INTRODUCTION
In the 1970s, Dr Demir from the University of Istanbul (Demir, Reference Demir1977) found an unusual species of Arcidae in Izmir Bay, Turkey. Those specimens were not similar to any of the Mediterranean native or alien species known at that time and resolved to be a new alien species for the Mediterranean Sea. The same species was then found in Greece (Zenetos, Reference Zenetos1994) and reached Italy a few years later (Morello & Solustri, Reference Morello and Solustri2001; Crocetta et al., Reference Crocetta, Renda and Colamonaco2008). Demir wondered whether the species could be Anadara transversa (Say, 1822) or Arca amygdalum Philippi, 1847. Demir had no clue on the original distribution of the species and this was another identification difficulty. Therefore, the shells were sent to the Muséum National d'Histoire Naturelle in Paris where they were identified as Arca amygdalum Philippi, 1847, a species from China.
Some years later Piani observed that the taxon Arca amygdalum had been used by Link (Reference Link1807) before Philippi and therefore proposed the nomen novum Scapharca demiri for this species. The taxon was later rearranged as Anadara demiri (Piani, Reference Piani1981).
No one questioned this identification in the later years when attention was driven more to the rapid expansion in other countries faced on the Mediterranean than to the correct taxonomy of the species. The Indo-Pacific origin of Anadara demiri was never questioned as well. Rinaldi (Reference Rinaldi2006) is the first author to underline the many similarities between Anadara demiri (Piani, Reference Piani1981) and Anadara transversa (Say, 1822) but his conclusions have been misguided by the assumption he was dealing with an Indo-Pacific species. Therefore, the following observations made the authors reconsider the origin and identification of Anadara demiri:
(1) the area of origin of this species has not been identified to date (Zenetos et al., Reference Zenetos, Gofas, Russo and Templado2004), despite Anadara are usually shallow water and common to abundant species, at least locally;
(2) there is no readily available record of Anadara species similar to Anadara demiri in the Indo-Pacific Ocean;
(3) there is a remarkable similarity of the specimens of Anadara demiri from the Mediterranean Sea to specimens of Anadara transversa (Say, 1822) from Florida, USA.
The present paper discusses the issues listed above. Throughout the discussion the taxon Anadara demiri will be used meaning the populations of the alien species of Anadara first discovered by Demir and later found in the Adriatic Sea.
MATERIALS AND METHODS
Forty-three lots of specimens identified as Anadara demiri and as Anadara transversa have been examined for morphological and molecular analysis. A complete list is given in Appendix I.
Morphological analysis was performed identifying and describing diagnostic characters and measuring antero-posterior length, height of specimens from umbo to ventral margin and the maximum width of valves. Specimens have been measured by a calliper to the lowest 0.1 mm.
Different ratios between measured variables were calculated and statistically treated with a Student's t-test to assess mean differences. Linear regression of ratios using natural log-transformed parameters was also performed using the equation ln Y = a + b ln X, the coefficient of determination (r 2) calculated, and Student's t-test was used to assess differences in the parameters of the regression (a and b).
The molecular analysis was conducted on specimens of A. demiri and A. inaequivalvis (Bruguière, 1789) from the Northern Adriatic Sea and on specimens of A. transversa from Florida.
DNA samples of the three species were extracted from specimens preserved in ethanol 95%. Total genomic DNA was isolated from a small mantle tissue section of each individual using the ChargeSwitch gDNA Tissue Kit (Invitrogen), following the protocol provided by the manufacturer and using a 150 µl volume of elution buffer. All the DNA extractions were verified by electrophoresis of 5 µl of DNA on 0.8% agarose gel stained with SBYR Safe (Invitrogen) in 1X TAE buffer, and visualized under UV light. All the DNA samples were subsequently stored at −20°C until used for analysis.
