INTRODUCTION
Besides the morphological variation with age and sex, some caprellid species show also considerable intraspecific variation. This is the case for Caprella penantis Leach, 1814, a world-wide distributed species of the Caprellidae, which could be a complex of different species in which it is difficult to understand if the morphological variation is intra- or interspecific (Guerra-García et al., Reference Guerra-García, Redondo-Gómez, Espina, Castillo, Luque, García-Gómez and Figueroa2006). Although there has been traditionally a gap in molecular studies on the Caprellidea, recently, different molecular approaches have been applied also for this group of amphipods. In this sense, Guerra-García et al. (Reference Guerra-García, Redondo-Gómez, Espina, Castillo, Luque, García-Gómez and Figueroa2006) showed, for the first time in caprellids, the validity of the RAPD technique as a tool for helping to solve taxonomic problems. Ito et al. (Reference Ito, Wada and Aoki2008) conducted the first molecular study to investigate the phylogenetic relationships among the Caprellidea based on the 18S rRNA. Ashton et al. (Reference Ashton, Stevens, Hart, Green, Burrows, Cook and Willis2008), using mitochondrial DNA, revealed multiple northern hemisphere introductions of the invader Caprella mutica Schurin, 1935. To our knowledge these are the only three works dealing with molecular tools and caprellids.
In the study of Guerra-García et al. (Reference Guerra-García, Redondo-Gómez, Espina, Castillo, Luque, García-Gómez and Figueroa2006), seven populations of Caprella penantis from the Strait of Gibraltar were morphologically and genetically compared among them and with other populations of the closest species Caprella dilatata Krøyer, 1843, to explore the intraspecific and interspecific genetic differentiation. Their results showed a clear separation between Caprella penantis and C. dilatata populations (only 8% similarity between them), supporting the morphological differences that indicate that both species are really different and valid species. However, all the populations of C. penantis from the Strait of Gibraltar were clustered together (85% similarity) indicating that they probably belong to the same species in spite of morphological variations among populations. Taking into account that in this previous study all the material was coming from the Strait of Gibraltar and that all the studied populations of C. penantis belonged to the form simulatrix, we have increased significantly the number of samples for the present study and we have considered populations from different areas of the world, not only from the Strait of Gibraltar, including also additional populations of C. dilatata and Caprella andreae Mayer, Reference Mayer1890.
MATERIALS AND METHODS
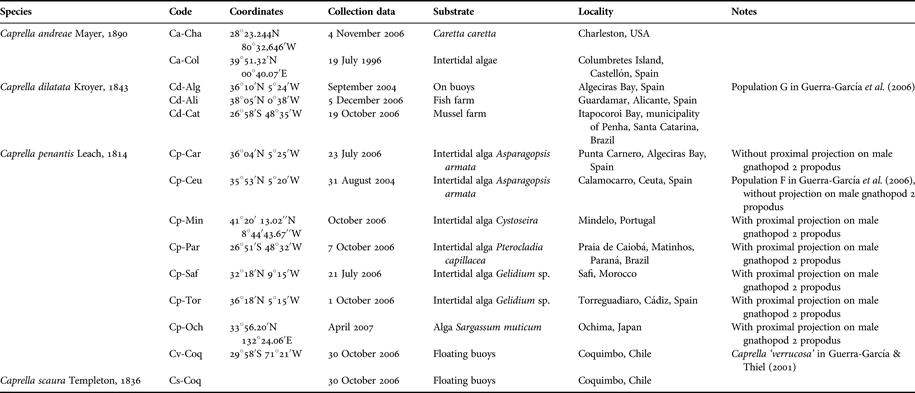
Specimens of eight populations of C. penantis, three populations of C. dilatata and two populations of C. andreae, were collected from different sites of the world (Table 1). For the genetic analysis, the species Caprella scaura Templeton, 1836, separate from the ‘acutifrons’ complex was also included as an ‘outgroup’. The caprellids were fixed in 95% ethanol. DNA was extracted using the DNeasy Blood and Tissue Kit (QIAGEN). Pooled individuals were used because relatively small quantities of DNA were available per individual and the aim of this study was to compare populations (Thomas et al., Reference Thomas, Blinn and Keim1997). Determination of the concentration and purity of the DNA, RAPD protocol and amplification conditions were the same as those of Guerra-García et al. (Reference Guerra-García, Redondo-Gómez, Espina, Castillo, Luque, García-Gómez and Figueroa2006). Amplification products were analysed by electrophoresis in 2.0% agarose (Seakem ® LE Agarose, Lonza) gels run at 90 V for 2.5 hours, stained with ethidium bromide and visualized by illumination with UV light. For band size determination, a 123 bp DNA ladder (Sigma-Aldrich) was loaded in lanes flanking groups of about 7 samples in each gel. All amplifications were repeated at least twice to check the stability of amplification products. Bands were scored as present (1) or absent (0) (Tansley & Brown, Reference Tansley and Brown2000; Costa et al., Reference Costa, Cunha, Neuparth, Theodorakis, Costa and Shugart2004a, Reference Costa, Neuparth, Theodorakis, Costa and Shugartb) by eye and only unequivocal bands were scored, with weak bands not being included. To ensure data accuracy, all samples were scored twice by the same individual, and the second round of scoring was conducted without reference to data from the initial round (Star et al., Reference Star, Apte and Gardner2003). The coefficient of Nei & Li (Reference Nei and Li1979), recommended to be employed in RAPD analysis (Lamboy, Reference Lamboy1994), and used most often in amphipods to estimate the degree of genetic differentiation between populations or species (Stewart, Reference Stewart1993; Culver et al., Reference Culver, Kane and Fong1995), was applied to calculate a similarity matrix (NTSYS-pc computer package, version 1.8 (Rohlf, Reference Rohlf1993)). The rest of the statistical treatment (UPGMA, SHAN, construction of the phenogram and the cophenetic value matrix) was conducted according to Guerra-García et al. (Reference Guerra-García, Redondo-Gómez, Espina, Castillo, Luque, García-Gómez and Figueroa2006).
