INTRODUCTION
The astonishing extent of Amazonian diversity has fascinated biologists since the 19th century (Bates Reference BATES1874, Wallace Reference WALLACE1854). However, the actual number of species occurring in Amazonia (Fouquet et al. Reference FOUQUET, GILLES, VENCES, MARTY, BLANC and GEMMELL2007, Funk et al. Reference FUNK, CAMINER and RON2012), and the processes responsible for their origins (Haffer Reference HAFFER1997, Hoorn et al. Reference HOORN, WESSELINGH, STEEGE, BERMUDEZ, MORA, SEVINK, SANMARTÍN, SANCHEZ-MESEGUER, ANDERSON and FIGUEIREDO2010) remain intensely debated. The first model proposed to explain animal diversity in Amazonia relied on the dissecting power of large Amazonian rivers acting as biogeographic barriers for many taxa (Wallace Reference WALLACE1854), a hypothesis actually formulated even earlier by de Castelnau (1851).
Since then, many hypotheses have been put forward to explain these high levels of diversity in Amazonia (reviewed in Leite & Rogers Reference LEITE and ROGERS2013, Noonan & Wray Reference NOONAN and WRAY2006). Two of these hypotheses invoke the barrier effect of the rivers to explain patterns of diversity, the riverine-barrier hypothesis (Bates Reference BATES1874, Capparella Reference CAPPARELLA1988, Mayr Reference MAYR1942, Wallace Reference WALLACE1854) and the river-refuge hypothesis (Ayres & Clutton-Brock Reference AYRES and CLUTTON-BROCK1992). The first proposes that riverine barriers separated once continuous populations leading to differentiation and, eventually, speciation. The second argues that Pleistocene forest refugia and rivers interacted with forest fragmentation driven by the cold, dry periods of the Quaternary to reinforce this isolation during and after forest expansion. Empirical support for these hypotheses comes from the observation that the boundaries of closely related species or subspecies often coincide with major Amazonian rivers (e.g. Primates (Ayres & Clutton-Brock Reference AYRES and CLUTTON-BROCK1992); birds (Haffer Reference HAFFER1997, Naka et al. Reference NAKA, BECHTOLDT, HENRIQUES and BRUMFIELD2012); lizards (Avila-Pires Reference AVILA-PIRES1995, Souza et al. Reference SOUZA, RODRIGUES and COHN-HAFT2013)).
However, few studies have used molecular data to explicitly test the barrier effect of rivers in Amazonia and they provided contrasting evidence for the effectiveness of rivers as barriers (e.g. frogs: Fouquet et al. Reference FOUQUET, LEDOUX, DUBUT, NOONAN and SCOTTI2012a, Funk et al. Reference FUNK, CALDWELL, PEDEN, PADIAL, DE LA RIVA and CANNATELLA2007, Gascon et al. Reference GASCON, LOUGHEED and BOGART1998, Kaefer et al. Reference KAEFER, TSUJI-NISHIKIDO, MOTA, FARIAS and LIMA2013, Lougheed et al. Reference LOUGHEED, GASCON, JONES, BOGART and BOAG1999; birds: Burney & Brumfield Reference BURNEY and BRUMFIELD2009, Naka et al. Reference NAKA, BECHTOLDT, HENRIQUES and BRUMFIELD2012, Ribas et al. Reference RIBAS, ALEIXO, NOGUEIRA, MIYAKI and CRACRAFT2011; mammals: Patton et al. Reference PATTON, SILVA and MALCOLM1994, Peres et al. Reference PERES, PATTON and SILVA1996). Though conclusions about the role of rivers on diversification vary among these studies, it is clear that the permeability of rivers to gene flow is influenced by (1) river size and course variation over time (Bates et al. Reference BATES, HAFFER and GRISMER2004) and (2) species' ability to disperse across them (Burney & Brumfield Reference BURNEY and BRUMFIELD2009).
