INTRODUCTION
Clytia Lamouroux, 1812 is a typical Campanulariidae (Cnidaria: Hydrozoa: Leptothecata) genus with many species and a near-cosmopolitan distribution. The life cycle of Clytia comprises a free swimming medusae stage and a sessile hydroid stage. Medusae of Clytia are frequently found in surface coastal waters, while hydroids of this genus are common in shallow-water benthic communities (Cornelius, Reference Cornelius, Barnes and Crothers1995; Madin et al., Reference Madin, Bollens, Horgan, Butler, Runge, Sullivan, Klein-Macphee, Durbin, Durbin, Keuren, Plourde, Bucklin and Clarke1996; Boero et al., Reference Boero, Camillo and Gravili2005; Lindner et al., Reference Lindner, Govindarajan and Migotto2011; Zhou et al., Reference Zhou, Zheng, He, Lin, Cao and Zhang2013). With growing attention focused on the ecological impact of jellyfish blooms, these tiny organisms are emerging as an important competitor and predators in the coastal marine ecosystem (Lucas et al., Reference Lucas, Williams, Williams and Sheader1995; Bouillon et al., Reference Bouillon, Gravili, Pagés, Gili and Boero2006; Gravili et al., Reference Gravili, D'Ambrosio, Di Camillo, Renna, Bouillon and Boero2008; Miglietta et al., Reference Miglietta, Rossi and Collin2008). Certain species, e.g. C. hemisphaerica (Linnaeus, 1767), have been firmly studied as a basal metazoan model organism to explore developmental mechanisms and resolve basic evolutionary questions (Houliston et al., Reference Houliston, Momose and Manuel2010).
However, species diagnosis in Clytia is challenging. Firstly, most species are based either on the hydroid (42 species) or the medusae form (30 species), with only 11 species having had their complete life cycle investigatedFootnote 1 (Mayer, Reference Mayer1910; Roosen-Runge, Reference Roosen-Runge1970; West & Renshaw, Reference West and Renshaw1970; Kubota, Reference Kubota1978a, Reference Kubotab; Cornelius, Reference Cornelius, Barnes and Crothers1995; Pagliara et al., Reference Pagliara, Bouillon and Boero2000; Lindner & Migotto, Reference Lindner and Migotto2002; Gravili et al., Reference Gravili, D'Ambrosio, Di Camillo, Renna, Bouillon and Boero2008; Lindner et al., Reference Lindner, Govindarajan and Migotto2011; Zhou et al., Reference Zhou, Zheng, He, Lin, Cao and Zhang2013). Secondly, diagnostic characters of some species in the genus were based on a single specimen, or sometimes even on an immature animal, e.g. C. ambigua (Agassiz & Mayer, Reference Agassiz and Mayer1899) and C. hexacanalis (Xu et al., Reference Xu, Huang, Chen, Hong, Qiu, Ruan and Hong1991). Thirdly, characters long thought to have taxonomic values tend to be variable between individuals of the same population, at least in certain species (Kubota, Reference Kubota1978a; Bouillon & Boero, Reference Bouillon and Boero2000). Finally, with the emerging of molecular techniques, cryptic species or even new species are being revealed (Lindner et al., Reference Lindner, Govindarajan and Migotto2011; Zhou et al., Reference Zhou, Zheng, He, Lin, Cao and Zhang2013).
Molecular-aided species diagnosis and phylogenetic analyses have made remarkable progress in species revisions and reconstruction of the evolution in the related taxa. DNA barcoding based on mitochondrial COI proved to be a useful molecular tool to identify species efficiently and reliably among many animal taxa (Hebert et al., Reference Hebert, Cywinska, Ball and deWaard2003a, Reference Hebert, Ratnasingham and deWaardb; Bucklin et al., Reference Bucklin, Hopcroft, Kosobokova, Nigro, Ortman, Jennings and Sweetman2010a, Reference Bucklin, Ortman, Jennings, Nigro, Sweetman, Copley, Sutton and Wiebeb, Reference Bucklin, Steinke and Blanco-Bercial2011; Ortman et al., Reference Ortman, Bucklin, Pagès and Youngbluth2010). In the genus Clytia, COI was also applied to identify ambiguous specimens (Laakmann & Holst, Reference Laakmann and Holst2014) and detect cryptic or new species (Lindner et al., Reference Lindner, Govindarajan and Migotto2011; Zhou et al., Reference Zhou, Zheng, He, Lin, Cao and Zhang2013). Recently, mitochondrial 16S was proposed as a candidate for hydrozoa barcoding (Moura et al., Reference Moura, Harris, Cunha and Rogers2008; Zheng et al., Reference Zheng, He, Lin, Cao and Zhang2014). Meanwhile mitochondrial 16S and COI, nuclear 18S, 28S and internal transcribed spacer 1 (ITS1) genes were used to resolve phylogenetic relationships at various taxonomic levels (Bridge et al., Reference Bridge, Cunningham, DeSalle and Buss1995; Schierwater & Ender, Reference Schierwater and Ender2000; Collins et al., Reference Collins, Winkelmann, Hadrys and Schierwater2005, Reference Collins, Schuchert, Marques, Jankowski, Medina and Schierwater2006; Govindarajan et al., Reference Govindarajan, Boero and Halanych2006; Schuchert, Reference Schuchert2014). In this study, morphological observations and molecular analyses were combined to validate the taxonomic position of a new Clytia species, Clytia gulangensis sp. nov., and thus to promote the revision of the genus Clytia.
