Introduction
Molecular phylogenetic investigations have led to the detection of many cryptic species amongst the parmelioid lichens. Examples include Parmelia ernstiae (Feuerer & Thell Reference Feuerer and Thell2002), P. serrana (Molina et al. Reference Molina, Crespo, Blanco, Lumbsch and Hawksworth2004), P. barrenoae (Divakar et al. Reference Divakar, Molina, Lumbsch and Crespo2005), P. encryptata (Molina et al. Reference Molina, Divakar, Millanes, Sanchez, Hawksworth and Crespo2011), and Melanelixia californica (Divakar et al. Reference Crespo, Ferencov, Perez-Ortega, Elix and Divakar2010a). This approach has been especially useful in Parmelina, where the combination of molecular and morphological information provided evidence of polyphyly of the genus: the Australasian species appear in a clade that is unrelated to the Northern Hemisphere species, and are now accommodated in the new genus Austroparmelina (Crespo et al. Reference Crespo, Ferencov, Perez-Ortega, Elix and Divakar2010). At the species level, as in other organisms, speciation in lichens has not always been accompanied by morphological variation. For instance, phylogenetic analysis and detailed morphological studies found two previously overlooked morphospecies within the widespread P. quercina: P. coleae and the re-established P. carporrhizans (Argüello et al. Reference Argüello, Del Prado, Cubas and Crespo2007).
At present, Parmelina includes seven species distributed in the temperate regions of the Northern Hemisphere (Elix Reference Elix1993; Argüello et al. Reference Argüello, Del Prado, Cubas and Crespo2007; Clerc & Truong Reference Clerc and Truong2008). Parmelina coleae is restricted to North America, P. gyrophorica is known only from southern China (Wang et al. Reference Wang, Chen and Elix2000), and five other species occur in western Europe, extending to Asia. Three of those five species always have apothecia: P. quercina, P. carporrhizans, and P. atricha (Poelt & Vězda Reference Poelt and Vězda1977; Nimis Reference Nimis1993; Clerc & Truong Reference Clerc and Truong2008). Parmelina tiliacea and P. pastillifera, characterized by the presence of isidia, are frequently sterile although individuals with apothecia are not unusual (Schauer Reference Schauer1965; Hale Reference Hale1976; Divakar & Upreti Reference Divakar and Upreti2005).
Parmelina tiliacea has a wide distribution, including Europe, northern Africa, the Middle East, Asia and the Indian subcontinent. It is a frequent epiphyte in south-western Europe, at c. 200–1500 m above sea level. However, this species is considered to be threatened in countries such as Denmark, The Netherlands, Poland, and Germany (Liška et al. Reference Liška, Palice, Dětinský, Vondrák, Lackovičová, Guttová, Lisická and Lizoň2006). Phylogenetic information about this species was, however, scarce and based on small samples. This study aimed to establish whether P. tiliacea was monophyletic or if cryptic species were hidden within its current circumscription, and to evaluate any implications for species conservation programmes. The phylogenetic relationships were assessed by analysis of two independent loci (nuclear ITS and mitochondrial LSU) with extensive sampling covering most of the distributional range. Samples of the isidiate species, P. pastillifera, found to be sister to P. tiliacea in previous studies (Argüello et al. Reference Argüello, Del Prado, Cubas and Crespo2007), were also included in the analysis. Morphological characters were re-evaluated and compared with the relationships found in the molecular analyses.
Material and Methods
Taxon sampling
Specimens of Parmelina tiliacea were collected from most of its distributional range, and from different altitudes and ecological areas. Locations included the Iberian Peninsula, the Canary Islands, Morocco, Turkey, France, Germany, Austria, Italy, Slovenia, Tunisia, and India. Details of localities, voucher specimens, and GenBank accession numbers are shown in Table 1. Samples of other Parmelina species were included in the phylogenetic analysis: P. carporrhizans from Turkey and the Canary Islands, P. coleae from the USA, P. atricha from France, P. pastillifera from the Iberian Peninsula and Turkey, and P. quercina from Turkey (Table 1). Two closely related Parmeliaceae, Myelochroa metarevoluta and M. galbina, were used as an outgroup (Blanco et al. Reference Blanco, Crespo, Ree and Lumbsch2006).