Major rivers undoubtedly impede dispersal for many terrestrial organisms, but these rivers may not be sufficiently impenetrable or long-lived to generate lasting or significant genetic structure and ultimately lead to speciation (Slatkin Reference SLATKIN1987). One of the reasons is meander loop cut-off (Hayes & Sewlal Reference HAYES and SEWLAL2004, Jackson & Austin Reference JACKSON and AUSTIN2013, Peres et al. Reference PERES, PATTON and SILVA1996), a common phenomenon of the large rivers of Amazonia, which are known to regularly change course. However, this is thought to be less common for the more channelled and stable clear-water rivers of the Brazilian and Guiana Shields (Ayres & Clutton-Brock Reference AYRES and CLUTTON-BROCK1992, Bates et al. Reference BATES, HAFFER and GRISMER2004, Lundberg et al. Reference LUNDBERG, MARSHALL, GUERRERO, HORTON, MALABARBA, WESSELINGH, Malabarba, Reis, Vari, Lucena and Lucena1998), making these regions ideal for the study of the role of rivers in speciation.
The ability to disperse across rivers is also expected to vary substantially among species, and can depend on life-history traits such as body size, habitat preference and reproductive mode (Fouquet et al. Reference FOUQUET, LEDOUX, DUBUT, NOONAN and SCOTTI2012a, Gascon et al. Reference GASCON, LOUGHEED and BOGART1998, Lampert et al. Reference LAMPERT, RAND, MUELLER and RYAN2003, Newman & Squire Reference LEITE and ROGERS2001, Richardson Reference RICHARDSON2012). In amphibians, species with large body size and free-living tadpoles deposited in lotic water are expected to disperse more readily than small-bodied, direct-developing species (Van Bocxlaer et al. Reference VAN BOCXLAER, LOADER, ROELANTS, BIJU, MENEGON and BOSSUYT2010, Wollenberg et al. Reference WOLLENBERG, VIEITES, GLAW and VENCES2011). Amphibians are thus particularly valuable models to investigate processes shaping genetic structure (Zeisset & Beebee Reference ZEISSET and BEEBEE2008), particularly in Amazonia where they display a high diversity of the aforementioned traits.
We hypothesize that genetic structure of anuran species across major rivers in the Guiana Shield is life-history dependent. Evaluating (1) the genetic structure and (2) the genetic distance among samples from opposite margins of the Oyapock River (a large, well-channelled river on Precambrian rock draining into the Atlantic Ocean), we tested whether variation in these two metrics are correlated with three traits that may influence dispersal: body size, habitat and development mode.
METHODS
Sampling
Samples (2–5 individuals from each of 28 species collected per locality, Appendix 1; taxonomy follows http://research.amnh.org/herpetology/amphibia/index.html) were gathered from opposite banks of the lower course of the Oyapock River, in the areas of St Georges (French Guiana) and Oiapoque (Amapá, Brazil) (Figure 1). In this area the Oyapock River width varies between 200 and 500 m. Its fast-flowing and well-channelled course is almost entirely without adjacent igapo (temporarily flooded areas) upriver of the coastal estuary, and as such is expected to have had a rather stable course over time (Bates et al. Reference BATES, HAFFER and GRISMER2004). Samples were also collected from localities within the interfluvium of the Oyapock and the Approuague rivers (Savane Virgine) and from Lourenço and Serra do Navio in Amapá (Figure 1). No other major obstacles such as patches of open habitat or mountains occur between the localities.

Figure 1. Map of the study area encompassing French Guiana and Amapá State (Brazil) with localities where frogs were sampled indicated with coloured circles and simplified drainage of the Oyapock River (yellow). Examples of two hypothetical topologies are illustrated on the top right corner indicative of a barrier effect of the river (topologies A and B) and three examples of topologies indicative of trans-riverine affinities (topology 0) are indicated below. Two dots in the same terminal indicate that the prediction will be the same regardless of the position of the samples.