For each species two DNA regions were amplified and sequenced: a fragment of about 530 base pair (bp) of the nuclear large subunit ribosomal DNA gene (nu-LSU rDNA) and a fragment of about 680 bp of the mitochondrial cytochrome oxidase subunit I gene (mt-COI gene). All the amplifications were performed via polymerase chain reactions (PCRs) using a GeneAmp PCR System 2700 thermal cycler (Applied Biosystems). The partial segment of the nuclear LSU rDNA gene was amplified in a single PCR reaction using the primers 28SA 5′–GACCCGTCTTGAAGCACG–3′ and 28SRD5B 5′–CCACAGCGCCAGTTCTGCTTAC–3′. PCR amplification was carried out using 1 µl of DNA from each genomic extraction as template in a final reaction volume of 25 µl containing 19.4 µl of water, 2.5 µl of 5 PRIME Taq buffer 10x (Eppendorf), 0.5 µl of MgCl2 25 mM (Eppendorf), 0.375 µl of dNTPs 100 mM (Promega), 0.5 µl of each primer 10 µM (Invitrogen) and 0.28 µl of 5 PRIME Taq DNA polymerase (Eppendorf). The PCR protocol consisted of an initial 4 minutes denaturation step at 94°C, 35 cycles of denaturation at 94°C for 30 seconds, annealing at 48°C for 1 minute and extension at 72°C for 1 minute, and a final 7 minutes extension step at 72°C. The partial segment of the mitochondrial COI gene was amplified in a single PCR reaction using the primers LCO1490 5′–GGTCAACAAATCATAAAGATATTGG–3′ and HCO2198 5′–TAAACTTCAGGGTGACCAAAAAA TCA–3′ (Folmer et al., Reference Folmer, Black, Hoeh, Lutz and Vrijenhoek1994). PCR amplification was carried out using 3 µl of DNA from each genomic extraction as template in a final reaction volume of 25 µl containing 17.4 µl of water, 2.5 µl of 5 PRIME Taq buffer 10x (Eppendorf), 0.5 µl of MgCl2 25 mM (Eppendorf), 0.375 µl of dNTPs 100 mM (Promega), 0.5 µl of each primer 10 µM (Invitrogen) and 0.28 µl of 5 PRIME Taq DNA polymerase (Eppendorf). The PCR protocol consisted of an initial 5 minutes denaturation step at 95°C, 35 cycles of denaturation at 95°C for 1 minute, annealing at 50°C for 1 minute and extension at 72°C for 1 minute, and a final 7 minutes extension step at 72°C. For each species and for each gene two separate PCRs were performed, in order to check the reproducibility of the experimental procedure and results. A negative and a positive control were also included in all reactions to verify the absence of external contamination in the samples and the success of PCRs respectively. The resulting double-stranded PCR products of both genes were checked by electrophoresis of 2.5 µl of DNA on 1.5% agarose gel in 1X TAE buffer, using SYBR Safe (Invitrogen) as a DNA gel stain and a known DNA 100 bp Ladder as a marker to estimate amplicons size, and visualized under UV light.
In those reactions producing a single band of the expected size, the remaining 22.5 µl of each amplicon were purified by the Qiaquick PCR purification kit (Qiagen). For the reactions producing multiple bands or that proved difficult to sequence directly, the remaining 22.5 µl were run for 1 hour at 80 mV on a 2% agarose gel. The resulting bands, containing the amplified DNA, were excised from gel under UV light and agarose was digested from bands using the PCR clean-up gel extraction (Macherey-Nagel) and the manufacturer's instructions, modified only by using double amount of buffer NT and double time of gel digestion. All the purified PCR products were checked and quantified by agarose gel electrophoresis of 2.5 µl of DNA (1.5% agarose, SBYR Safe gel stain, 1X TAE buffer) and subsequently stored at 4°C until sequencing reactions were carried out. Each purified PCR product was sequenced on both strands on an ABI Prism automated DNA sequencer using the amplifying primers.