Table 1. Specimens included in the phylogenetic study, with details of location, collectors, herbarium code and GenBank accession numbers

* Divakar et al. (2006),
† Blanco et al. (Reference Blanco, Crespo, Divakar, Esslinger, Hawksworth and Lumbsch2004)
DNA extraction and amplification
Small fragments of vegetative thallus from fresh or frozen herbarium specimens were ground with sterile glass pestles. Total DNA was extracted using the DNeasy Plant Mini Kit (Qiagen, Barcelona) according to the manufacturer's instructions, slightly modified as previously described (Crespo et al. Reference Crespo, Blanco and Hawksworth2001). Double-stranded DNA amplification of the two regions was performed using the primers: 1) ITS1-LM (Myllys et al. Reference Myllys, Lohtander, Källersjö and Tehler1999) and ITS2-KL (Lohtander et al. Reference Lohtander, Myllys, Sundin, Källersjö and Tehler1998) for the fungal nuITS rDNA region, and 2) ML3 and ML4 (Printzen Reference Printzen2002) for the fungal mtLSU rDNA region. For ITS amplification we used a reaction mixture of 50 µl, containing 5 µl of ×10 buffer with 2 mM MgCl2, 1 µl of dNTPs (10 mM of each base), 2·5 µl of each primer (10 µM), 1·25 µl of DNA polymerase (1 U µl−1), 27·75 µl of sterile water and 10 µl of dilute DNA template. Amplification of the mtLSU region was performed using PuRe Taq Ready-To-Go PCR Beads (GE Healthcare, UK) in a 25 µl volume containing 13·4 µl of sterile water, 2·5 U of puReTaq DNA Polymerase, 200 µM of each dNTP, BSA, buffer reaction and stabilizers (10 mM Tris-HCl pH 9.0, 50 mM KCl, 1·5 mM MgCl2), 1·5 µl of each primer and 5 µl of dilute DNA template.
The amplifications were run in an automatic thermocycler (Techne Progene) using the following parameters for nuITS rDNA: initial denaturation for 5 min at 94°C, followed by 35 cycles of 1 min at 94°C, 1 min at 56°C and 1·5 min at 72°C, and a final extension of 5 min at 72°C. Parameters for mtLSU rDNA were: initial denaturation for 10 min at 94°C, followed by 35 cycles of 1 min at 94°C, 1 min at 47°C and 3 min at 72°C, and a final extension of 10 min at 72°C.
PCR products were cleaned using a Bioclean Columns kit (Biotools, Madrid) according to the manufacturer's instructions. Sequencing was performed using the ABI Prism Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) with the amplification primers. Sequencing reactions were electrophoresed on a 3730 DNA analyzer (Applied Biosystem) at the Unidad de Genómica (Parque Científico de Madrid). Sequence fragments obtained were assembled and manually adjusted in BioEdit 7.0.9.0 (Hall Reference Hall1999).
Sequence alignments and phylogenetic analysis
Three separate matrices (one for each gene and another with both genes combined) were constructed and aligned using the Clustal W Multiple alignment program (Thompson et al. Reference Thompson, Higgins and Gibson1994). Ambiguously aligned regions were excluded manually from the matrices. The alignments were analyzed by Maximum Parsimony (MP) and a Bayesian Markov Chain Monte Carlo (B/MCMC) approach (Larget & Simon Reference Larget and Simon1999; Huelsenbeck et al. Reference Huelsenbeck, Rannala and Masly2000). The trees were rooted using M. galbina and M. metarevoluta as an outgroup.
Parsimony analyses were carried out using PAUP* 4.0b10 (Swofford Reference Swofford2003) performed at www.bioportal.uio.no, with equally weighted characters and gaps being interpreted as missing data. A heuristic search with 100 random taxon addition replicates was generated with tree-bisection-reconnection (TBR) branch-swapping and the MulTrees option in effect. Nonparametric bootstrap (Felsenstein Reference Felsenstein1985) was used to assess robustness of clades, running 4000 pseudoreplicates with the same settings as in the heuristic search. Only clades that received bootstrap support above 75% were considered to be strongly supported. To assess homoplasy levels, we calculated the consistency index (CI) and retention index (RI) from each parsimony search. Majority rule consensus trees were drawn using TREEVIEW (Page Reference Page1996).