Molecular analysis
Genomic DNA was extracted from tissue samples preserved in 95% ethanol using the Wizard Genomic DNA Extraction kit (Promega; Madison, WI, USA) (Appendix 1). A portion of the mitochondrial 16S rDNA locus was amplified by standard PCR techniques using previously described primers and PCR conditions (Hillis et al. Reference HILLIS, MORITZ, MABLE and OLMSTEAD1996, Salducci et al. Reference SALDUCCI, MARTY, FOUQUET and GILLES2005). Sequencing was performed using ABI Big Dye V3.1 (ABI, Foster City, CA, USA) and run on automated sequencers at Macrogen (Korea) and Beckman Coulter (UK). Sequences were edited and aligned with CodonCode Aligner v.3.5.2 (http://www.codoncode.com/aligner/download.htm). Novel sequences were deposited in GenBank (Appendix 1).
The 102 newly generated sequences were combined with 14 available sequences from GenBank and aligned with MAFFT v6 (Katoh & Standley Reference KATOH and STANDLEY2013) under default parameters and the E-INS-i strategy, which is designed for sequences with one conserved domain and long gaps. Our final alignment consisted of 520 base pairs (bp) that was incomplete only for the last c. 140 bp of 15 terminals (those sequenced using 16SF and 16SR).
We used the software jModeltest version 2.1.1 (Darriba et al. Reference DARRIBA, TABOADA, DOALLO and POSADA2012, Guindon & Gascuel Reference GUINDON and GASCUEL2003) to select the substitution model that best fit the data under the Bayesian Information Criterion. The resulting model (GTR + I + G) was employed in a Bayesian Analysis (BA) with MrBayes 3.2 (Ronquist & Huelsenbeck Reference RONQUIST and HUELSENBECK2003). The BA consisted of 20 × 106 generations and 10 Markov chains (one cold) sampled every 1000 generations. Adequate burn-in was determined by examining likelihood scores of the heated chains for convergence and stationarity in Tracer 1.5 (http://beast.bio.ed.ac.uk/Tracer) and effective sample size of values were >200. We also calculated pairwise (p) genetic distances between samples from the opposite margins of the Oyapock River (St Georges vs. Oiapoque only) with MEGA 5 (Tamura et al. Reference TAMURA, PETERSON, PETERSON, STECHER, NEI and KUMAR2011).
We assume that the absence of direct affinity and genetic distance >0.2% (minimal distance observed for species displaying reciprocal monophyly across the river) between the populations from opposite margins can be interpreted as a barrier effect of the Oyapock river. The most straightforward case will be species displaying reciprocally monophyletic lineages from each side (Figure 1; e.g. topology A). Alternative scenarios include species displaying monophyly of only one interfluvium which may result from gene flow in headwater areas (Figure 1; e.g. topology B). Results were coded as a binary character reflecting the presence (1) or absence (0) of genetic structure suggestive of a barrier effect. These two metrics (coded topologies and p-distances) were used as proxies of the extent to which the Oyapock River acts as a barrier.
For each species, we also coded three life-history traits: body size (max SVL: small = 10–30 mm; medium = 30–60 mm; large = >60 mm), habitat of adults (forest litter, arboreal and open habitats) and larval development (free exotrophic tadpoles in lentic or lotic waters vs. direct developing, endotrophic and phytotelmic tadpoles) (Figure 2). These data were collected from Lescure & Marty (Reference LESCURE and MARTY2001) and personal observations. For each of these traits we tested for an association with the presence/absence of genetic structure and genetic distance across the Oyapock River.