All the sequences obtained were entered in the BLAST program on NCBI site, to verify their identity, and aligned separately for the two genes using MEGA 4 (Tamura et al., Reference Tamura, Dudley, Nei and Kumar2007), set at default parameters. Additional sequence data from six species of Arcidae, two species of Ostreidae, two species of Mytilidae, and the species Unio pictorum (Linnaeus, 1758) (Unionidae) used as outgroup were retrieved from Genbank for both genes and included in the two alignments (Table 3). All the sequences were further analysed by visual inspection and manually refined. Thus all ambiguous bases were removed while all gaps were treated as missing data. A small number of hypervariable regions in COI alignment were not included in the analysis as they differ considerably and were especially difficult to align. The two unaligned external primer regions in both alignments were removed and hence excluded from the analysis. The resulting LSU alignment is 515 positions long while the aligned COI sequences spanned a total of 464 bp.
Three data sets (one containing only the LSU sequences, one containing only the COI sequences and one containing both sequences) were used to conduct the phylogenetic analysis so that in all 515 (LSU), 464 (COI) and 979 (LSU + COI) unambiguously aligned nucleotide bases were considered separately to deduce the phylogenetic relationships. Both LSU and COI dataset and the combined dataset (partitioned by gene) were analysed using the Bayesian inference (BI) (Alfaro et al., Reference Alfaro, Zoller and Lutzoni2003; Holder & Lewis, Reference Holder and Lewis2003; Huelsenbeck & Ronquist, Reference Huelsenbeck and Ronquist2001) which was performed using MrBayes 3.1.2 (Ronquist & Huelsenbeck, Reference Ronquist and Huelsenbeck2003) and the Markov Chain Monte Carlo (MCMC) tree sampling procedure (Yang & Rannala, Reference Yang and Rannala1997; Larget & Simon, Reference Larget and Simon1999). The analyses were conducted assuming the GTR + I + Г evolution model. The trees were constructed using 7,000,000 generations, with parameter values and trees calculated at every 100th step. The estimated log-likelihood scores were plotted against generation time using Microsoft Excel, to assess when the log-likelihood values reached the stationary and to determine the appropriate number of generations to exclude as burn-in. The log-likelihood scores were found to stabilize after 10,000 generations. Therefore, the first 10,000 generation trees were discarded and the remaining trees were used to draw a 50% majority rule consensus using sumt command. Support for trees nodes was determined based on the values of Bayesian posterior probability (BPP). Only BPP values equal or above 95% were considered significant (Erixon et al., Reference Erixon, Svennblad, Britton and Oxelman2003). Phylogenetic trees were drawn using Dendroscope 1.4 (Huson et al., Reference Huson, Richter, Rausch, Dezulian, Franz and Rupp2007).
DISCUSSION
1. Unknown original distribution of Anadara demiri
It is quite unusual for an alien species to have no clues on its area of origin. CIESM in its record of alien species into the Mediterranean (Zenetos et al., Reference Zenetos, Gofas, Russo and Templado2004) reports 137 species certainly recorded, but only Anadara demiri has the remarkable note: ‘original distribution uncertain’. It is stated ‘records in the Indian Ocean’ but no literature reference is provided.
The distribution of other alien species is not clearly described, but this is generally the case of very small species with complex systematics (e.g. Cerithiopsidae). However, at least the zoogeographical provenance is always known and an expected range indicated. It is hard to think of a species the size of Anadara demiri as having never been recorded from any place in the Indo-Pacific province. The mode of introduction is unknown as well, despite the reasonable hypothesis of being introduced by shipping, because no similar species are reported from the Red Sea and the eastern Atlantic Ocean.
2. No evidence of Indo-Pacific species similar to Mediterranean Anadara demiri
The locus typicus of Arca amygdalum Philippi, 1847 is China. But Anadara demiri has never been recorded in Chinese waters, nor in other areas in South-Eastern Asia. Even Demir (Reference Demir1977) highlighted that ‘This species is from the China Seas […], though it is not mentioned in “A Catalogue of Molluscan Shells from Taiwan (Formosa)” by Kuroda (Reference Kuroda1941) and in “Shells of the Western Pacific in colour, vol. 1” by Kira (Reference Kira1972), “vol. 2” by Habe (Reference Habe1971)’. A selected list of regional guides and monographs on the Indo-Pacific province has been considered and we comment about the records of Anadara species similar to Anadara demiri here.