MATERIALS AND METHODS
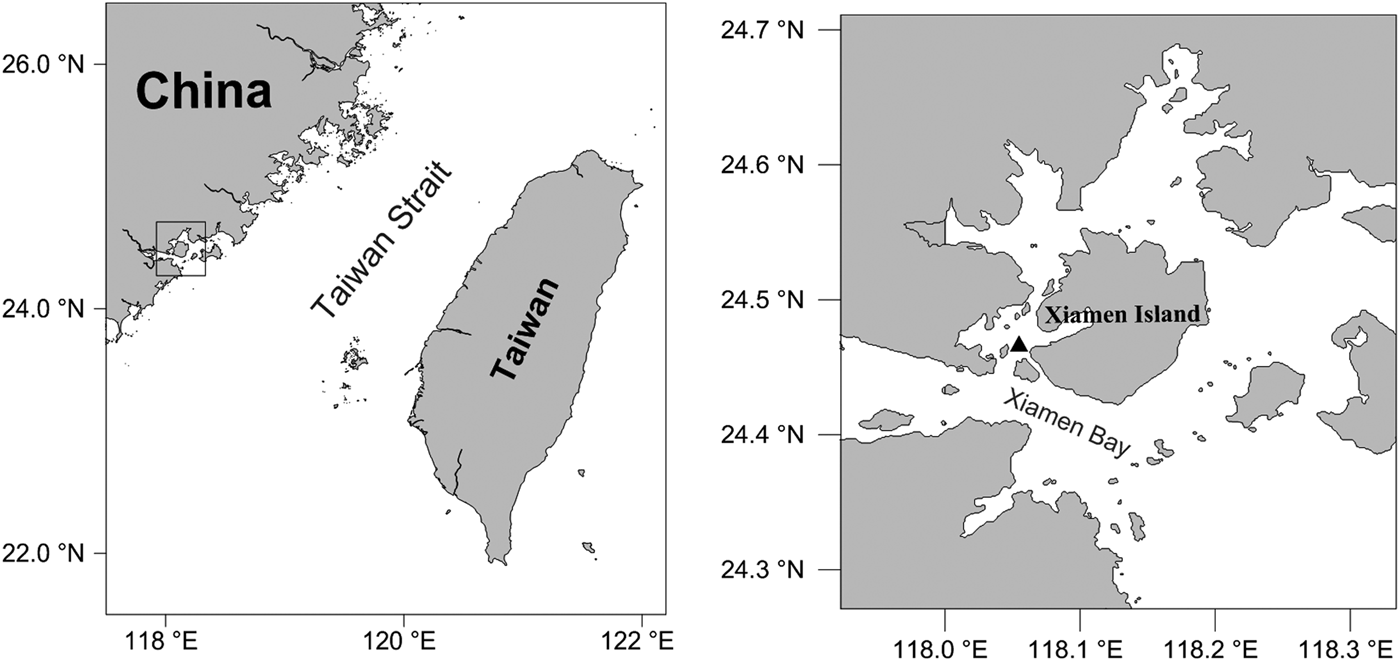
Medusae were collected from Xiamen Bay (24.4514°N 118.0753°E), East China Sea, using a plankton net with mesh size of 505 µm on 5 June 2012 (Figure 1). In the laboratory, 22 mature medusae with indistinguishable morphologies were kept in a 20 × 20 × 20 cm glass tank with filtered seawater (filter mesh size: 50 µm). Hydroids developed at the bottom of the aquaria after five days and they were then transferred onto a glass slide in a new tank. Colonies developed from two separate glass slides covered the whole tank surfaces eventually, and were used for life-cycle observation, respectively. Both medusae and hydroids were fed with Artemia sp. nauplii daily with water changed every other day after feeding. Water temperature was kept at 22 ± 3°C and salinity at 31 ± 2.

Fig. 1. Sampling site of Clytia gulangensis sp. nov.
Morphological measurements were accomplished with Zeiss SteREO Discovery V12 and Olympus BX51 microscope. Individuals intended for image documenting were acclimated with 3% MgCl2, specimens for morphological preservation were fixed in 5% formalin and those for DNA preservation were fixed in 95% ethanol. Fresh tissue was stimulated in 1% SDS (sodium dodecyl sulphate) for nematocysts type and distribution detection, and nematocysts nomenclature followed that of Östman (Reference Östman and Stepanjants1999, Reference Östman2000).
Total DNA was extracted from both medusa and hydroid with a modified phenol-chloroform extraction method (Zheng et al., Reference Zheng, Lin, Li, Cao, Xu and Huang2009); mitochondrial COI (primer: HCO2198-taaacttcagggtgaccaaaaaatca, LCO1490-gtcaacaaatcataaagatattgg; Folmer et al., Reference Folmer, Black, Hoeh, Lutz and Vrijenhoek1994) and 16S (primer: 16SH-cataattcaacatcgagg, 16SL-gactgtttaccaaaaacata; Ender & Schierwater, Reference Ender and Schierwater2003), nuclear 18S (primer: 18SF-gctgtatgtactgtgaaactgcg, 18SR-cacctacggaaaccttgttacgac; Leclère et al., Reference Leclère, Schuchert, Cruaud, Couloux and Manuel2009) and 28S (primer: 28S1F-tcccctagtaacggcgagtgaagcg, 28S1R-gagccaatccttwtcccgargtt, 28S2F-gacagcaggacggtggycatgg, 28S2R-ttcygacttagaggcgttcag; Medina et al., Reference Medina, Collins, Silberman and Sogin2001; Leclère et al., Reference Leclère, Schuchert, Cruaud, Couloux and Manuel2009) gene fragments were amplified according to the references herein. Purified PCR products were sequenced by Sangon Biotech on 3730xl DNA Analyzer with BigDye terminator v.3.1.
Sequences were checked manually based on chromatogram files, aligned using the NCBI Nucleotide Blast (BLASTn) program to confirm the validity, and submitted to the NCBI GenBank database. GenBank Accession numbers of C. gulangensis sp. nov. are KF962086–KF962090 (hydroid), KF962091–KF962095 (medusae from field), and KF962096–KF962100 (medusae from culture) for COI; KF962425–KF962429 (hydroid), KF962430–KF962434 (medusae from field), and KF962435–KF962439 (medusae from culture) for 16S; KF962218–KF962222 (hydroid), KF962223–KF962227 (medusae from field), and KF962228–KF962232 (medusae from culture) for 18S; and KF962318–KF962322 (hydroid) and KF962323–KF962327 (medusae from field), and KF962328–KF962332 (medusae from culture) for 28S, respectively.