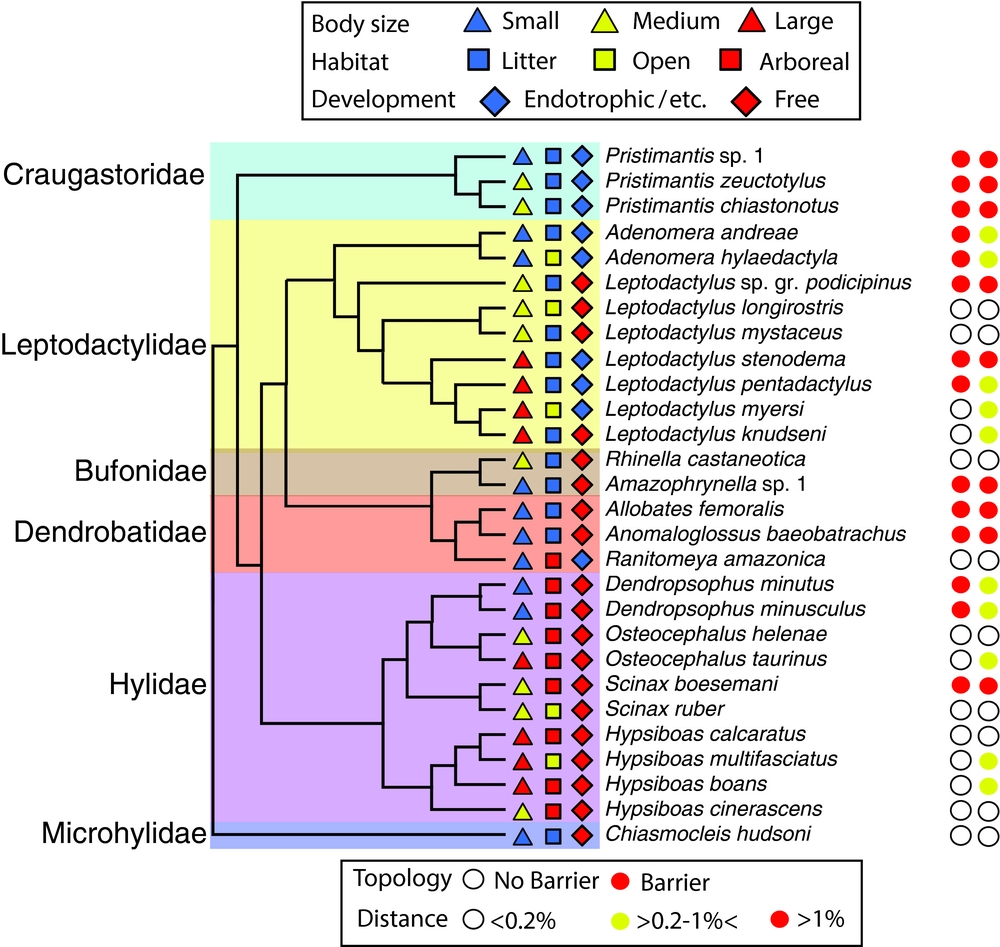
Figure 2. Phylogenetic relationship among the focal frog species sampled from opposite banks of the Oyapock River (from Pyron & Wiens (Reference PYRON and WIENS2011) and Fouquet et al. (Reference FOUQUET, BLOTTO, MARONNA, VERDADE, JUNCA, DE SA and RODRIGUES2013)). For each species, the coded life-history traits are indicated before the species name with coloured symbols (triangle, square, diamond) and the categorical results from the molecular analyses analyses are indicated after the species names with coloured circles.
Notably our study does not address whether this river currently acts as a barrier to dispersal, as this would require greater population-level sampling and multiple fast-evolving molecular markers. Instead, we used a single slow-evolving mtDNA locus, which will allow us to detect (robustly) historical events (e.g. fragmentation of the range). Observations of structure from our sampling can be the result of either the river acting as a primary barrier or secondary contact zone between demes originally isolated by other barriers. However, the maintenance of such structure is still informative with respect to the effects of rivers on gene flow.
Statistical analysis
We first tested whether p-distances display a significant phylogenetic signal by calculating Blomberg's K (Blomberg et al. Reference BLOMBERG, GARLAND and IVES2003). This statistic is a representation of the phylogenetic signal in the tip data relative to the expectation for a trait evolving by Brownian motion along the specified phylogeny. Values near 0 indicate a lack of a phylogenetic signal, and values approaching 1 indicate Brownian character evolution (i.e. a tendency for closely related species to display similar trans-riverine p-distance). Significance of K was assessed by 1000 replicates of randomly shuffling trans-riverine p-distance values among species.