In the Red Sea there are no comparable species recorded (Sharabati & Sharabati, Reference Sharabati and Sharabati1984; Oliver, Reference Oliver1992; Zuschin & Oliver, Reference Zuschin and Oliver2003).
In the Indian Ocean, the only species which share the elongated form of Anadara demiri are A. ehrenburgi (Dunker, 1868) and Scapharca indica (Spengler, 1789). The former is much bigger (up to 70 mm), has the beaks in front of midline while A. demiri has them in the anterior third. The latter has a too much elongated shape (Bosch et al., Reference Bosch, Dance, Moolenbeek and Oliver1995). There are no comparable species recorded in the Seychelles (Jarrett, Reference Jarrett2000) or India (Lutaenko, Reference Lutaenko2006).
South-Eastern Asia is the most interesting area, since the type locality of Arca amygdalum is China. However, there are not any species similar to Anadara demiri in Vietnam (Evseev & Lutaenko, Reference Evseev and Lutaenko1998; Thach, Reference Thach2005), the Philippines (Springsteen & Leobrera, Reference Springsteen and Leobrera1986), China (Zhongyan, Reference Zhongyan2004; Fengshan & Suping, Reference Fengshan and Suping2008) and Korea (Min, Reference Min2004). Only Swennen et al. (Reference Swennen, Moolenbeek, Ruttanadakul, Hobbelink, Dekker and Hajisamae2001) illustrate ‘Anadara’ jousseaumei (Lamy, 1907) from Thailand, a species which has a shape similar to A. demiri but interspaces are much smaller than ribs, while in Anadara demiri they are of the same size. The recent research by the University of Catania in the Gulf of Thailand (Robba et al., Reference Robba, Di Geronimo, Chaimanee, Negri and Sanfilippo2002, Reference Robba, Di Geronimo, Chaimanee, Negri and Sanfilippo2004, Reference Robba, Di Geronimo, Chaimanee, Negri and Sanfilippo2007) lead to the illustration and description of hundreds of species of molluscs, but no Anadara shows any relation to A. demiri. The closest one is Anadarinae sp. 2 (Robba et al., Reference Robba, Di Geronimo, Chaimanee, Negri and Sanfilippo2004) which is clearly different because of the high number of radial ribs: 50 against 30–38 in A. demiri.
Many works have dealt with the Japanese fauna (Habe, Reference Habe1965, Reference Habe1971; Kuroda et al., Reference Kuroda, Habe and Oyama1971; Kira, Reference Kira1972; Okutani, Reference Okutani2000) but none report any species similar to A. demiri.
The Australian bivalve fauna has been studied extensively (Lamprell & Healy, Reference Lamprell and Healy1998). Anadara gubernaculum (Reeve, 1844) and Anadara jurata Iredale, 1939 are the most similar species to Anadara demiri and both share the same size-range. However, the former has ‘flattened bifurcate ribs’, a character never found in A. demiri, the latter has the umbo too anteriorly and has interstices ‘crossed by dense ridges’ while interstices in A. demiri have very fine growth striae, sometimes barely visible. Also New Zealand (Powell, Reference Powell1979) does not host comparable species.
In the Pacific Ocean Cernohorsky (Reference Cernohorsky1972) and Kay (Reference Kay1979) do not report any comparable species. Keen (Reference Keen1971) illustrates Lunarca brevifrons (Sowerby, 1833) which has the same size and overall appearance of Anadara demiri, but differs in having bifurcating ribs.
3. Remarkable similarity of Mediterranean Anadara demiri specimens with Anadara transversa (Say, 1822)
More than 120 specimens of Anadara transversa Say, 1822 from the north-western Atlantic Ocean have been examined. A wide range of variability has been observed. However, the similarities between Anadara demiri and the specimens of Anadara transversa from the southern part of its range (Sanibel Island, Florida, in particular) are striking (Figures 4 & 5).