Multiple sequences were aligned using ClustalX V2.1 (Larkin et al., Reference Larkin, Blackshields, Brown, Chenna, McGettigan, McWilliam, Valentin, Wallace, Wilm, Lopez, Thompson, Gibson and Higgins2007), genetic distance was determined by MEGA 5.2 (Tamura et al., Reference Tamura, Peterson, Peterson, Stecher, Nei and Kumar2011) with Kimura-2-Parameter model, and phylogenetic analyses were performed using PAUP* 4.0b10 (Swofford, Reference Swofford2002) and MEGA 5.2 with GTR + G + I model which was suggested as optimal substitution model by the built-in model test module. For DNA barcoding purpose, Kimura 2-parameter genetic distance and neighbour-joining tree were generated from all Clytia COI sequences available in GenBank, with Leptothecata Aequorea conica Browne, 1905 and Gangliostoma guangdongensis Xu, 1983 selected as outgroups. For phylogenetic analyses of Clytia, maximum likelihood and maximum parsimony analyses were conducted for 16S and 18S individually and for both genes combined; sequences of both genes from all Clytia species and other representative campanulariids in GenBank were included; and Calycella syringa Linnaeus, 1767 and Opercularella pumila Clark, 1875 (accepted as Campanulina pumila Clark, 1875) were selected as outgroups (Govindarajan et al., Reference Govindarajan, Boero and Halanych2006; Lindner et al., Reference Lindner, Govindarajan and Migotto2011). We preferred the results obtained with likelihood analyses because of known problems of parsimony with rate variation (Govindarajan et al., Reference Govindarajan, Boero and Halanych2006). Though nuclear 28S rDNA was proved to be an informative marker for both species revision and evolution analysis purpose (e.g. Evans et al., Reference Evans, Lindner, Raikova, Collins and Cartwright2008; Leclère et al., Reference Leclère, Schuchert, Cruaud, Couloux and Manuel2009), it was not used in this study as not enough data from Clytia species were available. Taxa employed in this study and GenBank accession numbers are listed in Table S1.
RESULTS
SYSTEMATICS
Phylum CNIDARIA
Class HYDROZOA Owen, 1843
Order LEPTOTHECATA Cornelius, 1992
Family CAMPANULARIIDAE Johnston, 1836
Genus Clytia Lamouroux, 1812
Clytia gulangensis sp. nov. Jinru He & Lianming Zheng
(Figures 2–4; Tables 1–3)

Fig. 2. Hydroids of Clytia gulangensis sp. nov.: (A) colony; (B) hydranth; (C) gonotheca with medusae buds; (D) hydrothecal margin with cusps; (E) illustration of a trophotheca; (F) illustration of a gonotheca. Scale bars: A–C, 0.5 mm; D, 0.1 mm; E, 0.5 mm; F, 0.25 mm.

Fig. 3. Medusae of Clytia gulangensis sp. nov.: (A) newly released medusa, side view, white arrow: gonad; (B) newly released medusa, oral view; (C) mature medusae (60 days), side view; (D) mature medusae (35 days), oral view; (E) mature medusae (35 days), side view; (F) margin of mature medusa, white arrow: statocysts with statoliths; (G, H) illustration of newly released medusa; (I, J) illustration of mature medusa. Scale bars: A, B, 0.2 mm; C–E, 1.5 mm; F, 0.5 mm; G, H, 0.2 mm; I, 0.3 mm; J, 0.2 mm.

Fig. 4. Nematocysts of Clytia gulangensis sp. nov.: (A) A-type microbasic mastigophores from hydranth; (B) B-type microbasic mastigophores from hydranth; (C) A-type microbasic mastigophores from newly released medusae; (D) Ic-type isorhiza from newly released medusae; (E) Ic-type isorhiza from mature medusae; (F) A-type microbasic mastigophores from mature medusae. Scale bars: A–F, 10 µm.
Table 1. Measurements (mean ± standard deviation (range)) of colonies of Clytia gulangensis sp. nov. (N = 30).

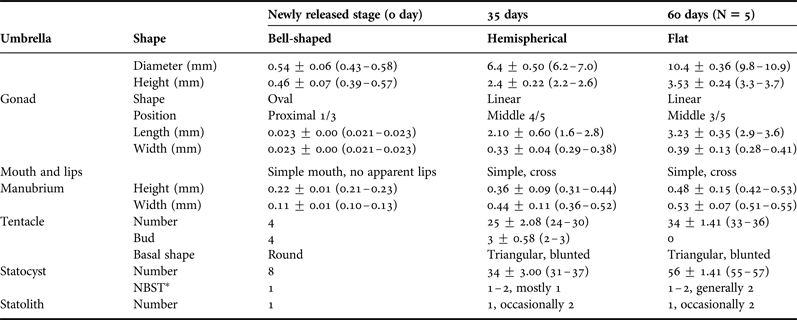
Table 2. Comparison of morphology (mean ± standard deviation (range)) of Clytia gulangensis sp. nov. at successive developing medusae stages, in mm (N = 30 unless otherwise mentioned).

NBST*, number between successive tentacles.
Table 3. Measurements (mean ± standard deviation (range)) of microbasic mastigophore nematocysts of Clytia gulangensis sp. nov., in μm. (N = 50).

MATERIAL EXAMINED
Holotype: XMBCG01, male medusa, diameter 6.43 mm, height 2.36 mm.
Paratypes: XMBCG02, male medusa, diameter 6.82 mm, height 2.62 mm; XMBCG03, male medusa, diameter 10.70 mm, height 3.62 mm; XMBCG20, polyp with both trophosome and mature gonosome.
Nontype material: XMBCG07, female medusa, diameter 5.78 mm, height 2.54 mm; XMBCG14, female medusa, diameter 6.00 mm, height 2.52 mm; XMBCG05, polyp with both trophosome and immature gonosome.
The medusae specimens XMBCG01 and XMBCG02 were collected in Xiamen Bay, China, 24°27′5″N 118°4′31″E, 5 June 2012. XMBCG03, XMBCG07, XMBCG14 (medusa) and XMBCG05, XMBCG20 (polyps) were obtained from individuals cultured in the laboratory. All type specimens are deposited in the Department of Marine Biological Science and Technology, College of Ocean and Earth Sciences, Xiamen University, China.
ETYMOLOGY
Clytia gulangensis sp. nov. is named after Gulang Island around which the specimens were collected.
DIAGNOSIS
The stolonal polyps, the campanulate hydrothecae, and free medusae with a normal velum, without cirri or excretory papillae identify the animals as members of the genus Clytia.
Clytia gulangensis sp. nov. is distinguished from its congeners and other campanulariids by the combination of the following characters:
Polyp: stems monosiphonic, sometimes polysiphonic, branching irregularly 2–3 times. Hydrotheca elongate campanulate, about three times as long as wide, with 8–12 blunt, triangular cusps, slightly asymmetrical, separated by deep, rounded embayments, without inward folds. Gonotheca on hydrorhiza and pedicel, club-shaped, somewhat pod-like, with neck, stalk short with indistinct 1–3 annulations, with smooth walls, forming one to two rows of up to six medusae buds. Mature ones with neck just below the aperture. B-type microbasic mastigophores 7.98–8.48 µm long and 2.16–2.21 µm wide.
Adult medusa: umbrella flatter than a hemisphere, 6.2–10.5 mm in diameter, up to 36 tentacles, with 1–2 statocysts between successive tentacles, each containing a single statolith, rarely two. Gonads linear, more wavy band-like when mature, covering 3/5–4/5 of the radial canal, leaving spaces at both ends. Ic-type isorhizas 7.21–7.50 µm long and 2.54–2.58 µm wide.