Using the binary character states resulting from the phylogenetic analysis (presence/absence of genetic structure across Oyapock River), we tested whether the proportion of species for which the river represents a barrier is correlated with (1) body size (2) habitat and (3) larval development using Chi-square tests.
We also tested whether p-distances between samples from opposite margins are dependent on species' (1) body size (non-parametric Kruskal–Wallis test) (2) habitat (non-parametric Kruskal–Wallis test) and (3) mode of development (non-parametric Wilcoxon test). Non-parametric tests were used because p-distance were not normally distributed and samples sizes were low. All statistical analysis were performed with the R software v 2.15.3 (http://cran.r-project.org/) using the package Picante (Kembel et al. Reference KEMBEL, COWAN, HELMUS, CORNWELL, MORLON, ACKERLY, BLOMBERG and WEBB2010).
RESULTS
Phylogenetic analysis and genetic distances
Relationships among conspecific samples ranged from deeply structured to uniform (Appendix 2). Thirteen species displayed genetic structure consistent with isolation by the Oyapock River (e.g. either reciprocal monophyly of the A/O interfluvium and Oiapoque/Amapà populations and p distance >1%; or monophyly of only the A/O interfluvium with genetic distances >0.2%) (Figure 2). The topologies recovered within the other 15 species did not possess genetic structure reflective of a barrier effect of the Oyapock River. Genetic distance between conspecific samples from St Georges and Oiapoque ranged from 0 in nine species to 2.7% in Pristimantis chiastonotus and Anomaloglossus baeobatrachus (Table 1).
Table 1. Table summarizing the results from the molecular analyses and the coding of the life history traits for each focal frog species sampled on each bank of the Oyapock River: species identification (sample size = n), the three trait modalities (Development, Habitat, Body size), p-distances between samples from opposite margins, topology recovered from the phylogenetic analysis (A and B are indicative of a genetic structure matching the course of the river while 0 are not).

Analyses of life-history traits
Genetic distances did not display a significant phylogenetic signal (Blomberg's K, P = 0.16, N = 28 species). Smaller-bodied species were not found to display higher genetic distance across the Oyapock River than medium or large body-sized species (Kruskal–Wallis, K = 1.83, P = 0.40, df = 2). Nor did they display more cases of genetic structure matching the course of the river (Chi-square test, χ2 = 3.5, P = 0.18, df = 2). However, forest-litter species did display greater genetic differentiation across the Oyapock River than arboreal and open-habitat species (Figure 3, Kruskal–Wallis, K = 5.8, P = 0.05, df = 2) and were more prone to display genetic structure across the river (Figure 3, Chi-square test, χ2 = 7.0, P = 0.03, df = 2). Similarly, species with free-living larvae displayed lower genetic differentiation across the river than species with terrestrial larvae (Figure 3, Wilcoxon test, W = 45.5, P = 0.05, df = 1), but the proportion of species displaying structure across the river did not differ significantly among larval types (Figure 3, Chi-square test, χ2 = 3.5, P = 0.06, df = 1).

Figure 3. Box plots comparing genetic distances to developmental (classical in red and direct/endo./phyt. in blue) (a) and habitat traits (arboreal in red, forest litter in blue and open habitat in yellow) (b); and genetic structure classification to developmental (c) and habitat traits (d) of focal frog species sampled from opposite banks of the Oyapock River. The proportion of species with barrier effect is indicated with solid colours and the proportion of species with no barrier effect in white (c, d).
DISCUSSION
The lower Oyapock River is a barrier for many taxa
The lower course of the Oyapock River corresponds to phylogeographic breaks in 13 of 28 species examined, a pattern similar to the findings of previous studies of amphibians in the region (Fouquet et al. Reference FOUQUET, LEDOUX, DUBUT, NOONAN and SCOTTI2012a, Reference FOUQUET, NOONAN, RODRIGUES, PECH, GILLES and GEMMELL2012b) and birds along the Branco and Negro rivers (Naka et al. Reference NAKA, BECHTOLDT, HENRIQUES and BRUMFIELD2012). The fact that such structure was not recovered in frogs (Scinax, Scarthyla, Allobates, Engystomops) studied in central Amazonia (Gascon et al. Reference GASCON, LOUGHEED and BOGART1998, Lougheed et al. Reference LOUGHEED, GASCON, JONES, BOGART and BOAG1999) is likely due to the highly dynamic nature of the courses of Amazonian floodplain rivers, allowing populations to shift from one bank to the other via recurrent cutting-off of meanders (Jackson & Austin Reference JACKSON and AUSTIN2013). Guiana Shield rivers on the other hand, while not as large, are channelled on Precambrian rocks and are more stable over time (Bates et al. Reference BATES, HAFFER and GRISMER2004).