Table 1. List of species used for the molecular study using RAPD.
RESULTS AND DISCUSSION
Thirteen primers yielded a total of 154 consistently well-amplified DNA fragments with an average of 11.85 bands per primer (Table 2). Overall, 23 of these bands were monomorphic (15%) and 131 were polymorphic (85%). Primers 5 and 10 had the highest percentage of polymorphic fragments between samples (both 100%). Cophenetic correlation analysis strongly supported the reliability of the phenogram based on the original distance matrix (r = 0.845). The phenogram (Figure 1) separated clearly C. dilatata and C. andreae from the populations of C. penantis, supporting the validity of these species, traditionally considered altogether under the old ‘acutifrons’ complex. Populations of C. penantis from Spain, Portugal, Morocco, Japan and Brazil were clustered together in the RAPD analysis, indicating that, probably, all the specimens of C. penantis could belong to the same species. From the eight populations of this species included in the analysis, only the population of Punta Carnero (Cp-Car) and Ceuta (Cp-Ceu) belonged to the form simulatrix (without proximal projection on male gnathopod 2 propodus). The remaining populations were all provided with a proximal projection belonging, at least, to the forms testudo and lusitanica. Interestingly, the cluster did not show the populations with projection within the same group. Population Cp-Ceu was more similar to the Japanese and Brazilian specimens, that were provided with proximal projection, and Cp-Car was more similar to the population of Mindelo (Cp-Min) and to the population of Torreguadiaro (Cp-Tor), also provided with projection in male gnathopod 2 propodus. According to these results, it seems that the presence/absence of this proximal projection should not be strong enough as morphological difference for separating species, and that the different forms of C. penantis could be explained by intraspecific variation. The only population which showed higher genetic differentiation within the C. penantis complex was the form gibbosa from Coquimbo, Chile (Mayer, Reference Mayer1890, p. 52). This form was referred as Caprella verrucosa Boeck, 1871, in Guerra-García & Thiel (Reference Guerra-García and Thiel2001) and Thiel et al. (Reference Thiel, Guerra-García, Lancellotti and Vásquez2003) due to the presence of abundant dorsal tubercles on pereonites, especially on pereonites 5, 6 and 7, and the robustness of antenna 1 (figure 4 in Guerra-García & Thiel, Reference Guerra-García and Thiel2001). However, the morphology of the specimens changes considerably along the coast of Chile (Thiel et al., Reference Thiel, Guerra-García, Lancellotti and Vásquez2003). Further molecular and morphological phylogenetic studies are necessary to elucidate if the material from Coquimbo is really belonging to Caprella penantis, C. verrucosa or to a different undescribed species. One of the limitations of the RAPD molecular approach is that we cannot conclude which genetic difference as measured by the Nei & Li coefficient is necessary to establish specific differences. Consequently, this method should be used with care to make definite statements about the limits considered to distinguish species, and future studies based on mtDNA, 18S rRNA, ISSR should be necessary to consider if Caprella penantis f. gibbosa should be erected as a new species differing from C. penantis and C. verrucosa. Unfortunately, material of the real C. verrucosa from California or Japan were not available in the present study to be compared with the ‘C. verrucosa’ (C. penantis f. gibbosa) from Coquimbo.

Fig. 1. Phenogram of the fourteen populations studied, based on similarities generated by UPGMA clustering of RAPD band scores.
Table 2. Number of amplification products (bands) obtained per primer in the random amplified polymorphic DNA (RAPD) analysis of the cosmopolitan Caprella penantis and related species.