DESCRIPTION
Hydroid
Colonies stolonal or with erect stems branching 2–3 times irregularly. Branches given off upwardly from stem; pedicel up to 5.9 mm high, smooth, with 9–17 proximal and 5–10 distal annuli. Creeping hydrorhiza slightly annulated occasionally at the junction where branches occurred (Figure 2A, E).
Hydrothecae elongate campanulate, with thin perisarc and smooth walls, about 3 times as long as wide (0.53–1.02 mm long and 0.18–0.33 mm wide at aperture); rim with 8–12 blunt, slightly asymmetrical, triangular or pyramidal cusps, separated by deep, rounded embayments, without inward folds. Hydrothecal diaphragm thin, near base of hydrotheca; basal chamber 46–89 µm long and 78–114 µm wide at diaphragm. Pedunculated hypostome spherical or oval in oral view. Hydranth with 14–24 filiform tentacles, 0.5–0.8 mm in length (Figure 2B, D; Table 1). Coenosarc whitish.
Gonotheca on stolons and pedicels, or directly on branches, nearly cylindrical in shape. And sometimes pod-like. Gonothecae smooth, about three times as long as wide (0.79–0.90 mm long and 0.26–0.29 mm wide at distal end); with one side of the wall nearly straight and opposite side contracting down, with constriction below the truncated distal margin, with short stalk, slightly annulated 1–3 times. Up to 6 medusae of 1–2 rows in each gonangium (Figure 2C, F; Table 1).
A- and B-type microbasic mastigophores on hydranth and coenosarcs. Capsules of discharged A-type microbasic mastigophores 6.76 ± 0.60 (6.07–7.82) μm long and 2.00 ± 0.25 (1.75–2.40) μm wide, and B-type microbasic mastigophores 8.23 ± 0.35 (7.98–8.48) μm long and 2.19 ± 0.04 (2.16–2.21) μm wide in vivo, respectively. Tubes of discharged A-type microbasic mastigophores form an obtuse angle to the long axis of the capsule, but sometimes coincide with the direction of the latter. Shaft of discharged B-type microbasic mastigophores wider and longer than other types, and the angle between shaft and long axis more obvious (Figure 4A, B; Table 3).
Newly-released medusae
Umbrella bell-shaped, somewhat cubic, 0.43–0.58 mm in diameter and 0.39–0.57 mm in height; with ring canal and four radial canals; four prominent perradial bulbs with tentacles and four small interradial developing bulbs; eight adradial statocysts, each containing a single statolith. Gonads on proximal 1/3 of radial canals, oval in shape. Manubrium quadrate, half the height of bell cavity, with slightly recurved lips. Velum broad (Figure 3A, B; Table 2).
Development
Medusae two days after release with eight tentacles and eight statocysts. Umbrella flattened with diameter increasing, gonads extending along radial canals and turned wavy band-like three weeks since release. Tentacle number increased to 30 or more in about 20 days, and statocyst number increased from one to two between two successive tentacles in most cases during growth.
Mature medusae
Medusae grew mature about 35 days after liberation as judged by sperm or egg release. Umbrella flatter than a hemisphere, 6.2–7.0 mm in diameter and 2.2–2.6 mm in height. Jelly thin and flexible. Tentacles 24 to 30 in number, well-developed, with mediate, rounded, basal bulbs. Statocysts 31 to 37 in number, alternate in position with the tentacles, rarely two between successive tentacles, each containing a single, spherical concretion. Velum narrow, four straight, slender radial canals and a narrow circular canal. Manubrium short and quadratic in cross-section, four slightly recurved lips. Four gonads situated very close to circular canal, and stretched to gastro-vescular cavity, occupied almost entire radial canals when fully developed, also became more band-like and somewhat contorted. The medusa is transparent with the exception of the manubrium, gonads, and tentacle bulbs, which are somewhat flesh-colour (Figure 3D–F; Table 2).
Further rearing produced medusae with extended umbrella diameter and increased statocyst number. Few medusae with longevity about 60 days measured maximum diameter of 10.9 mm and height of 3.7 mm. Tentacle numbers up to 36 without rudimentary bulbs, and statocysts of 55–57 in total and mostly two between successive tentacles, statolith remained single in each statocyst, rarely two. The wavy banded linear gonads extended about 3/5 of the radial canals (Figure 3C; Table 2).
Variation of nematocysts size among individuals is insignificant and discharged capsule size from young and adult medusae reveal little differences. A-type (7.58 ± 0.40 µm long and 2.26 ± 0.17 µm wide) microbasic mastigophores from medusae are larger than those from hydroid. And Ic-type isorhizas (7.40 ± 0.47 µm long and 2.56 ± 0.03 µm wide) are frequently found in medusae (Figure 4C–F; Table 3).
DISTRIBUTION
This species is only collected in large numbers on the surface in Xiamen Bay, East China Sea. Further field investigations on its distribution and seasonal variation remain unresolved.
BIOLOGICAL NOTES
This new species can be easily reared under conditions described above. And medusae release in large numbers in about every 20 days. When the temperature is maintained at 12–30°C, salinity at 25–35, and fed every 1–3 days, hydroids can survive with at least 1/3 hydranth extending and feeding, while budding could be affected by delayed release. The polyps seem to be rather tolerant to salinity variation (25–40) induced by water evaporation and replenishment of fresh water, while starvation (longer than 5 days) and low temperature (below 10°C) would be a devastating induction to hydranth resting. For medusae, warm temperature (25–30°C), optimal salinity (28–32) and daily feeding would be necessary, and water quality should be secured to avoid unexpected death.