Fouquet et al. (Reference FOUQUET, NOONAN, RODRIGUES, PECH, GILLES and GEMMELL2012b) hypothesized that the congruent phylogeographic structure observed in frogs of the region originated during periods of climatic instability. Cold/dry periods of the Pleistocene are thought to have aridified the central Guiana Shield restricting wet-forest patches to the coastal plain, where the lower courses of the rivers would have fragmented species' distributions. During these times, the narrower headwaters would not have been traversable as they lay in inhospitably arid, savanna habitat. This is the basis of a hypothesis analogous to the river refuge hypothesis (Ayres & Clutton-Brock Reference AYRES and CLUTTON-BROCK1992), but shaped and scaled to the unique hydrology of the eastern Guiana Shield (not all of Amazonia). Notably, these Pleistocene events are likely to have produced the type of intraspecific structure (not speciation, Rull Reference RULL2011) we see here. Our results suggest this hypothesis has significant explanatory power for forest-litter-dwelling species with terrestrial larval development. This hypothesis is further supported by the suggestion of upstream dispersal in several species (e.g. Amazophrynella sp. and Leptodactylus pentadactylus) where the Oiapoque population is sister to all others (Figure 1).
While the slowly evolving nature of the locus employed here and the limited within-population sampling precludes estimates of gene flow across the river per se, our data do provide insight into the historical isolation of anuran populations across the Oyapock River. In other words, the Oyapock River may represent a barrier for species in which this structure is not detectable with the evolving mtDNA used here. It is also possible that gene flow may go undocumented in some species for which we report significant structure across the river due to our sampling limitations. This latter, type-two error, seems unlikely given the results of previous phylogeographic studies (Fouquet et al. Reference FOUQUET, LEDOUX, DUBUT, NOONAN and SCOTTI2012a, Reference FOUQUET, NOONAN, RODRIGUES, PECH, GILLES and GEMMELL2012b) which report a very low incidence of polyphyletic populations.
A limitation of our dataset is the limited sampling from each locality and we thus cannot rule out the occurrence of polyphyletic populations that we recovered as monophyletic. Nevertheless, this is the case only for 12 species out of 28 because more extensive datasets are already available in that area for 16 species included herein (Fouquet Reference FOUQUET2008, Reference FOUQUET, LEDOUX, DUBUT, NOONAN and SCOTTI2012a, Reference FOUQUET, NOONAN, RODRIGUES, PECH, GILLES and GEMMELL2012b; Funk et al. Reference FUNK, CAMINER and RON2012, Gehara et al. Reference GEHARA, CRAWFORD, ORRICO, RODRIGUEZ, LÖTTERS, FOUQUET, BALDO, BARRIENTOS, BRUSQUETTI, CASTROVIEJO-FISHER, DE LA RIVA, ERNST, FAIVOVICH, GAGLIARDI URRUTIA, GLAW, GUAYASAMIN, HÖLTING, JANSEN, KOK, KWET, LINGNAU, LYRA, MORAVEC, PADIAL, POMBAL, ROJAS-RUNJAIC, SCHULZE, SEÑARIS, SOLÉ, RODRIGUEZ, TWOMEY, HADDAD, VENCES and KÖHLER2014, Jungfer et al. Reference JUNGFER, FAIVOVICH, PADIAL, CASTROVIEJO-FISHER, LYRA, BERNECK, IGLESIAS, KOK, MACCULLOCH, RODRIGUES, VERDADE, TORRES GASTELLO, CHAPARRO, VALDUJO, REICHLE, MORAVEC, GVOŽDÍK, GAGLIARDI-URRUTIA, ERNST, DE LA RIVA, MEANS, LIMA, SEÑARIS, WHEELER and HADDAD2013, Peloso et al. Reference PELOSO, STURARO, FORLANI, MOTTA and WHEELER2014) which confirmed the existence or absence of phylogeographic breaks matching the lower course of the Oyapock river. Among these 12 species, only four displayed obvious trans-riverine genetic structure; i.e. occurrence of undetected polyphyly that would indicate recent dispersal across the river cannot be ruled out for only four species. However, these four species are represented by pairs of populations sampled on each side of the Oyapock river. Additionally, even though undetected polyphyletic populations may occur in our sampling, the very existence of such structure remains meaningful even if secondarily admixed. We therefore choose to favour the taxonomic sampling (i.e. more species but fewer individuals) to assess whether the genetic structure of a greater number of frog species corresponds to this river.