The considered characters are here listed and commented upon. It is highlighted whether the character is considered of diagnostic importance for the genus Anadara.
(1) valves outline (diagnostic);
(2) mean length/height ratio (L/H) (diagnostic);
(3) mean length/width (L/W); this character is not considered a diagnostic character in the literature for species discrimination, however it was considered another useful parameter to understand the affinities between the two samples;
(4) position of the umbos (diagnostic);
(5) size, this is not considered a truly diagnostic character since adult specimen size is variable from 20 to almost 40 mm, however, the overall small size has to be considered a diagnostic character differentiating these species from other Anadara groups;
(6) inequivalvity in the adult stage;
(7) number of ribs, this character may be affected by the difficulties in precisely counting ribs near the umbos or because of valve erosion; it is considered a diagnostic character despite that it may be affected by a ±10–15% error;
(8) rib sculpture (diagnostic);
(9) rib size (diagnostic);
(10) chevrons on ligament, despite other authors considering this an important character, we have observed high variation of this pattern in specimens from the same locality, possibly the same population; chevrons tend to be more visible if the ligamental area is particularly wide;
(11) external colour, this is generally dull white in Anadara, showing very little variation. The presence of shadows of different colours (e.g. yellow, rose and cream) near the umbo is usually not a diagnostic character.
Table 1 contains a summary of a comparative morphological study between two sets of specimens: 50 specimens of Anadara demiri from the Adriatic Sea and 50 specimens of Anadara transversa from Florida. The groups of specimens analysed show a close relationship.
The size ratios similarity are impressive since Anadara tend to be quite variable in shape. These data have been statistically treated with a Student's t-test (α = 0.05). Dataset t value for L/H ratio is 0.779 while t values are 1.982 for α = 0.05 assessing that there is not a significant difference between the two data sets for α < 0.05. The same analysis was performed for the L/W ratio. Dataset t value for L/W ratio is 1.010. Again there is not a significant difference between the two datasets for the probabilities α < 0.05. To further support these results, a linear regression of ratios using natural log-transformed parameters was also performed. Numerical results are reported in Table 2. Very high values of r2 were found for the L/H ratio while slightly lower values of r2 were found for the L/W ratio. However, in both cases the Student's t-test assessed the lack of significant differences between the Y-axis intercept, a, and the slope, b, parameters (α < 0.05).
Table 1. Morphological comparison between Anadara demiri and Anadara transversa.

Table 2. Linear regression parameters of natural log transformed morphometric ratios of Anadara demiri and Anadara transversa.

Table 3. List of species from GenBank analysed in this study.

*numbers correspond to sequence fragments or to accession entries from GenBank; **base pairs numbers refer to the COI position in the complete mitochondrial genome.
Valves sculpture does not show significant differences. The different frequencies of the presence of chevrons are not considered of diagnostic importance.
The molecular analysis, based on nuclear LSU and mitochondrial COI sequences data, highlights that A. demiri and A. transversa are closely related. Although the Bayesian LSU tree (Figure 1) cannot resolve the relationships within the Arcidae, it shows clearly that A. demiri and A. transversa constitute a monophyletic group completely separated from all the other members of Arcidae included in the analysis, and it is supported by a 100% value of PP. The lack of resolution among Arcidae was not a surprise as LSU is a slow-evolving gene and in fact all the LSU analysed sequences are very similar. However, this low variation among LSU sequences was sufficient to separate A. demiri and A. transversa from the other Arcidae with strong support. The phylogenetic trees obtained from Bayesian analysis of COI sequences and COI–LSU combined dataset place A. demiri and A. transversa in a distinct group separated from other Arcidae as well, again highly supported (PP = 100%) as monophyletic. Their topology, identical except for a few different PP values, is illustrated in Figures 2 & 3, and shows clearly two highly supported (PP = 100%) major lineages within the family of Arcidae: one included A. demiri and A. transversa and the other five of the seven species of the family included in the analysis (Scapharca subcrenata (Lischke, 1869), Scapharca satowi (Dunker, 1882), Tegillarca granosa (Linnaeus, 1758) and Anadara inaequivalvis (Bruguière, 1789)). Arca ventricosa (Lamarck, 1819) and Barbatia lacerata Gray, 1842 appear more distant, while the position of Scapharca broughtoni (Schrenck, 1867) is probably due to an artefact of the tree-sampling. The analysis also shows that a very low genetic differentiation exists between A. demiri and A. transversa, as neither the COI tree nor the LSU–COI tree can distinguish between them. This represents further evidence of the close relationships between the two species.

