INTRODUCTION
The family Pectinidae is one of the most conspicuous groups of Bivalvia and an important component of ecosystems, constituting the support of artisanal and industrial fisheries (Waller, Reference Waller and Shumway1991). The phylogenetic relationships of pectinids have been explored using morphological (e.g. Thiele, Reference Thiele1935; Hertlein, Reference Hertlein, Moore and Teichert1969; Waller, Reference Waller1986, Reference Waller and Shumway1991, Reference Waller, Shumway and Parsons2006) and molecular data (e.g. Canapa et al., Reference Canapa, Barucca, Marinelli and Olmo2000; Barucca et al., Reference Barucca, Olmo, Schiaparelli and Canapa2004; Saavedra & Peña, Reference Saavedra and Peña2006; Puslednik & Serb, Reference Puslednik and Serb2008; Alejandrino et al., Reference Alejandrino, Puslednik and Serb2011; Feng et al., Reference Feng, Li, Kong and Zheng2011; Krause & von Brand, Reference Krause, von Brand, Shumway and Parsons2016). The most accepted classification system derived from morphological characters is the one of Waller (Reference Waller, Shumway and Parsons2006), who divided the family Pectinidae into three subfamilies: Camptonectinae, Chlamydinae and Pectininae, using microsculptural shell features and morphological characteristics of the juveniles. Waller's hypothesis of subfamily relationships generally matched the molecular phylogeny of Alejandrino et al. (Reference Alejandrino, Puslednik and Serb2011), the molecular phylogenetic study with the most comprehensive taxon sampling to date, with only the placement of Aequipectini as a major difference (Serb, Reference Serb, Shumway and Parson2016). This molecular study and other contributions published in the last two decades (see Waller, Reference Waller, Shumway and Parsons2006; Krause & von Brand, Reference Krause, von Brand, Shumway and Parsons2016; Serb, Reference Serb, Shumway and Parson2016), although extensive, did not include taxa from the south-western Atlantic.
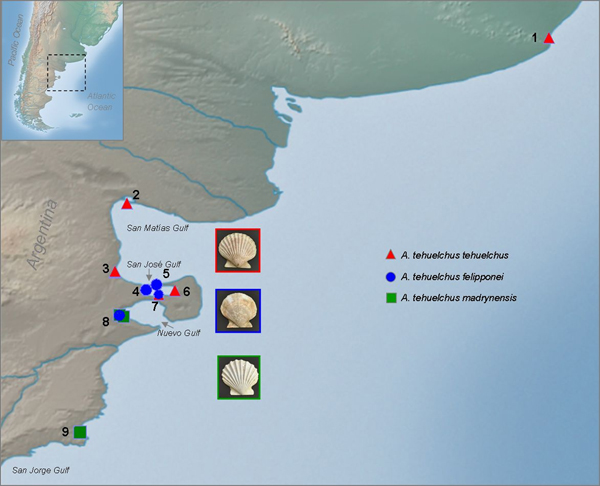
In the south-western Atlantic Ocean, scallops are represented by Aequipecten (Chlamys) tehuelchus (d'Orbigny 1842), Austrochlamys natans (Philippi 1845), Cyclopecten falklandicus Dell, 1964, Flexopecten felipponei (Dall 1922) and Zygochlamys patagonica (King & Broderip 1832). Only two of these species, Z. patagonica and the Tehuelche scallop A. tehuelchus, are commercially exploited in the south-western Atlantic (Orensanz et al., Reference Orensanz, Pascual, Fernandez and Shumway1991). The Tehuelche scallop is the target of small inshore fisheries that operate within the northern Patagonian gulfs, involving dredging and commercial diving, and they are of considerable significance for the local economies (Orensanz et al., Reference Orensanz, Pascual, Fernandez and Shumway1991; Soria et al., Reference Soria, Orensanz, Morsán, Parma, Amoroso, Shumway and Parsons2016). Despite their importance, the status and phylogenetic relationships of these taxa have not been fully elucidated.