The heterogeneity in phylogenetic signal between the two data partitions was examined by MP and Bayesian approach (Wiens Reference Wiens1998; Buckley et al. Reference Buckley, Arensburger, Simon and Chambers2002; Divakar et al. Reference Crespo, Ferencov, Perez-Ortega, Elix and Divakar2010b). The level of bootstrap support and posterior probabilities were used to detect significance levels of localized incongruence between the two gene partitions and the concatenated analysis. The set of topologies reaching ≥ 75% bootstrap under parsimony and the 0·95 posterior probabilities for the Bayesian approach were estimated (Hillis & Bull Reference Hillis and Bull1993). If no conflict was evident, it was assumed that the two data sets were congruent and could be combined.
Bayesian analyses were carried out using the program MrBayes 3.1.2 (Huelsenbeck & Ronquist Reference Huelsenbeck and Ronquist2001). Posterior probabilities were approximated by sampling trees using a Markov Chain Monte Carlo (MCMC) method. The posterior probabilities of each branch were calculated by counting the frequency of trees that were visited during the course of the MCMC analysis. The analysis was performed assuming the general time reversible model (Rodriguez et al. Reference Rodriguez, Oliver, Marin and Medina1990) including estimation of invariant sites and assuming a discrete gamma distribution with six rate categories (GTR + I + G) for the single-gene and the combined analyses. A run with 2 million generations, starting with a random tree and employing 8 simultaneous chains, was executed. Every 200th tree was saved to a file. We plotted the log-likelihood scores of sample points against generation time using TRACER version 1.0 (http://evolve.zoo.ox.ac.uk/software.html?id=tracer) and determined that stationarity had been achieved when the log-likelihood values of the sample points reached a stable equilibrium value (Huelsenbeck & Ronquist Reference Huelsenbeck and Ronquist2001). The first 2000 trees were discarded as burn-in before stationarity was reached. Unlike nonparametric boostrap values (Felsenstein Reference Felsenstein1985), these are estimated probabilities of the clades under the assumed model (Rannala & Yang Reference Rannala and Yang1996), and hence posterior probabilities ≥ 95% were considered significant support.
Hypothesis testing
As the results of the phylogenetic analysis are incongruent with the current species concept of Parmelina tiliacea, we examined whether our data were sufficient to reject the monophyly of P. tiliacea. Two methods were employed: 1) the Shimodaira–Hasegawa (SH) test (Shimodaira & Hasegawa Reference Shimodaira and Hasegawa1999); 2) the expected likelihood weight (ELW) test following Strimmer and Rambaut (Reference Strimmer and Rambaut2002). We compared the ML tree constrained to have P. tiliacea as monophyletic and the unconstrained ML tree. The SH and ELW tests were performed using Tree-PUZZLE 5.2 (Schmidt et al. Reference Schmidt, Strimmer, Vingron and von Haeseler2004) with the combined dataset from a sample of 200 unique trees (the best trees agreeing with the null hypotheses) and the unconstrained ML tree. These trees were inferred in Tree-PUZZLE employing the GTR + I + G nucleotide substitution model.
Genetic distances
Pairwise maximum likelihood distances, given as the number of nucleotide substitutions per site, between the ITS rDNA sequences were calculated with TREE-PUZZLE 5.2 (Schmidt et al. Reference Schmidt, Strimmer, Vingron and von Haeseler2004) using the HKY+G (Hasegawa et al. Reference Hasegawa, Kishino and Yano1985) model of nucleotide substitution with between-site variation, and assuming a discrete gamma distribution with six rate categories. Ambiguous characters and repeated haplotypes were removed in the matrix as explained in Del-Prado et al. (Reference Del-Prado, Cubas, Lumbsch, Divakar, Blanco, Amo De Paz, Molina and Crespo2010).
Morphological and chemical studies
All specimens included in the molecular analysis were morphologically revised (Table 1). The size and shape of the ascospores were studied in the specimens bearing apothecia. Sections of apothecia were hand-cut with a razor blade under a binocular microscope, and mounted in distilled water. Ascospores and longitudinal sections of apothecia were observed and photographed under a light microscope (Leitz DMRB). Measurements of the length and width were taken in 20 well-developed ascospores per sample at ×1000 magnification. The description of spore shape follows Kirk et al. (Reference Kirk, Cannon, Minter and Stalpers2008). Statistical tests were performed using STATGRAPHICS Plus 5.1.