Fig. 1. Fifty per cent majority rule consensus tree from Bayesian analysis (GTR + I + Г) of nuclear LSU rDNA gene dataset, rooted with Unio pictorum. Numbers above branches represent posterior probability values (only values above 95% are statistically significant). The numbers at the end of the name of the Anadara demiri and Anadara transversa sequences used refer to the samples names used in this study.

Fig. 2. Fifty per cent majority rule consensus tree from Bayesian analysis (GTR + I + Г) of mitochondrial COI gene dataset, rooted with Unio pictorum. Numbers above branches represent posterior probability values (only values above 95% are statistically significant). The numbers at the end of the name of the Anadara demiri and Anadara transversa sequences refer to the samples names used in this study.

Fig. 3. Fifty per cent majority rule consensus tree from Bayesian analysis (GTR + I + Г) of combined dataset from concatenated sequences of the nuclear LSU rDNA gene and mitochondrial COI gene, rooted with Unio pictorum. Numbers above branches represent posterior probability values (only values above 95% are statistically significant). The numbers at the end of the name of the Anadara demiri and Anadara transversa sequences refer to the samples names used in this study. The unusual position of Scapharca broughtoni is probably due to an artefact of tree-building.
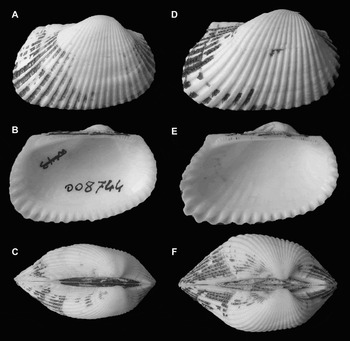
Fig. 4. Comparison between Anadara transversa (Say, 1822) and Anadara demiri (Piani, Reference Piani1981) showing infraspecific variability. (A–B–C) A. transversa, Sanibel Island, Florida, USA, length 24.9 mm, height 16.9 mm (MZB 8744); (D–E–F) A. demiri, Chioggia (Venezia), Italy, length 27.7 mm, height 18.3 mm.

Fig. 5. Comparison between Anadara transversa (Say, 1822) and Anadara demiri (Piani, Reference Piani1981) showing infraspecific variability. (A–B–C) A. transversa, Sanibel Island, Florida, USA, length 28 mm, height 18 mm; (D–E–F) A. demiri, Giulianova (Teramo), Italy, length 31.5 mm, height 21.7 mm.

Fig. 6. Sculpture detail of the anterior part of the left valve. (A) Anadara transversa (Say, 1822), Sanibel Island, Florida, USA, length 28.7 mm, height 20.5 mm (MZB 8744); (B) Anadara demiri (Piani, Reference Piani1981), Rimini, Italy, length 20.4 mm, height 13.8.
CONCLUSIONS
The history of the first identification of A. demiri, the morphological evidence and the molecular data led us to the conclusion that the alien species we find in the Adriatic Sea, Greece and Turkey is an allochthonous population of A. transversa. The taxon Scapharca demiri Piani, Reference Piani1981 has hence to be considered a junior synonym of Anadara transversa (Say, 1822). Morphological comparison may suggest the origin of the Mediterranean population is from the southern part of the range of A. transversa, e.g. Gulf of Mexico coasts of Florida (Sanibel Island specimens seem to be the morphologically closest to the Adriatic ones).