Riverine barriers are life-history dependent
Our results suggest that the extent to which the Oyapock River acts as a barrier depends on species-specific life-history traits. These ecological differences are reflected in our recovered patterns of genetic structure and, consequently, the amount of genetic variation between demes. Fouquet et al. (Reference FOUQUET, NOONAN, RODRIGUES, PECH, GILLES and GEMMELL2012b) hypothesized that idiosyncrasies among the phylogeographic patterns of 12 forest-litter frog species of the Eastern Guiana Shield were partly due to variation in specific traits such as body size, implying differences in generation time and population size. Wollenberg et al. (Reference WOLLENBERG, VIEITES, GLAW and VENCES2011) also established a link between small body size, low dispersal and high speciation rate in Malagasy frogs. Body size has also been shown to be positively correlated with dispersal ability and geographic distribution (Van Bocxlaer et al. Reference VAN BOCXLAER, LOADER, ROELANTS, BIJU, MENEGON and BOSSUYT2010), again revealing a role of life-history and morphology in population structure. Our findings reveal no significant link between body size and isolation across the Oyapock River. This may be due to the fact that our sampling is phylogenetically highly heterogeneous (seven families) rather than focusing on one specific clade, as in former studies. This may also highlight the distinction between the dispersal ability over distance through homogeneous landscape and across a barrier such as a river. The latter may be more dependent on life-history traits other than the body size. Our study instead highlights the importance of habitat and reproductive mode.
Studies of frog species sampled along the Juruá River found greater genetic differentiation among species associated with terra firme than flooded areas, though this structure was not coincident with the course of the river (Gascon et al. Reference GASCON, LOUGHEED and BOGART1998, Lougheed et al. Reference LOUGHEED, GASCON, JONES, BOGART and BOAG1999). Fouquet et al. (Reference FOUQUET, GILLES, VENCES, MARTY, BLANC and GEMMELL2007) also demonstrated that open-habitat and aquatic species display less genetic structure/divergence than forest-dwelling species over vast distances in Amazonia. The extent to which population structure of birds is explained by rivers has also been demonstrated to be highly influenced by ecology, with canopy species exhibiting lower genetic divergence across Amazonian rivers than understorey birds (Burney & Brumfield Reference BURNEY and BRUMFIELD2009). Similarly, arboreal species in our study display lower genetic distances across the river and fewer of these species possess genetic structure concordant with the Oyapock than forest-litter species. In frogs at least, arboreal species may be more prone to cross rivers via tree fall. Higher dispersal ability of arboreal species may also be due to a greater tolerance to temperature variation and desiccation inherent to this microhabitat than forest-litter species for which the hygrometry and temperature of the forest floor is more stable. Therefore, arboreal species may be able to disperse more easily through the dry, or at least drier, climate areas of the central Guiana Shield (Gond et al. Reference GOND, FREYCON, MOLINO, BRUNAUX, INGRASSIA, JOUBERT, PEKEL, PRÉVOST, THIERRON and TROMBE2011). The only arboreal species in our study to display evidence of isolation across the river utilize stagnant water at the edge of forest (Dendropsophus minusculus, Scinax boesemani).