REMARKS
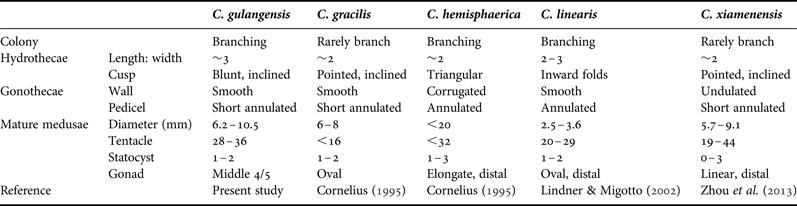
Hydroids of C. gulangensis sp. nov. resemble C. delicatula (Thornely, Reference Thornely and Willey1900), C. elongata (Marktanner-Turneretscher, 1890), C. elsaeoswaldae (Stechow, 1914), C. gracilis (Sars, 1850), C. gregaria (Agassiz, Reference Agassiz1862), C. linearis (Thorneley, Reference Thornely and Willey1900) and C. tottoni (Leloup, 1935) as all of them have elongate campanulate hydrothecae and smooth gonothecal walls. Both C. gregaria and C. linearis show signs of inward folds at the hydrothecal embayments (Roosen-Runge, Reference Roosen-Runge1970; Lindner & Migotto, Reference Lindner and Migotto2002; Schuchert, Reference Schuchert2003), which is not the case in our species; C. elsaeoswaldae, C. gracilis and C. tottoni are all similar to C. gulangensis sp. nov. in having inclined hydrothecal cusps, while C. elsaeoswaldae have slightly undulated gonothecal walls, C. gracilis have tilted cusps with one side almost vertical and the other oblique, with also slightly everted embayment margin, and C. tottoni have cusps projecting inwardly, instead of inclined pyramidal cusps and smooth embayments in our species (Cornelius, Reference Cornelius, Barnes and Crothers1995; Schuchert, Reference Schuchert2003; Galea, Reference Galea2010; Lindner et al., Reference Lindner, Govindarajan and Migotto2011); the ratio of hydrothecal length to width is about 3 (2.75–3.28) in our species, while hydrothecae of C. delicatula, C. elongata, and C. gracilis are all about 2 times as long as wide, moreover, C. delicatula have deeply-cut, acute cusps (Hiro, Reference Hiro1939; Kubota, Reference Kubota1978a), hydranth of C. elongata have just about 10 tentacles, instead of 14–24 in our species (Vervoort & Watson, Reference Vervoort and Watson2003). The B-type microbasic mastigophores of C. gulangensis sp. nov. (7.98–8.48 × 2.16–2.21 µm) is much smaller in size compared to other Clytia species (10.0–24.0 × 2.5–5.5 µm) reported, with the exception of C. noliformis (McCrady, 1859) (6.5–7.0 × 2.0–2.5) (Östman, Reference Östman1979a, Reference Östmanb, Reference Östman and Stepanjants1999) (Table 3).
Medusae of the present species resemble those of C. attenuata (Calkins, 1899), C. brunescens (Bigelow, Reference Bigelow1904), C. gregaria, C. hemisphaerica, C. languida (Agassiz, Reference Agassiz1862), C. lomae (Torrey, Reference Torrey1909), C. macrogonia (Bouillon, Reference Bouillon1984), C. malayense (Kramp, Reference Kramp1961) and C. uchidai (Kramp, Reference Kramp1961) in having about 30 marginal tentacles and 1–2 statocysts between successive tentacles, but differ in shape and situation of gonads. Clytia attenuata is considered conspecific with C. gracilis (Calder, Reference Calder1991), or C. hemisphaerica (Cornelius, Reference Cornelius1982; Bouillon et al., Reference Bouillon, Gravili, Pagés, Gili and Boero2006), and a life-cycle study reported the gonads of mature C. attenuata to be oval to sacciform (West & Renshaw, Reference West and Renshaw1970), while redescription of C. gracilis from north-west European waters revealed that medusae of this species have up to 16 tentacles (Cornelius, Reference Cornelius, Barnes and Crothers1995). Clytia brunescens bears thick and prominent gonads which are nearly hemispherical and occupying proximal third of radial canal (Bigelow, Reference Bigelow1904); C. gregaria, C. hemisphaerica, C. lomae, C. malayense and C. uchidai all have oval to linear ovaries which extend distal half of radial canal (Agassiz, Reference Agassiz1862; Torrey, Reference Torrey1909; Kramp, Reference Kramp1961; Kubota, Reference Kubota1978b); C. languida bears linear ovaries nearly covering the entire radial canal, but statocysts between every two tentacles are 2–3 in number (Agassiz, Reference Agassiz1862); C. macrogonia has cylindrical gonads extending almost entire radial canal which is much more prominent than those of our species, and its manubrium is cruciform with rounded perradial lobes which are absent in C. gulangensis sp. nov. (Bouillon, Reference Bouillon1984; Bouillon et al., Reference Bouillon, Medel, Pagès, Gili, Boero and Gravili2004; Du et al., Reference Du, Xu, Huang and Guo2012) (Table 4).
Table 4. General features of some similar species of Clytia.

Though sharing a similar shape of umbrella and the approximate number of tentacles in medusae, C. gulangensis sp. nov. is also different from C. xiamenensis (Zhou et al., Reference Zhou, Zheng, He, Lin, Cao and Zhang2013), which was described recently from the same area from both hydroid and medusae stages. In C. xiamenensis, gonothecal walls are undulated, statocysts being 0–3 in number, and gonads occupying distal half of radial canal (instead of smooth gonothecal walls, 1–2 nematocysts between successive tentacles, and elongated wavy gonads in our species); the novel LA-type microbasic mastigophores from C. xiamenensis are never found in our individuals, and B-type microbasic mastigophores of C. gulangensis sp. nov. are much smaller than that of C. xiamenensis (Zhou et al., Reference Zhou, Zheng, He, Lin, Cao and Zhang2013) (Table 4).
