rocks, whereas the species of Bryobilimbia are primarily bryophilous (over various substrata). However, L. ahlesii and two undescribed species known to us occur on damp siliceous rocks and are also referable to Bryobilimbia.
Lecidea berengeriana. It is currently unclear which species formerly considered as belonging to the ‘Lecidea hypnorum group’ (see below) are referable to Bryobilimbia and which are more closely related to L. berengeriana. However, it appears that L. berengeriana and closely related species are distinguished morphologically from Bryobilimbia by having a thick tartareous to subsquamulose thallus, capitate paraphyses to 6 µm wide, and ellipsoid conidia. This last character is also shared with Romjularia, supporting the relationship revealed by the molecular analysis. The mature ascospores also lack any perispore or gelatinous coat and there are rarely any blue-violet (K+ green) granules in apothecial sections, a feature that is characteristic of Bryobilimbia. There is also some evidence that apothecium development is gymnocarpous in L. berengeriana whereas it is hemiangiocarpous in Bryobilimbia (A. M. Fryday, unpublished data).
Lecidoma. The single species, L. demissum, differs from Bryobilimbia most significantly in having a chlorococcoid photobiont that divides into 2–4 daughter cells, and a hyaline hypothecium (always red-brown in Bryobilimbia). The apothecia also lack the blue-violet (K+ green) granules in section that are characteristic of Bryobilimbia, and the mature ascospores lack any perispore or gelatinous coat. The species is restricted to acid soils in alpine regions.
Romjularia. The single species, R. lurida, differs from Bryobilimbia in having a thallus composed of squamules up to 5 mm across that are rounded at the apices, apothecia with a pale brown hypothecium, and ellipsoid conidia. The apothecia lack the blue-violet (K+ green) granules in section that are characteristic of Bryobilimbia, and the mature ascospores lack any perispore or gelatinous coat. The species is also restricted to calcareous soils associated with limestone.
Key to Bryobilimbia and related genera
This is a preliminary key to the genera of Lecideaceae s. lat. that appear closely related to Bryobilimbia. A full key to the genera of Lecideaceae s. lat. is in preparation and will be published elsewhere (Fryday & Hertel Reference Fryday and Hertel2014).
-
1 Exciple thick and carbonaceous throughout, clearly separated from the paler hypothecium ... Farnoldia
Excipulum not carbonaceous throughout ... 2
-
2(1) Saxicolous. Thallus usually crustose, rarely squamulose ... 3
Muscicolous, terricolous or on moribund Diapensia. Thallus crustose to squamulose; if directly on rock then thallus squamulose ... 6
-
3(2) Thallus distinctly squamulose. On limestone (rare occurrences for this usually terricolous species) ... Romjularia
Thallus crustose. On siliceous or calcareous rocks ... 4
-
4(3) Hypothecium and exciple usually dark brown (Arnoldiana-brown), if orange-brown (Superba-brown) then ascospores >20 µm long; olivaceous pigments (Cinereorufa-green, K+HCl+ blue) usually present in epithecium and exciple cortex. Ascospores with well-developed gelatinous coat. Conidia borne apically ... Porpidia
Hypothecium and exciple orange-brown (Superba-brown); usually lacking olivaceous pigments internally, if present then Baglietoana-green (K+HCl+ violaceous) in exciple, medulla and/or lower hymenium and hypothecium. Ascospores <18 µm long with poorly- or well-developed gelatinous coat ... 5
-
5(4) On damp siliceous rocks; brown or olivaceous pigments present Conidia borne apically ... Bryobilimbia
On calcareous rocks; hypothecium paler than exciple, K−, only brown pigments present internally. Conidia borne laterally and apically ... Clauzadea
-
6(2) Terricolous. Thallus either distinctly squamulose or areolate-squamulose with distinct marginal lobes ... 7
Muscicolous on rocks, trees or soil. Thallus crustose or minutely squamulose without distinct marginal lobes ... 8
-
7(6) On acid alpine soils. Thallus areolate-squamulose with wide marginal lobes; hypothecium hyaline; paraphyses simple, thick (3–4 µm) distinctly capitate (5–7 µm) ... Lecidoma
On calcareous soils, often in limestone crevices. Thallus squamulose; hypothecium pale brown; paraphyses thinner ... Romjularia
-
8(6) Conidia bacilliform, c. 1 µm wide. Paraphyses only slightly swollen at apex. Thallus inconspicuous ... Bryobilimbia
Conidia ellipsoid, >2 µm wide. Paraphyses distinctly swollen at apex (to 6 µm). Thallus granular to minutely squamulose ... L. berengeriana-group
Key to species of Bryobilimbia s. str.
In most cases the species currently placed in the new genus are easily separated and, where there is doubt, good descriptions are provided by Aptroot et al. (Reference Aptroot, Gilbert, Hawksworth, Coppins, Smith, Aptroot, Coppins, Fletcher, Gilbert, James and Wolseley2009) and Kantvilas et al. (Reference Kantvilas, Messuti and Lumbsch2005). The exception is B. diapensiae, for which no good, modern, English description exists, although descriptions of this species are provided by Foucard (Reference Foucard2001; Swedish) and Fries (Reference Fries1874; Latin). Bryobilimbia diapensiae is distinguished by its habitat of moribund Diapensia and the presence of an olivaceous subhymenium.