The Tehuelche scallop is a warm-temperature species endemic to the Argentine biogeographic province, inhabiting shallow shelf bottoms from Rio de Janeiro, Brazil (23°S) to the north of the San Jorge Gulf, Argentina (45°S) (Castellanos, Reference Castellanos1971). It exhibits two geographic variants which differ in shell morphology (Figure 1). The ‘tehuelchus’ phenotype is distributed in the warm-temperate Argentine biogeographic province (north of 42°30′S), typically has 14–18 ‘squamous’ and relatively rounded ribs, and is the only one that forms large banks which support important fisheries mostly confined to the northern Patagonian gulfs (Orensanz et al., Reference Orensanz, Pascual, Fernandez and Shumway1991). The ‘madrynensis’ phenotype is distributed in the cold-temperate Magellanic province (42°30′–45°S), presents 11–14 comparatively softer and more pronounced ribs (the number of ribs is not correlated with the size of the valve) and lives at low densities (Castellanos, Reference Castellanos1971). This southern geographic variant has been described as a separate species, Aequipecten madrynensis (Lahille 1906) (Castellanos, Reference Castellanos1971) or as a subspecies, A. tehuelchus madrynensis (Bavay 1906) (Orensanz et al., Reference Orensanz, Pascual, Fernandez and Shumway1991). The correlation between morphological and genetic differences of the two forms was explored by Real et al. (Reference Real, Julio, Gardenal and Ciocco2004), who found no evidence of interspecific genetic differentiation using allozymes. Another related species that lives in sympatry with tehuelchus and madrynensis is the non-commercial pectinid Flexopecten felipponei (Dall 1922) (Figure 1), which has rarely been recorded in the south-western Atlantic Ocean (Waller, Reference Waller and Shumway1991), distributed in rocky and sandy bottoms from 36°S to San Matías and Nuevo Gulfs (43°S) (Castellanos, Reference Castellanos1970, Reference Castellanos1971). Orensanz et al. (Reference Orensanz, Pascual, Fernandez and Shumway1991) proposed that F. felipponei would be a phenotypic variant of A. tehuelchus, based on examination of soft tissue of numerous individuals with intermediate characters and the presence of simultaneous hermaphroditism. The phylogenetic relationships among the two geographic variants of the Tehuelche scallop and F. felipponei remains unknown.

Fig. 1. Phenotype variation of the Tehuelche scallop. To the left: Flexopecten ‘felipponei’ of two different sizes; to the right at the top, Aequipecten tehuelchus ‘tehuelchus’ and below, Aequipecten tehuelchus ‘madrynensis’.
The main goal of this study was to place the two putative subspecies (tehuelchus and madrynensys), as well as the sympatric F. felipponei, within a partial phylogenetic framework of the family Pectinidae and to investigate their taxonomic status using four molecular markers. The phylogenetic reconstruction will allow us to infer relationships among these taxa within the pectinid family and to explore the hypothesis that A. t. tehuelchus, A. t. madrynensys and F. felipponei correspond to different phenotypes of the same species. This study utilized the phylogenetic species concept (Nixon & Wheeler, Reference Nixon and Wheeler1990).
MATERIALS AND METHODS
Sample collection
Scallops were obtained from the subtidal of 10 localities distributed along the south-western Atlantic Ocean (Figure 2, Table 1) covering the distribution of the three taxa. All Aequipecten scallops collected north of 42°30′S were of the ‘tehuelchus’ variant while all collected from Golfo Nuevo to the south (up to 45°S) were of the ‘madrynensys’ variant. All specimens were fixed in 96% ethanol.