Open-habitat species also exhibit little if any genetic structure coincident with the river course. In fact, none of the open-adapted species displays structure across the river except A. hylaedactyla, which has endotrophic larvae. This may be the result of recent historical expansion of open landscapes during the Quaternary allowing these species to disperse, possibly in the headwater regions (Fouquet et al. Reference FOUQUET, NOONAN, BLANC and ORRICO2011). Open landscape connections have been hypothesized in the interior of the Guiana Shield, but expansion may also have occurred along the coast during periods of lowered sea levels. The coastal region of the Guiana Shield is currently a patchwork of savannas, mangroves, swamps and forests facing a very shallow 100-km continental shelf. During cold/dry periods of the Quaternary, this area was likely prone to harbour a vast extent of open habitat which may have favoured dispersal, as exemplified by the occurrence of relictual savannas peppered throughout the coast of the region and the distribution of open habitat species. As they are more tolerant of climatic conditions of open areas, open habitat specialists may be also more prone to dispersal during the dry season across a reduced-width river (e.g. across rocky rapids).
Reproductive mode also seems to be an important factor determining dispersal ability across the river. This pattern may be directly related to larval dispersal, at least for the species with large tadpoles that breed directly in the rivers (Hypsiboas boans, Osteocephalus helenae) or in flooded areas (Leptodactylus mystaceus). On the other hand, all species with direct developing (Pristimantis) or endotrophic larvae (Adenomera) display strong differentiation across the river. Though there have been few studies that explicitly examine barrier effects of multiple taxa with varying larval development strategies, such characteristics have been shown to affect the distribution and connectivity of populations (Van Bocxlaer et al. Reference VAN BOCXLAER, LOADER, ROELANTS, BIJU, MENEGON and BOSSUYT2010).
This study represents the first attempt to test the riverine barrier hypothesis with a large multitaxon dataset. In the Guiana Shield, the historical stability of the river courses has allowed us to explore the influence of life-history traits on the dispersal ability of anurans across a river barrier. Our findings clearly demonstrate that these traits predict the extent to which the Oyapock River, and likely other stable, major rivers in the tropics, acts as a barrier to dispersal in anurans.
ACKNOWLEDGEMENTS
We thank Damien Davy, Johann Tascon, Antonia Cristinoi for their assistance. This work has benefited from a grant of Observatoire Homme Milieux Oyapock (CNRS) and an ‘Investissements d'Avenir’ grant managed by Agence Nationale de la Recherche (CEBA, ref. ANR-10-LABX-25-01). We also thank Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Secretaria de Estado da Ciencia e Tecnologia do Amapá (SETEC), and the following institutions for providing collecting permits: Instituto brasileiro do meio ambiente e dos recursos naturais (IBAMA; 2001.007142/2006-17), Conselho Nacional de Desenvolvimento Cientifico e Tecnologico (CNPq; EXC 035/06-CR) and Instituto Chico Mendes de Conservação da Biodiversidade (ICMBIO; 30309-1, 19754-5 and 10126-1). We finally thank the Institut Pasteur de Guyane and particularly V. Lacoste for letting us use the laboratory.
Appendix 1. Frog samples from French Guiana (left bank of the Oyapock River) and Amapá State (Brazil) (right bank of the Oyapock River) included in the molecular analyses: species identification, field numbers, locality, geographic coordinates and GB accession numbers.


Appendix 2. Subtrees obtained from a Bayesian analysis of a single alignment of a 16S fragment for each frog species sampled at the border between French Guiana and Amapà State, Brazil, i.e. on each side of the Oyapock River. Posterior probabilities (×100) of each branch are indicated, with asterisks when posterior probability = 1. Coloured symbols correspond to the localities (cf. Figure 1). Species names in red (left) are the ones displaying trans-riverine population structure (topologies A, B). Species names in blue (right) are the ones with no trans-riverine population structure.