Caprella penantis is regarded as one of the most problematic caprellids throughout the world, since this species has been recorded under several species or subspecies names from the temperate regions of the world and the need for genetic studies to determine its nomenclatural status at each locality has been pointed out in most of the taxonomic caprellids studies (McCain, Reference McCain1968; Laubitz, Reference Laubitz1972). In Mayer's monographs (Reference Mayer1890, Reference Mayer1903) he described nineteen forms of the ‘acutifrons’ group (forms typica, minor, tabida, tibada, neglecta, gibbosa, andreae, carolinensis, virginia, lusitanica, natalensis, porcellio, simulatrix, testudo, angusta, incisa, verrucosa, borealis and cristibrachium). Several of these forms have already been given specific rank. Forms typica and minor have been assigned to Caprella dilatata (McCain, Reference McCain1968) and probably forms tabida and tibada also belong to C. dilatata, although its taxonomic status under Caprella penantis is still under discussion (Guerra-García et al., Reference Guerra-García, Redondo-Gómez, Espina, Castillo, Luque, García-Gómez and Figueroa2006). Caprella neglecta Mayer, Reference Mayer1890, was also considered as a valid species (Vassilenko, Reference Vassilenko1967; Laubitz, Reference Laubitz1972), form andreae was assigned to Caprella andreae (McCain, Reference McCain1968), forms natalensis and porcellio to Caprella natalensis Mayer, Reference Mayer1903 (Laubitz, Reference Laubitz1972), forms incisa, verrucosa, borealis and cristibrachium to Caprella incisa Mayer, Reference Mayer1903, C. verrucosa, C. borealis Mayer, Reference Mayer1903, and C. cristibrachium Mayer, Reference Mayer1903, respectively (Utinomi, Reference Utinomi1943; Dougherty & Steinberg, Reference Dougherty and Steinberg1953; McCain, Reference McCain1968; Laubitz, Reference Laubitz1972). The form angusta was considered as Caprella angusta Mayer, Reference Mayer1903, by Dougherty & Steinberg (Reference Dougherty and Steinberg1953) and Laubitz (Reference Laubitz1970); but Laubitz (Reference Laubitz1972) questioned the validity of this species. Consequently, its position is still unclear. The remaining forms gibbosa, carolinensis, virginia, lusitanica, testudo (all these forms with proximal projection in the propodus of male ganthopod 2) and simulatrix (without projection) are, consequently, still classified under the species Caprella penantis (McCain, Reference McCain1968; Laubitz, Reference Laubitz1970, Reference Laubitz1972; Krapp-Schickel, Reference Krapp-Schickel and Ruffo1993). The present study shows that, using RAPD, all C. penantis forms seem to be very similar from the molecular point of view, suggesting that there are no clear evidences to separate them as different species. The presence/absence of the projection seems not to be related with a species differentiation but with phenotypical intraspecific variation. The morphological variation among populations of C. penantis could be mainly influenced by the ecological characteristics of the habitat. In this sense, Bynum (Reference Bynum1980) conducted a morphometric study using C. penantis from coastal and estuarine sites in North Carolina and found a gradient of forms related to the degree of exposure to turbulence. Body parts and appendages associated with grasping the substrate of the same caprellid species are capable of modification depending on the degree of wave exposure, thus exhibiting ecological plasticity or ‘ecotopic variation’ (Caine, Reference Caine1989; Guerra-García, Reference Guerra-García2001). Caprellid species in environments with strong wave action tend to have a more robust body than those living in calmer places (Hirayama & Kikuchi, Reference Hirayama and Kikuchi1980).
The RAPD analysis (Welsh & McClelland, Reference Welsh and McClelland1990; Williams et al., Reference Williams, Kubelik, Livak, Rafalski and Tingey1990) requires no prior knowledge of DNA sequence (Hadrys et al., Reference Hadrys, Balick and Schierwater1992) and is faster and cheaper than many other molecular techniques (Costa et al., Reference Costa, Neuparth, Theodorakis, Costa and Shugart2004b), but this method also has some limitations and low reproducibility (Williams et al., Reference Williams, Kubelik, Livak, Rafalski and Tingey1990; Pérez et al., Reference Pérez, Albornoz and Domínguez1998). In fact, in spite of using the same methodology and similar laboratory conditions, the number of bands obtained per primer in the study carried out by Guerra-García et al. (Reference Guerra-García, Redondo-Gómez, Espina, Castillo, Luque, García-Gómez and Figueroa2006, p. 102; Table 2) differs from the number of the present study (see Table 2). However, although RAPD analysis must be interpreted with caution, the results of the present study based on a wide range of samples throughout the world support the preliminary observations made by Guerra-García et al. (Reference Guerra-García, Redondo-Gómez, Espina, Castillo, Luque, García-Gómez and Figueroa2006) based exclusively on material from the Strait of Gibraltar. Additional sampling and future studies based on a larger set of molecular markers and techniques (e.g. mtDNA, 18S rRNA, ISSR, etc.) are necessary to confirm the patterns obtained with RAPD analyses.
ACKNOWLEDGEMENTS
Financial support for this work was provided by the Ministerio de Educación y Ciencia (Project CGL2007-60044/BOS) co-financed by FEDER funds, and by the Consejería de Innovación, Ciencia y Empresa, Junta de Andalucía (Project P07-RNM-02524). Special thanks to M. Thiel, S. Masunari, R. King, A. Engelen, J. Templado and M. Vázquez-Luis for providing some of the specimens used in the present study. We are very grateful also to M.P. Cabezas-Cabezas for revising the English in the manuscript.