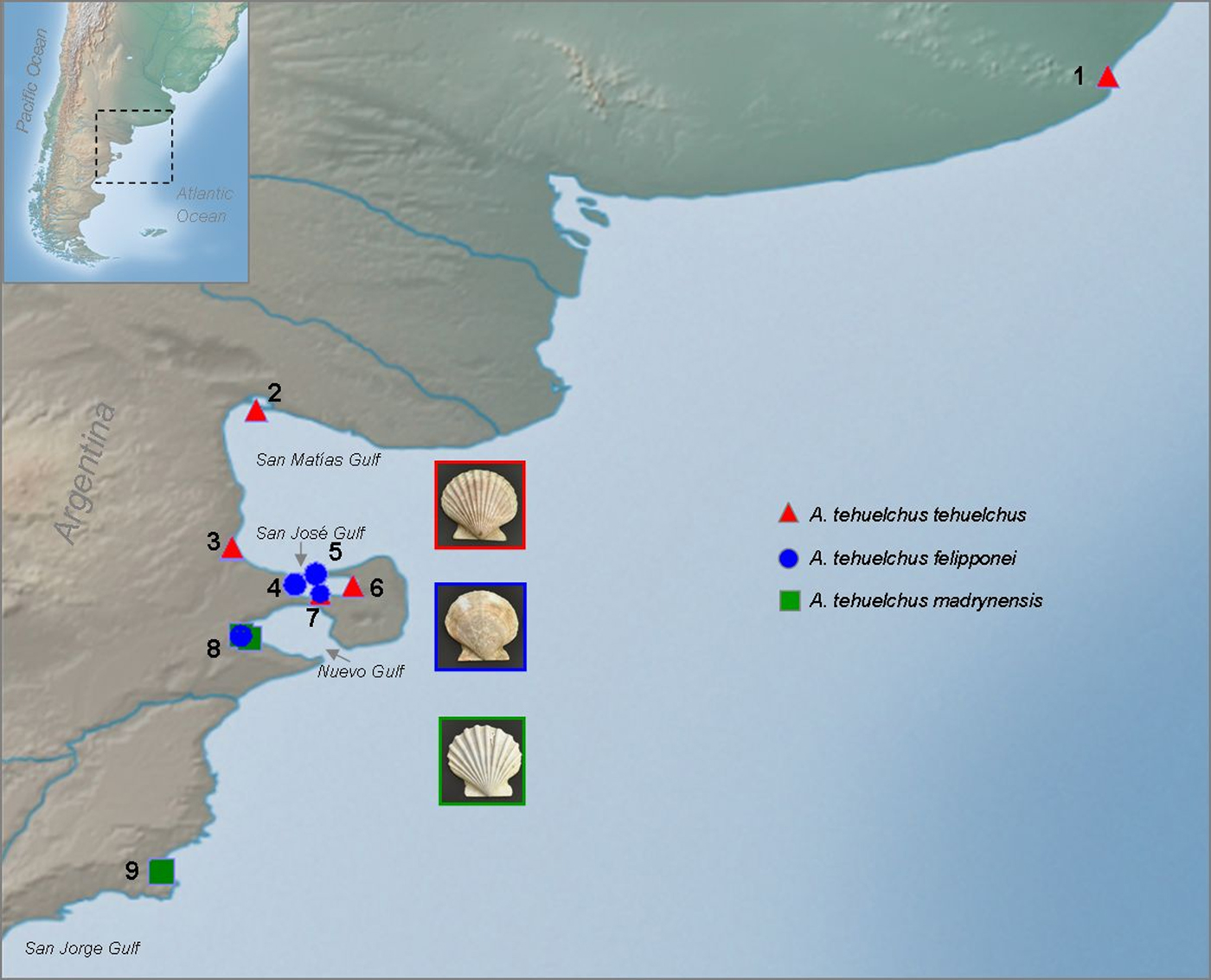
Fig. 2. Sampling localities of each phenotype along the south-western Atlantic coast: (1) Mar del Plata; (2) Villarino beach; (3) Puerto Lobos, (4) La Tapera; (5) Larralde beach; (6) Punta Conos; (7) Punta Buenos Aires; (8) Puerto Madryn I, (9) Puerto Madryn II and (10) Camarones (for details see Table 1).
Table 1. General information on samples analysed as part of this study.

a Golfo San Matías, Río Negro: Specimens were collected in sublittoral zone during ‘Holberg 02’ campaign of Patagonian scallop prospection and by-catch with a bottom trawl.