Anadara demiri is considered one of the 100 worst invasive alien species in the Mediterranean Sea by Streftaris & Zenetos (Reference Streftaris and Zenetos2006). This is the first alien species of molluscs in the Mediterranean Sea of North American origin with such an invasive potential, despite approximately 216 alien species are now present in the Mediterranean Sea (Zenetos et al., Reference Zenetos, Meriç, Verlaque, Galli, Boudouresque, Giangrande, Çinar and Bilecenoğlu2008).
ACKNOWLEDGEMENTS
The present work required the help of many friends and institutions. Kevin Czaja, Phil Fallon, Flavio Favero, Marlo Krisberg, Cristina Mazziotti, Ken Piech and Daniele Trono provided specimens. Christopher R. Garvie, He Jing, Lothar Maithas (Museum für Naturkunde, Berlin), Ron Noseworthy, Francisco Welter-Schultes provided references and translations. Kathie Way (British Museum Natural History, London), Virginie Héros (Muséum National d'Histoire Naturelle, Paris), Yves Finet (Muséum d'Histoire Naturelle, Genève), Paul Callomon (Academy of Natural Sciences, Philadelphia), Thomas von Rintelen (Museum für Naturkunde, Berlin) sent information on type material stored in the collections under their responsibility. Marco Passamonti, Federico Plazzi and Francesco Zaccanti, Department of Experimental Evolutionary Biology at the University of Bologna helped in discussion of the molecular and morphometric analysis results. Stefano Palazzi, Department of Geology, Section of Oceanology and Palaeoecology, University of Catania, patiently replied to questions and helped resolve doubts. Two anonymous referees gave important suggestions to improve the manuscript.
APPENDIX I
List of material examined.
Anadara demiri (Piani, Reference Piani1981):
Turkey
– Aegean Sea, Izmir, fishermen's nets; 1996 (coll. Albano; No. 6231): 2 specimens
Greece
– Thermaikos Gulf (north-western Aegean Sea), near Thessaloniki, –8.5 m on muddy bottom; December 2002 (coll. Albano; No. 6261): 2 specimens
Italy (listed from northern to southern Italy)
– Friuli Venezia Giulia, Grado (Gorizia), fishermen's nets; January 2004 (coll. Albano; No. 6393): 1 specimen
– Veneto, Chioggia (Venezia), Ca' Roman, beached; December 2001 (coll. Albano; No. 5539): 1 specimen
– Emilia Romagna, Cesenatico (Forlì-Cesena), beached; (coll. Albano; No. 5697): 1 specimen
– Emilia Romagna, between Cervia and Cesenatico (Forlì-Cesena), beached; April 2001 (coll. Albano; No. 5167): 4 specimens
– Emilia Romagna, Porto Garibaldi (Ferrara), fishermen's nets; July 2002 (coll. Albano; No. 6650): 3 specimens
– Emilia Romagna, Porto Corsini (Ravenna), beached; February 2003 (coll. Albano; No. 6142–6151): 8 specimens
– Emilia Romagna, Cesenatico (Forlì-Cesena), beached; February, 2003 (coll. Albano; No. 6155): 2 specimens
– Emilia Romagna, Cesenatico (Forlì-Cesena), Zadina Pineta, beached; February, 2003 (coll. Albano; No. 6219): 1 specimen
– Emilia-Romagna, Rimini, beached; December 2003 (coll. Albano; No. 6421): 30 specimens
– Emilia-Romagna, Rimini, fishing boats; December 2003 (coll. Albano; No. 6429): 20 specimens
– Marche, Ancona, fishing boats; February 2004 (coll. Albano; No. 6640): 1 specimen
– Marche, San Benedetto del Tronto (Ascoli Piceno), fishermen's nets; October 2005 (coll. Albano; No. 7250): 4 specimens
– Abruzzo, Giulianova (Teramo), fresh beached; March 2003 (coll. Albano; No. 6267): 100 specimens