-
1 On damp siliceous rocks ... 2
Muscicolous on rocks, trees or soil ... 4
-
2(1) Ascospores 3·0–4·5(–6·0) µm wide (rare occurrences for this usually muscicolous species) ... B. sanguineoatra
Ascospores (5–)6–7(–9) µm wide ... 3
-
3(2) Apothecia with brown pigments only ... B. ahlesii var. ahlesii
Apothecia with brown and olivaceous green pigments ... B. ahlesii var. nemoralis
-
4(1) Mature apothecia forming large, blackberry-like clusters; exciple blue-black (K+ aeruginose). Terricolous in the southern cool temperate zone ... B. australis
Mature apothecia remaining single; exciple brown. Ecology and distribution various ... 5
-
5(4) Ascospores 4·5–6·0(–7·0) µm wide with a warted perispore, often 1-septate. Over bryophytes on calcareous rocks and trees ... B. hypnorum
Ascospores narrow ellipsoid, <5 µm wide, smooth, simple. Over bryophytes on trees or directly on moribund Diapensia plants ... 6
-
6(5) Only brown apothecial pigments present. Growing over bryophytes, usually on trees but occasionally on siliceous rocks; rarely directly on bark or siliceous rock. Cool temperate to boreal ... B. sanguineoatra
Subhymenium olivaceous-green. On moribund Diapensia in Arctic regions ... B. diapensiae
Excluded Species
Mycobilimbia austrocalifornica (Zahlbr.) K. Knudsen
Opuscula Philolichenum 2: 36 (2005).—Lecidea austrocalifornica Zahlbr., Cat. Lich. Univ. 3: 738 (1925), nomen novum pro Lecidea subplebeia Nyl. in Hasse non Lecidea subplebeia Vain. (1890).
Lecidea subplebeia Nyl. in Hasse (non Lecidea subplebeia Vain.), Bull. Torrey Bot. Club 24: 447 (1897); type: USA, California, Los Angeles Co., Santa Monica Range, on earth, near Soldier's Home, November 1896, H. E. Hasse s.n. (H-Nyl 12067—lectotype, designated by Knudsen Reference Knudsen2005).
The type collection is Carbonea latypizodes (Nyl.) Knoph & Rambold and the other collections mentioned by Knudsen (Reference Knudsen2005) are Placynthiella hyporhoda (Th. Fr.) Coppins & P. James (C. Printzen, unpublished data).
In addition, the following species, which are traditionally included in the ‘Lecidea hypnorum group’, are not here included in Bryobilimbia. Our analysis shows that Lecidea berengeriana is not congeneric with B. hypnorum, and the other species are morphologically and anatomically closer to Lecidea berengeriana.
Lecidea berengeriana (A. Massal.) Nyl.
Lecidea berengeriana (A. Massal.) Nyl., Not. Sällsk. Fauna Fl. Fenn. Förh. 8: 144 (1866).—Biatora berengeriana A. Massal., Ric. auton. lich. crost.: 128 (1852).—Mycobilimbia berengeriana (A. Massal.) Hafellner & V. Wirth, in Wirth, Die Flechten Baden-Württembergs. Verbreitungsatlas: 511 (Reference Wirth1987).
Lecidea diplotypa Vain.
Lecidea diplotypa Vain., Étud. class. lich. Brésil, 11: 30 (1890).—Mycobilimbia diplotypa (Vain.) Kalb, Lichenes Neotropici, Fascicle IX (nos 351–400) (Neumarkt): 11, no. 382 (1986).
Lecidea fissuriseda Poelt
Lecidea fissuriseda Poelt, Mitt. Bot. Staatssamml. München 4: 181 (1961).—Mycobilimbia fissuriseda (Poelt) Poelt & Hafellner, in Hafellner, Herzogia 8: 56 (Reference Hafellner1989).
Lecidea holopolia (Tuck.) Zahlbr.
Biatora holopolia Tuck, Syn. N. Amer. Lich. 2: 26 (1888).—Lecidea holopolia (Tuck.) Zahlbr., Cat. Lich. Univ. 3: 782 (1925).
Mycobilimbia olivacea Aragón, Sarrion & Hafellner, Lichenologist 35: 3 (Reference Sarrión, Aragón, Hafellner, Rico and Burgaz2003).
Lecidea strasseri Zahlbr.
Lecidea strasseri Zahlbr., Verh. Zool.-Bot. Ges. Wien 48: 357 (1898).
Lecidea subfilamentosa (Zahlbr.) Fryday
Phyllopsora subfilamentosa Zahlbr., Ann. Mycol. 33: 44 (1935).—Fuscidea subfilamentosa (Zahlbr.) Brako, in Egan, Bryologist 90: 163 (1987).—Lecidea subfilamentosa (Zahlbr.) Fryday, Lichenologist 39: 322 (Reference Fryday2008).
Mycobilimbia meridionalis Kantvilas
Mycobilimbia meridionalis Kantvilas, Lichenologist 37: 255 (2004).
Mycobilimbia parvilobulosa Aragón et al.
Mycobilimbia parvilobulosa Aragón, Sarrión & Hafellner, Lichenologist 35: 6 (Reference Sarrión, Aragón, Hafellner, Rico and Burgaz2003).
We thank the curators of B, F and O for the loan of collections in their care, the curators of M and W for searching their collections for relevant material, Zdeněk Palice (Průhonice) for confirming that the lectotype of Lecidea hypnorum is housed in PRA, not PR as stated in the protologue, and Sergio Pérez-Ortega (Madrid) for providing collections of Romjularia lurida and Bryobilimbia australis for DNA isolation.