DNA BARCODING AND PHYLOGENETICS
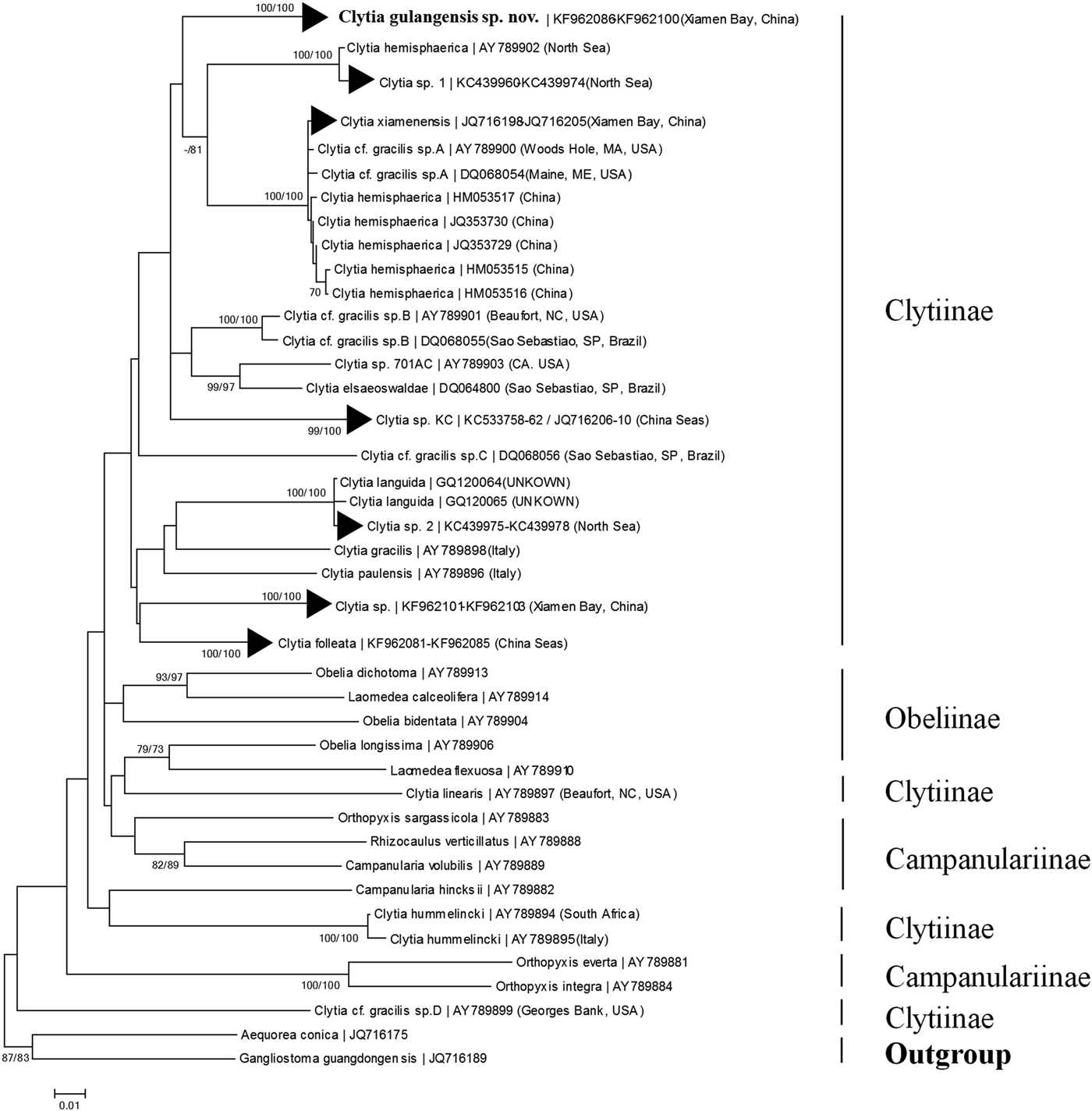
The COI alignment for DNA barcoding analysis included 95 operational taxonomic units (OTUs) and 669 base pairs. Sequence divergence (measured as K2P genetic distance) of COI between individuals of C. gulangensis sp. nov. ranged from 0 to 0.003. For the genus Clytia, intra-specific genetic distance varied from 0 (multiple species, e.g. C. folleata (McCrady, 1859)) to 0.014 (C. hemisphaerica); inter-specific genetic distance ranged from 0.049 (between C. elsaeoswaldae and Clytia sp. 701AC) to 0.257 (between Clytia cf. gracilis sp. D and C. hummelincki (Leloup, 1935)). For C. gulangensis sp. nov., minimum genetic distance (0.062) was observed in comparison to Clytia cf. gracilis sp. B; maximum genetic distance (0.198) was observed in comparison to Clytia cf. gracilis sp. D (Table S2). Thus, a barcoding gap (Meyer & Paulay, Reference Meyer and Paulay2005) was confirmed both in the genus Clytia and between C. gulangensis sp. nov. and all the other Clytia species, respectively. In the present study, genetic distance between C. gulangensis sp. nov. and the other four species collected in sympatry, ranged from 0.071 to 0.110, also showed obvious barcoding gaps (details in Tables S1 and S2). Neighbour-joining topology of all Clytia and other representative Campanulariidae failed to recover a monophyletic Clytia as certain sequences of C. linearis, C. hummelincki and Clytia. cf. gracilis sp. D were placed outside the Clytiinae clade, but still revealed an independent clade of C. gulangensis sp. nov., which supported the validity of this new species (Figure 5).

Fig. 5. Neighbour-joining clustering of Campanulariidae based on mitochondrial COI sequences. Branch support values given as bootstrap values higher than 70 obtained from maximum likelihood and neighbour-joining analyses are shown close to each branch.
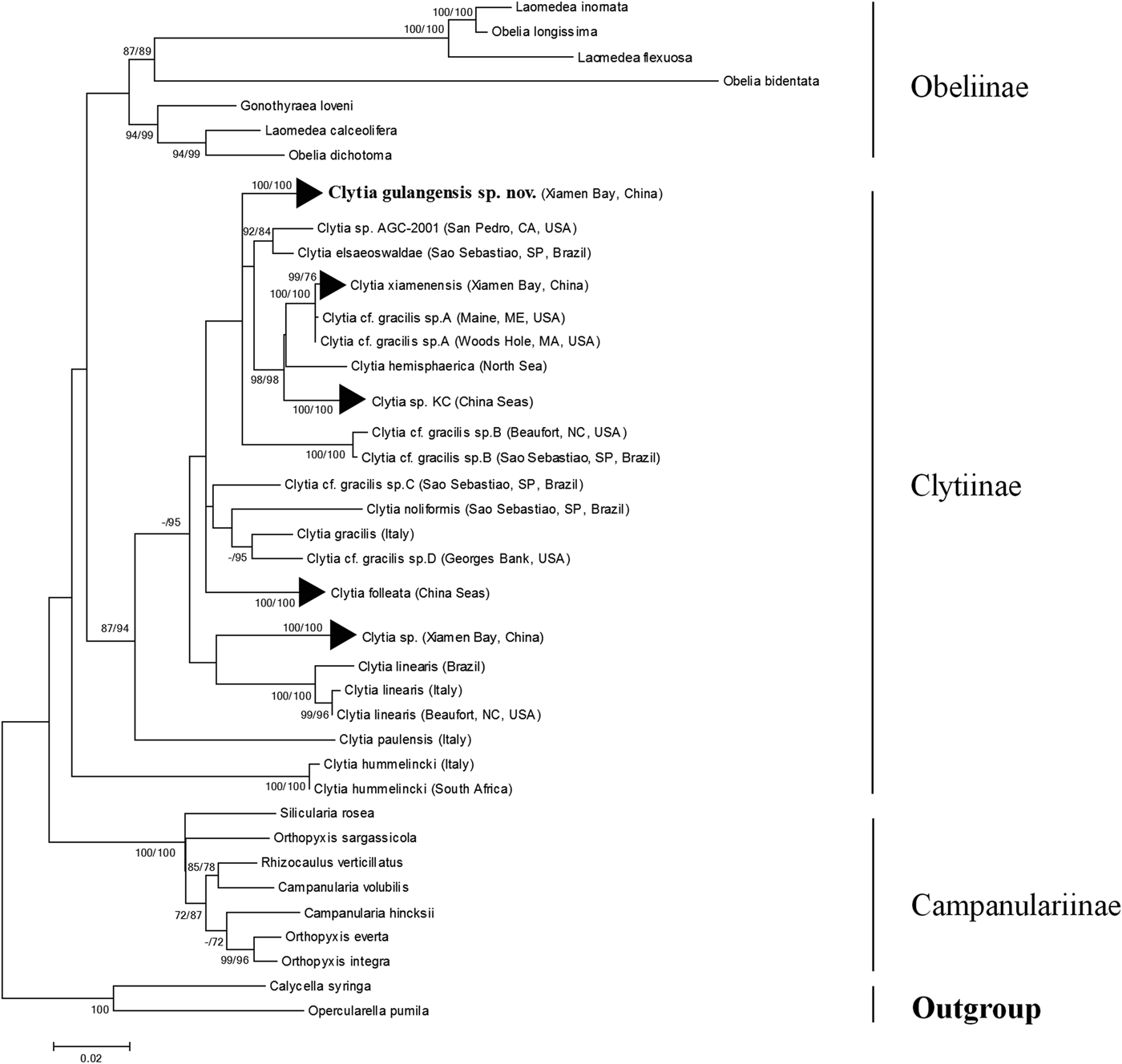
Maximum likelihood topology based on the 16S (Figure S1), 18S (Figure S2) and 16S plus 18S (Figure 6) dataset all suggested a monophyletic C. gulangensis sp. nov. clade, which further confirmed the separation from all known Clytia sequences. Taxa incorporated into the Campanulariidae phylogenetic analysis were adjusted to include representatives from subfamily Obeliinae and Campanulariinae, along with all Clytiinae (genus Clytia) sequences available on GenBank. Though two sequences (C. hemisphaerica FJ550601; C. noliformis EU272611) which have only 18S entries and five sequences (C. elsaeoswaldae DQ068064; C. hemisphaerica EU999221; C. hemisphaerica HM053545; Clytia sp. AY800195; C. viridicans (Leuckart, 1956) AY346365) which have only 16S entries were excluded in the final alignment, 11 Clytia species with morphological descriptions were included. Maximum likelihood analyses for 16S failed to recover a monophyletic Clytiinae, and 18S failed to reveal a monophyletic Obeliinae, respectively. The 16S plus 18S alignment for phylogenetic analysis, which included 71 OTUs and 2252 base pairs, revealed both monophyletic Obeliinae and Campanulariinae, and thus better resolved the relationships among Campanulariidae relatively. With C. hummelincki turning out to be a sister taxon to Obeliinae plus Clytia (with the only exception of C. hummelincki) clade, Clytiinae failed to form a monophyletic lineage yet. And the monophyletic C. gulangensis sp. nov. clade was deeply rooted in the main Clytia clade (Figure 6).