Chemical analyses were performed by thin-layer chromatography using standard methods (Culberson Reference Culberson1972; Elix & Ernst-Russell Reference Elix and Ernst-Russell1993; Orange et al. Reference Orange, James and White2001) in all the specimens analyzed.
Results
Phylogenetic analyses
Seventy-six new sequences were generated for this study, including 38 new nuclear ITS rDNA and 39 new mitochondrial LSU rDNA sequences. The aligned matrices had 436 unambiguous nucleotide positions in the nuITS and 668 in the mtLSU. Comparison of the topology of MP and Bayesian trees based on the individual genes showed no supported conflicts (results not shown), thus both regions were combined to increase the resolution of the clades.
The MP analysis of the combined data matrix resulted in the 30 most parsimonious trees (tree length = 259 steps, CI = 0·8147, RI = 0·9159). Thirty-six positions in the matrix were parsimony uninformative and 145 were informative. For the Bayesian analysis the likelihood parameters had the following average values [± one standard deviation (SD)]: likelihood (lnL) = −3095·525 (0·2307), base frequency π(A) = 0·2097 (0·0003), π(C) = 0·2829 (0·0003), π(G) = 0·2644 (0·0003), π(T) = 0·243 (0·0003), rate matrix r(AC) = 0·1256 (0·0007), r(AG) = 0·1365 (0·0007), r(AT) = 0·1667 (0·0009), r(CG) = 0·0764 (0·0005), r(CT) = 0·4096 (0·001), r(GT) = 0·0851 (0·0005), the gamma shape parameter α = 90·139 (2·8764) and the pinvar = 0·5757 (0·0045). Since the topologies of combined dataset analyses using MP and B/MCMC approaches showed no supported conflicts, only the 50% majority rule consensus tree of Bayesian tree sampling is shown (Fig. 1).
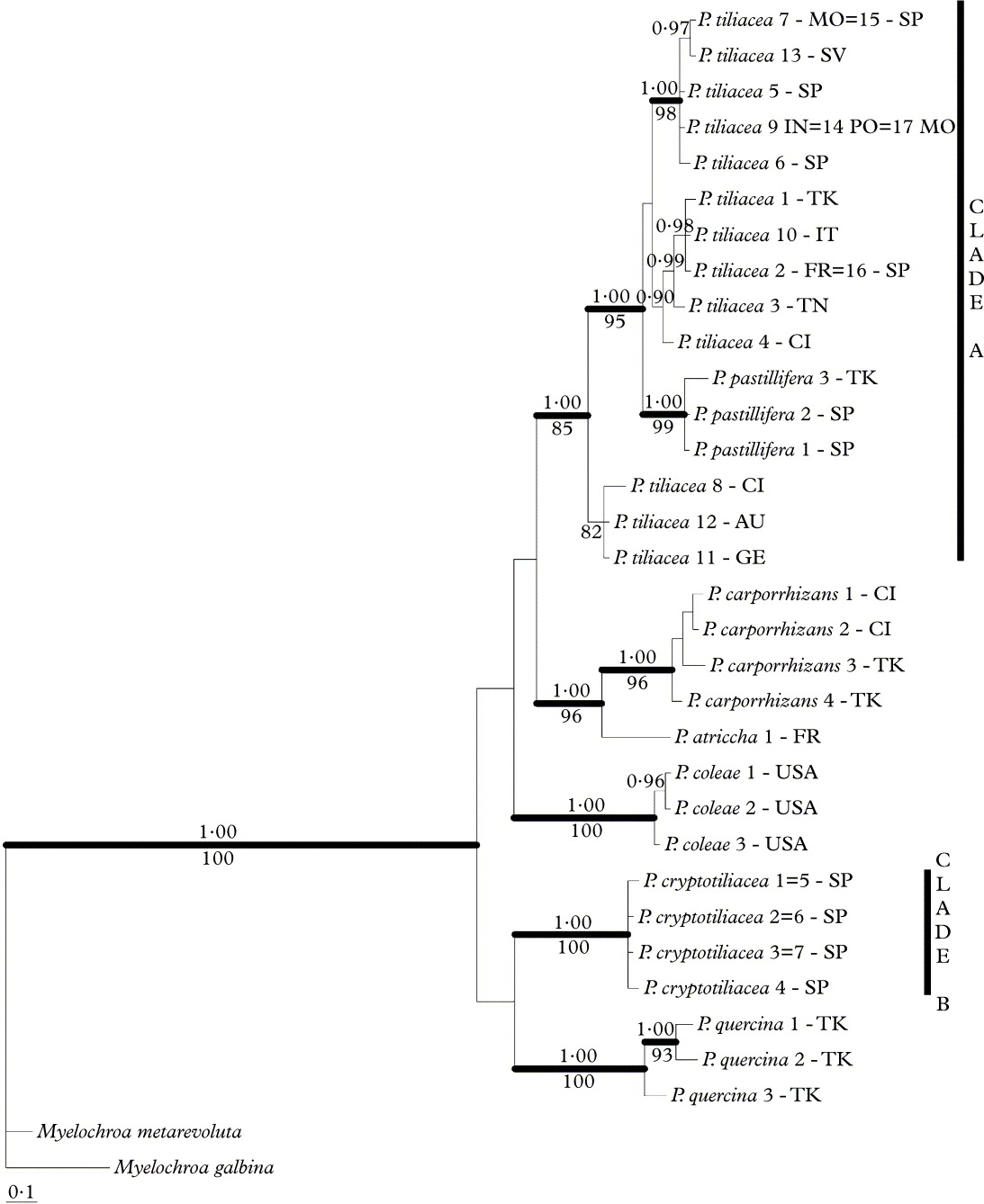