– Puglia, San Isidoro (Lecce); –30 m, September 2003 (coll. Trono): 6 specimens
Anadara transversa (Say, 1822):
United States, Massachusetts
– Inset Beach (Buzzards Bay), July, 1983, M. Taviani legit, (Zoological Museum of the University of Bologna (ZMB), No. 009051): 1 valve
– Hyannis, Monument Beach, October 2002, K. Czaja legit (coll. Albano; No. 9381): 6 specimens
– off Wood's Hole, brought up with lobster traps, April 2003, K. Czaja legit (coll. Albano; No. 9385): 4 specimens
– Mattapoisett, Nye's Cove, March, 2005, K. Czaja legit (coll. Albano; No. 9386): 7 specimens
United States, Rhode Island
– Newport, Easton Beach, June, 2005, K. Czaja legit (coll. Albano; No. 9380): 12 valves
United States, New York
– Napeague Harbor; May 1998, P. Fallon legit (coll. Albano; No. 9150): 4 specimens
– East Marion, Orient Harbor, October 1996, P. Fallon legit (coll. Albano; No. 9151): 1 specimen
– Orient Point, K. Czaja legit (coll. Albano; No. 9383): 2 specimens
– off Orient Point, brought up with lobster traps, December 2003, K. Czaja legit (coll. Albano; No. 9384): 8 specimens
United States, Florida
– Captiva Island; P. Fallon legit (coll. Albano; No. 9152): 6 specimens
– Sanibel Island, March 1963, J. Zager legit (Zoological Museum of the University of Bologna, No. 008744): 3 specimens
– Siesta Key, Sarasota; February, 2007, K. Piech legit (coll. Albano; No. 9153): 20 specimens
– Sanibel Island, March 1998, K. Piech legit (coll. Albano; No. 9154): 20 specimens
– Sanibel Island, June 2005; K. Czaja legit (coll. Albano; No. 9382): 3 specimens
– Brevard County, Port Canaveral; September 1998, M. Krisberg legit (coll. Albano No. 9155): 2 specimens
– inside Fort Pierce inlet; July 1995, M. Krisberg legit (coll. Albano No. 9156): 2 specimens
– Brevard County, inside Sebastian inlet; February 1998, M. Krisberg legit (coll. Albano No. 9157): 1 specimen
– Hillsborough County, Hurricane Pass; October 1997, M. Krisberg legit (coll. Albano No. 9158): 1 specimen
– Pinellas County, Fort Desoto Park; February 1997, M. Krisberg legit (coll. Albano No. 9159): 2 specimens
– Volusia County, Ponce inlet; September 1998, M. Krisberg legit (coll. Albano No. 9160): 4 specimens
– Collier County, Pompano Hump; June 2006, M. Krisberg legit (coll. Albano No. 9161): 2 specimens
– Sarasota County, Sarasota, New Pass, Quick Point Park; M. Krisberg legit (coll. Albano No. 9162): 5 specimens
Molecular work has been based on the following material:
Anadara demiri (Piani, Reference Piani1981)
– Italy, Emilia Romagna, Porto Garibaldi (Ferrara); fishing-boats harbour, October 2007, F. Favero legit: 6 specimens;
– Italy, Emilia Romagna, off Porto Garibaldi, 44°39′69″N–12°17′28″E, legit C. Mazziotti, ARPA Emilia-Romagna, Struttura Oceanografica Daphne; July 2007: 24 specimens;
Anadara transversa (Say, 1822)
– United States, Massachusetts, Edgartown, Martha's Vineyard at Sengenkontacket Pond, 41°23′N–70°30′W, August 2007, K. Czaja legit: 2 specimens;
– United States, Florida, St Johns County, St Augustine Inlet, 29°54′30″N–81°17′05″W, August 2007, M. Krisberg legit: 8 specimens;
Anadara inaequivalvis (Bruguière, 1789)
– Italy, Emilia Romagna, off Porto Garibaldi, 44°39′69″N–12°17′28″E, legit C. Mazziotti, ARPA Emilia-Romagna, Struttura Oceanografica Daphne; July 2007: 20 specimens;