Fig. 6. Maximum likelihood phylogenetic analysis of Campanulariidae based on mitochondrial 16S and nuclear 18S rDNA. Branch support values given as bootstrap values higher than 70 obtained in maximum parsimony and maximum likelihood analyses are shown close to each branch.
DISCUSSION
Life-cycle investigations are essential for species diagnosis and taxonomic revision in the genus Clytia. Life-cycle studies provide valuable information on variations of taxonomic characters by investigating the developmental process (e.g. Kubota, Reference Kubota1978b), and helped uncover cryptic or new species by supplementing novel characters in another stage (Lindner et al., Reference Lindner, Govindarajan and Migotto2011; Zhou et al., Reference Zhou, Zheng, He, Lin, Cao and Zhang2013). Currently, 61 Clytia species have been recognized, of which 60 species are listed in the World Register of Marine Species (WoRMS) (Schuchert, Reference Schuchert2013) and C. xiamenensis was described recently (Zhou et al., Reference Zhou, Zheng, He, Lin, Cao and Zhang2013). Among all those species recorded, only 11 species have their life cycle investigated, but for C. hummelincki the mature medusa remains unknown (Gravili et al., Reference Gravili, D'Ambrosio, Di Camillo, Renna, Bouillon and Boero2008). With most species being morphologically diagnosed either by their polyp or medusae stage, the gap between diagnoses based on only one of the stages definitely contributed to the inflation of synonyms in this genus (e.g. Calder, Reference Calder1991; Schuchert, Reference Schuchert1998). In the present study, the fixed hydrothecal length to width ratio, blunt and asymmetric cusps, smooth gonothecal walls, in association with linear, extended gonads and smaller B-type microbasic mastigophores supported C. gulangensis sp. nov. to be unique compared to all the existing Clytia species. However, diagnosis characters for either medusae or hydroid alone are quite ambiguous. With tentacle numbers ranged from 24 to 36 and statocysts between successive tentacles about 1–2, medusae of C. gulangensis sp. nov. from field plankton specimens can be easily miss-identified to other related Clytia, e.g. C. hemisphaerica, C. gracilis and C. linearis. Meanwhile, pieces of colony bearing a single campanulated hydrotheca with blunt, triangular cusps or a smooth, cylindrical gonotheca cannot be attributed exclusively to an exact species, either. Thus, careful investigations on life cycle description would be essential to uncover detailed diagnosis characters both from hydroid and medusae to detect novel species and help resolve species revision (Zhou et al., Reference Zhou, Zheng, He, Lin, Cao and Zhang2013).