Fig. 1. Majority-rule consensus tree based on 18 000 trees from B/MCMC tree sampling procedure from a combined data set of nuITS rDNA and mtLSU rDNA sequences. Posterior probabilities ≥ 0·95 in the Bayesian analysis are indicated above the branches and MP boostrap values ≥ 0·75 below branches. Branches with significant support in both analyses are in bold. (AU = Austria, CI = Canary Islands, FR = France, GE = Germany, IN = India, IT = Italy, MO = Morocco, PO = Portugal, SP = Spain, SV = Slovenia, TK = Turkey, TN = Tunisia, USA = United States of America).
The topology of the tree (Fig. 1) showed that the samples of P. carporrhizans, P. coleae and P. quercina form well-supported monophyletic groups. Parmelina atricha is sister to P. carporrhizans but other relationships lacked statistical support. However, the samples of P. tiliacea separated into two independent groups (clades A and B) with strong statistical support. Clade A included P. tiliacea specimens from a wide geographic area and also a nested clade with all the P. pastillifera specimens examined. Clade B included only samples from central Spain. As the relationships between the Parmelina species are not well supported, the monophyly of P. tiliacea as a single species (including clades A and B) was tested and significantly rejected (P < 0·002 in the SH and ELW tests). The same results were obtained irrespective of whether P. pastillifera was included in clade A.
The pairwise genetic distances between the ITS haplotypes were also calculated to estimate the genetic divergence of clades A and B (Table 2). When all the samples were assigned to a single species, the intraspecific mean was remarkably high (0·030 ± 0·006 s/s), with a maximum distance of 0·093 s/s. On the other hand, when specimens of clades A and B are considered as two separate species, the maximum values of their intraspecific genetic distances fall to 0·0169 s/s for clade A and 0·002 for clade B, and the interspecific distances range from 0·071–0·093. Similar values were obtained irrespective of whether P. pastillifera specimens were included in clade A (Table 2).
Table 2. Pairwise genetic distances between ITS haplotypes

Morphology and chemistry
Morphological analysis of apothecia of both groups A and B revealed subtle differences in the outer hyphae forming the true exciple. The cells of the exciple in group B had thinner walls than those of group A (Fig. 2B). Measurements of the ascospores of representative specimens of each clade showed that both clades had spores of similar length (clade A, ![]() ± SD = 10·53 µm ± 1·17, range = 8–15 µm; clade B,
± SD = 10·53 µm ± 1·17, range = 8–15 µm; clade B, ![]() = 10·66 µm ± 0·94; range = 9–13 µm) that did not show a statistically significant difference (P = 0·4130). On the contrary, however, the mean width of the ascospores of both clades differed significantly (P = 0·0000): those from group A (
= 10·66 µm ± 0·94; range = 9–13 µm) that did not show a statistically significant difference (P = 0·4130). On the contrary, however, the mean width of the ascospores of both clades differed significantly (P = 0·0000): those from group A (![]() ± SD = 5·69 ± 0·58, range = 5–7 µm) being wider than those of group B (
± SD = 5·69 ± 0·58, range = 5–7 µm) being wider than those of group B (![]() ± SD = 4·51 ± 0·53, range = 3–5 µm). Spores of clade A (length/breadth ratio of 1·85) can be termed as elongate while those of clade B (length/breadth ratio of 2·36) are cylindrical. Chemical analysis revealed the presence of lecanoric acid and atranorin in all species of Parmelina studied.
± SD = 4·51 ± 0·53, range = 3–5 µm). Spores of clade A (length/breadth ratio of 1·85) can be termed as elongate while those of clade B (length/breadth ratio of 2·36) are cylindrical. Chemical analysis revealed the presence of lecanoric acid and atranorin in all species of Parmelina studied.
Taxonomy
Parmelina tiliacea was first described as Lichen tiliaceus by Hoffmann in 1784, and was almost certainly collected in southern Germany although no locality was indicated. Based on the results of our phylogenetic analysis, taxonomic changes are made to reflect that the current name P. tiliacea encompasses two separate lineages, separated by a large genetic distance, that merit formal recognition. Specimens of clade A are assigned to P. tiliacea because it includes samples from central Europe (Germany, Austria, Italy and Slovenia), and the application of that name is fixed here by designation of a sequenced epitype. Specimens of clade B (which included samples from Spain) are recognized as the new species Parmelina cryptotiliacea sp. nov. The new species has a large genetic distance, a more restricted distributional area, and small morphological differences compared with P. tiliacea.
Parmelina cryptotiliacea A. Crespo & Núñez-Zapata sp. nov
MycoBank No.: 561685
Similis Parmelina tiliacea sed differte in ascosporis angusta (3–5 µm latis), cellulis in hyphis excipulis cum muris attenuatis, et in sequencis molecularis ITS et mtLSU.
Typus: Spain, Extremadura, Parque Natural de Monfragüe, 39°49′37·9″N 06°03′27·5″W, on Quercus ilex subsp. ballota, alt. 207 m, June 2005, H. T. Lumbsch, A. Green, P. K. Divakar & A. Argüello (MAF-Lich Reference Jørgensen16454— holotypus).
(Fig. 2)

Fig. 2. Parmelina cryptotiliacea. A, habit (MAF-Lich 16454, holotype). B & C, comparison of exciple cells and ascospores; B, P. cryptotiliacea (MAF-Lich 16453); C, P. tiliacea (MAF-Lich 15350); the graph shows the mean (+), the range of the middle 50% of the data and the full range of the width of the ascospores in samples of both species. Mean differences between species are statistically significant (P = <0·0001). See details of samples in Table 1. Scales: A=50 mm; B & C=20 µm (exciples) and 10 µm (ascospores).
Thallus adnate on bark, pale mineral grey to mineral grey; lobes irregularly branched, sublinear-elongate, often imbricate, rounded at the apices, 2–7 mm wide, the margins more or less crenate and undulate, not ciliate; upper surface more or less shiny, without maculae, usually pruinose, irregularly cracked, densely isidiate; medulla white; lower surface black with brown edge, moderately to densely rhizinate, rhizines black, simple, 1–2 mm long. Isidia cylindrical, short 0·5–1·5 mm, rarely branched, usually blackening at the tips.
Apothecia frequent, adnate, to 4 mm diam. Asci 8-spored. Ascospores cylindrical, length 9–13 µm (10·66 ± 0·94 µm), width 3–5 µm (4·51 µm ± 0·53 µm).
Pycnidia not seen.
Chemistry. Upper cortex K+ yellow; medulla K−, C+ red, KC+ red, P−. Containing atranorin and lecanoric acid, but no fatty acids.
Distribution and ecology. At present the species is known only from four localities in Spain: National Park of Monfragüe (Extremadura), El Pardo (Madrid), Puertollano, and San Quintin mine (both Castilla – La Mancha). It grows on tree trunks and rocks in relatively lowland areas (alt. 250–700 m asl) with a low relative humidity. At low elevations the new species is sympatric with P. tiliacea.