DNA barcoding offers great help for Clytia biodiversity studies by identifying species easily and quickly. While morphological studies offer great details about taxonomic placement for type specimens, immature individuals or fragments cannot be identified reliably using morphology alone (Bouillon & Boero, Reference Bouillon and Boero2000). For 17 Clytia species recorded from China seas, medusae of C. hemisphaerica, C. languida, C. linearis and C. xiamenensis all have equivalent number of tentacles and oval to linear gonads extending distal half of radial canals, which would show great resemblance with immature medusae of C. gulangensis sp. nov. as well (Huang, Reference Huang2008; Huang & Lin, Reference Huang and Lin2012; Zhou et al., Reference Zhou, Zheng, He, Lin, Cao and Zhang2013). However, COI based DNA barcoding effectively separated these ambiguous individuals by adequate genetic distance and monophyletic clustering (Figure 5; Table S2), as was addressed by Laakmann & Holst (Reference Laakmann and Holst2014) to attribute unsorted morphological groups to Clytia and Obelia species. Moreover, medusae of four other species collected in the same region as the present study are also distinctly identified through COI barcoding (Figure 5; Table S2). Due to its accuracy and regardless of morphological variation, DNA barcoding would be the first choice to achieve a better understanding about Clytia biodiversity from field zooplankton specimens.
Taxonomic position of C. gulangensis sp. nov. in typical Clytia clade were confirmed with both 16S and 18S genes. In the present maximum likelihood phylogenetic tree based on 16S plus 18S dataset, C. gulangensis sp. nov. was more closely related to C. elsaeoswaldae, C. xiamenensis, and C. hemisphaerica than other Clytia species (Figure 6). While both C. elsaeoswaldae and C. xiamenensis share inclined hydrothecal cusps with C. gulangensis sp. nov., the former two differ from the latter in having undulated gonothecal wall and distally placed gonads. Clytia hemisphaerica has rounded, symmetric cusps, but its medusae have the same tentacle numbers as C. gulangensis sp. nov. Clytia hemisphaerica has also the same undulated gonothecal wall as C. elsaeoswaldae and C. xiamenensis (Table 4). As was shown, diagnostic characters such as hydrothecal cusps, gonothecal outline, and medusae tentacles cannot fully explain the phylogenetic relationships among those species. Clytia hummelincki, C. paulensis and C. linearis all separated far from the C. gulangensis sp. nov. clade (Figure 6). Additionally, morphological characters such as the cup-like hydrotheca in C. hummelincki (Fraser, Reference Fraser1944), bibbed cusps in C. paulensis (Millard, Reference Millard1975) and inward folds in C. linearis (Thornely, Reference Thornely and Willey1900) allow to distinguish them from C. gulangensis sp. nov.
Phylogenetic reconstruction revealed complicated relationships among Clytia species. Maximum likelihood topology of 16S plus 18S phylogenetic reconstruction placed C. hummelincki as a sister clade to Clytiinae plus Obeliinae lineage. Unlike other Clytia species, C. hummelincki possesses a sub-hydrothecal spherule and a Campanulariinae-like colony growth pattern (Fraser, Reference Fraser1944; Boero et al., Reference Boero, Bouillon and Piraino1996; Kelmo & Attrill, Reference Kelmo and Attrill2003) and its taxonomic position was found to be basal to Campanulariinae and Clytiinae (Govindarajan et al., Reference Govindarajan, Boero and Halanych2006; Gravili et al., Reference Gravili, D'Ambrosio, Di Camillo, Renna, Bouillon and Boero2008). The remaining Clytia, viz. C. paulensis, C. linearis, C. folleata, C. gracilis, C. gulangensis sp. nov., C. elsaeoswaldae, C. hemisphaerica, and C. xiamenensis formed a well supported clade (Figure 6). Species with particular morphological characters, like bibbed cusps in C. paulensis and inward folds in C. linearis do not group together, which is in accordance with the observations made in phylogenetic analysis of C. gracilis-like species (Lindner et al., Reference Lindner, Govindarajan and Migotto2011).
In the context of C. gracilis-like species, the combined morphological and molecular evidences available tend to support these distinct genealogical lineages as separate species, rather than a single species with a strong population stratification. Firstly, though recognized by inclined hydrothecal cusps and smooth gonothecal walls, C. elsaeoswaldae, C. tottoni and C. xiamenensis are well morphologically described as distinct valid species from C. gracilis (Cornelius, Reference Cornelius, Barnes and Crothers1995; Schuchert, Reference Schuchert2003; Galea, Reference Galea2010; Lindner et al., Reference Lindner, Govindarajan and Migotto2011; Zhou et al., Reference Zhou, Zheng, He, Lin, Cao and Zhang2013). In the present study, the asymmetrical pyramidal cusps, smooth embayment margin, pod-like gonothecae and medusae with linear, extended gonads and tentacle number around 30 still reveal diagnostic differences from typical C. gracilis. Secondly, our phylogenetic results based on COI, 16S and 18S sequences also supported the polyphyletic C. gracilis-like clades, which are differently related to other Clytia species with distinct morphological characteristics, as was stressed by Govindarajan et al. (Reference Govindarajan, Boero and Halanych2006) and Lindner et al. (Reference Lindner, Govindarajan and Migotto2011). Finally, in a global molecular phylogeny of C. gracilis-like species, the found clades are not geographically delimited lineages. Individuals of Clytia cf. gracilis across the northern Atlantic Ocean (from Brazil and USA) showed minimal variations (e.g. Clytia cf. gracilis sp. B, Lindner et al., Reference Lindner, Govindarajan and Migotto2011). And C. xiamenensis, described as a new species from the same region as the present species, has also been found in Woods Hole and Maine, USA (Zhou et al., Reference Zhou, Zheng, He, Lin, Cao and Zhang2013) (Figures 5 and 6).
The problem of appropriate species concepts in Hydrozoa and the recognition of cryptic species vs mere sub-species level lineages has recently been brought into discussion by Schuchert (Reference Schuchert2014). Our data on Clytia species contribute to this ongoing debate by providing data on morphological and molecular variations in a local population of a putative cosmopolitan species complex. By adding similar data from many other populations, this will allow in future a more comprehensive view on the global level of both species diversity and phylogenetic relationships in the genus Clytia.
Supplementary materials and methods
The supplementary material referred to in this article can be found online at journals.cambridge.org/mbi.
ACKNOWLEDGEMENTS
We are grateful to Dr Peter Schuchert for offering original references. We also thank Weidi Yang for helping produce photograph, and Dangnni Zhang for preparing illustrations. Additionally, we thank Open and Sharing Platform of Equipment and Technology (OSPET) of COE of Xiamen University for providing equipments and technical support.
FINANCIAL SUPPORT
This work was supported by the National Natural Science Foundation of China (Grant No. 41006078), the Fundamental Research Funds for the Central Universities (Grant No. 2010121037), the Public Science and Technology Research Funds Projects of Ocean (Grant No. 201005012-3 and 201005015-5), and Marine Science Base Project for Scientific Research Training and Capacity Enhancement-Xiamen University (Grant No. J1210050).