Remarks. Parmelina cryptotiliacea is a cryptic morph of P. tiliacea. When apothecia are present, P. cryptotiliacea can be morphologically differentiated from P. tiliacea by its narrower ascospores (width 3–5 µm, compared with 5–7 µm in P. tiliacea) and the thinner walls of the exciple cells. However, for samples lacking apothecia, a comparison of the ITS sequences is the only reliable way to distinguish this cryptic species.
Parmelina tiliacea (Hoffm.) Hale
Phytologia 28: 481 (1974).—Lichen tiliaceus Hoffm., Enum. Lich.: 96 (1784); type: Europe, sine loc., op. cit.: tab. 16 fig. 2 (—lectotype designated by Jørgensen Reference Jørgensen1972); Germany, Bavaria, Oberfranken, Kteis Forchheim, Ehrenburg, alt. 450 m, on Tilia platyphyllos, 27 September 2009, W. & G. Brachel (MAF-Lich 16485—epitypus hic designatus).
For further synonyms see Hale (Reference Hale1976) and Dobson & Hawksworth (Reference Dobson and Hawksworth1976).
Remarks. As no material collected and named by Hoffmann prior to 1784 has been located, Jørgensen (Reference Jørgensen1972) designated the original illustration as lectotype for this name. There is Hoffmann material in the Moscow herbarium (MW) which was studied by Peter W. James in 1975 and which belongs to the current concept of the species, but as it is unlocalized and was probably collected after 1804 this cannot be treated as original material appropriate for lectotypification (Dobson & Hawksworth Reference Dobson and Hawksworth1976). In order to fix the application of Hoffmann's epithet in the sense it is used in the present paper, we therefore designate a sequenced collection from southern Germany as an epitype for the name.
Discussion
The presence of cryptic species in widely or disjunctly distributed species has often been found in Parmeliaceae and seems to be a rather common phenomenon, as noted above. The cryptic species found here within P. tiliacea s. lat., was not detected in previous investigations due to the restricted sampling used in earlier phylogenetic studies (Blanco et al. Reference Blanco, Crespo, Divakar, Esslinger, Hawksworth and Lumbsch2004, Reference Blanco, Crespo, Ree and Lumbsch2006; Thell et al. Reference Thell, Feuerer, Kärnefelt, Myllys and Stenroos2004; Argüello et al. Reference Argüello, Del Prado, Cubas and Crespo2007).
The present investigation, based on more extensive sampling throughout the geographical range of the species, has detected haplotype variability between and within populations of P. tiliacea s. lat. The topology of the tree, based on two independent loci nuITS and mtLSU, and the SH and ELW hypothesis tests, indicates that specimens of P. tiliacea do not form a monophyletic group but fall in two independent strongly supported clades (clades A and B; Fig.1), named here as P. tiliacea s. str. and P. cryptotiliacea, respectively. The genetic distances between the ITS haplotypes also supports the separation of the two independent clades at the specific level: assignment of all the samples to a single species gave an extremely high intraspecific mean (0·030 ± 0·006 s/s), with a maximum distance (0·093 s/s) that is four to five times larger than the values found in other Parmelina species (e.g. P. quercina mean = 0·007 ± 0·004 s/s; maximum distance = 0·013 s/s; Del Prado et al. Reference Del-Prado, Cubas, Lumbsch, Divakar, Blanco, Amo De Paz, Molina and Crespo2010). When clades A and B are considered as separate species (P. tiliacea and P. cryptotiliacea), their intraspecific distances fall within the species range found in other Parmeliaceae (≤ 0·017 s/s; Del Prado et al. Reference Del-Prado, Cubas, Lumbsch, Divakar, Blanco, Amo De Paz, Molina and Crespo2010).
It should be noted that the genetic distances between ITS haplotypes of clade A, fall within the general intraspecific range even if the haplotypes of P. pastillifera are included. These data suggest that P. pastillifera is genetically very close to P. tiliacea, despite the morphological distinctive features (button-like isidia versus the cylindrical isidia of P. tiliacea; Dobson & Hawksworth Reference Dobson and Hawksworth1976) and the different geographical ranges (P. pastillifera is much more common in humid western European areas than P. tiliacea). A comprehensive population study is being carried out to establish the phylogenetic relationships between P. tiliacea and P. pastillifera, and no formal taxonomic changes are made pending the outcome of that on-going investigation.
Despite the genetic and small anatomical differences, P. tiliacea and P. cryptotiliacea do not have clear ecological differences. Parmelina cryptotiliacea was collected near the Central System and in the south central area of the Iberian Peninsula between 250 and 700 m above sea level, where climatic conditions are characterized by low summer rainfall. However, P. tiliacea is also present in the same localities where P. cryptotiliacea was collected. Parmelina tiliacea has a more extensive distribution and altitudinal range (see Table 1), and therefore grows under a wider range of climatic conditions. The known distribution of P. cryptotilacea shows that it grows sympatrically with P. tiliacea at low elevations and shares the same habitats.
Specimens of P. tiliacea and P. cryptotiliacea cannot be easily differentiated by morphology alone in the absence of apothecia. Types of perforations of the thallus surface that allow gas exchange, have been regarded as a key character for interspecific characters in parmelioids (e.g. Parmelina in Argüello et al. Reference Argüello, Del Prado, Cubas and Crespo2007, Melanelia in Blanco et al. Reference Blanco, Crespo, Divakar, Esslinger, Hawksworth and Lumbsch2004, parmotremoid groups in Blanco et al. Reference Blanco, Crespo, Divakar, Elix and Lumbsch2005). However, the presence of pruina on the upper surface of the specimens examined prevented us from critically analyzing this feature. Ascospores of parmelioid lichens have not been routinely studied, mainly due to the frequent absence of ascomata, but Argüello et al. (Reference Argüello, Del Prado, Cubas and Crespo2007) showed a correlation between ascospore sizes and phylogenetic hypothesis, especially in the Parmelina group. Although the difference in width of ascospores has statistical support in the present study, it is not strong and could be misleading if the degree of maturity of the apothecia and spores is not taken into account. Internal structures of ascoma have also not been considered in any depth in parmelioid lichens (Hale Reference Hale1976); the other difference between P. tiliacea and P. cryptotiliacea (the thickness of the cell wall of the exciple) is also subtle and, due to the frequent absence of apothecia in P. tiliacea, this structure has been scarcely studied in the past. Chemistry does not provide any diagnostic characters because all species in the genus Parmelina share the same compounds (Hale Reference Hale1976; Diaz-Guerra & Manrique Reference Diaz-Guerra and Manrique1984).
Conclusion
As found in other cosmopolitan species of lichenized fungi, molecular phylogeny based on nuITS and mtLSU rDNA regions has revealed that the traditional concept of Parmelina tiliacea includes two separate lineages: one corresponding to P. tiliacea s. str., a widespread lichen from Europe, and another cryptic, genetically separated lineage that is formally described here as a new species, P. cryptotiliacea. Parmelina cryptotiliacea can be considered a cryptic species because it can be morphologically separated from P. tiliacea only by small anatomical differences in the apothecium and ascospores, and a high proportion of populations lack apothecia. In this and similar cases, molecular data are the most reliable way to distinguish such morphologically close species. Our data indicate that P. tiliacea s. str. has a wide distribution, growing in Europe, Middle-East Asia and the Indian subcontinent, while P. cryptotiliacea has only been found in Spain, where it is sympatric with P. tiliacea at low elevations. The restricted area of P. cryptotiliacea suggests that it could be a threatened endemic species, and its conservation status should be evaluated independently of P. tiliacea even if molecular tools are necessary for its identification.
We would like to express our sincere thanks to all the collectors and herbaria listed in Table 1 for sending us fresh material for examination, and Phil Mason for his comments and suggestions. We thank two anonymous referees and the editor for critical comments on the manuscript. Sequencing was carried out in the Centro de Genómica (Parque Científico de Madrid). This work was supported by the Spanish Ministry of Science and Innovation (CGL2010 – 21646/BOS), Ramón y Cajal grant (RYC02007-01576) to PKD, FPI grant to JN-Z, and undertaken while DLH was also in receipt of a Spanish Ministry of Science and Innovation grant (CGL 2008-01600).