DNA extraction, amplification and sequencing
Genomic DNA was isolated from the adductor muscle using the phenol-chloroform protocol (Sambrook et al., Reference Sambrook, Fritsch and Maniatis1989). We amplified fragments of two mitochondrial ribosomal genes, 12S (domain III) and 16S (domain IV and V) rRNA, of 394 bp and 465 bp (aligned length), and two nuclear genes, the large ribosomal subunit (28S), of 745 bp (aligned length), and the Histone H3 of 295 bp (aligned length). We chose these genes due to the high availability of related species sequences to compare in the GenBank database. In addition, to efficiently distinguish these three taxa, we tried hard to amplify, with numerous combinations of primers and conditions, the cytochrome c oxidase subunit I (COI), but we did not have success. The primers used for both amplification and sequencing of the selected genes are listed in Table 2. When possible, we sequenced two specimens per locality for each gene for a total of 93 sequences (26 for 16S rRNA, 23 for 12S rRNA, 20 for 28S rRNA and 24 for H3). To amplify the genes we used Tsg DNA polymerase (Bio Basic Inc., Canada). The protocol used included an initial denaturing temperature of 94°C for 1 min; followed by 30 cycles of 94°C for 30 s; an annealing temperature of 50°C for 30 s; 72°C for 1 min; and a final extension at 72°C for 10 min. After extraction and amplification the DNA was visualized by UV transillumination in 1% agarose gels stained with green gel (BIOTUM). Extractions, amplifications, purification of PCR products and sequencing of both strands of DNA samples were performed in the Laboratory of Molecular Biology (IDEAus – CONICET, Argentina). DNA sequences were edited in CodonCode Aligner v 2.0.4 and aligned using default parameters with Clustal W version 1.75 (Thompson et al., Reference Thompson, Higgins and Gibson1994). All DNA sequences were deposited in GenBank and their accession numbers (KY055443-KY055527, KY070308-KY070314) are detailed in Table 1. The estimates of evolutionary divergence over sequence pairs within and between A. ‘tehuelchus’, A. ‘madrynensis’ and F. ‘feliponei’ were calculated using ‘p-distance’ (Kimura, Reference Kimura1980) in MEGA v5 (Tamura et al., Reference Tamura, Peterson, Peterson, Stecher, Nei and Kumar2011).
Table 2. Primers used in this study.

Phylogenetic analysis
To assess the degree of saturation of nucleotide saturations of mitochondrial gene regions, a test of substitution saturation (Xia & Lemey, Reference Xia, Lemey, Lemey, Salemi and Vandamme2009) was performed in DAMBE v5 (Xia, Reference Xia2013). To determine if different partitions of the data have significantly different signals and following the concatenation of all genes, a partition homogeneity test with 1000 replications in PAUP (Swofford, Reference Swofford1998) was carried out. Sequences of related species were downloaded from GenBank (Table 3). Zygochlamys amandi Soot-Ryen 1959, Z. patagonica (King 1832) and Veprichlamys jousseaumei (Bavay, 1904) (Pectinoidea: Pectinidae) were selected as outgroups. This selection was based on the robust phylogeny of Alejandrino et al. (Reference Alejandrino, Puslednik and Serb2011).
Table 3. DNA sequences used in this study.

Three methods were utilized for phylogenetic reconstruction of each gene (Supplementary material SM1) and also of the concatenated dataset: Maximum parsimony (MP), Maximum pikelihood (ML) and Bayesian inference (BI).
Maximum parsimony analysis was carried out using TNT (Goloboff et al., Reference Goloboff, Farris and Nixon2008). We used the traditional search, including a tree bisection–reconnection (TBR) as swapping algorithm and 10 trees saved by replication. Then, a strict consensus tree was calculated. Branch support was evaluated through bootstrapping (500 replicates).
The Akaike Information Criterion (AIC), implemented in jModelTest v 2.1.10 (Darriba et al., Reference Darriba, Taboada, Doallo and Posada2012), was applied to each dataset to find the models of evolution that best fit the data. The General Time Reversible model (GTR + I + G, Tavaré, Reference Tavaré1985) was the best-fit substitution model in the four datasets (Supplementary material SM1, Table 1). The selected model was used in ML analysis on the concatenated dataset, conducted with RAxML 7.4.2 (Stamatakis, Reference Stamatakis2006) implemented in raxmlGUI 1.3 (Silvestro & Michalak, Reference Silvestro and Michalak2012), with a rapid bootstrapping analysis (500 replicates).
The phylogeny reconstructed with BI was estimated with different substitution (HKY + G + I, Hasegawa et al., Reference Hasegawa, Kishino and Yano1985, and GTR + G + I, Tavaré, Reference Tavaré1985) and clock (strict and relaxed) models (Supplementary material SM1). The marginal-likelihood scores of the posterior distributions were compared using Bayes factors (BFs, Kass & Raftery, Reference Kass and Raftery1995) with two different methods: harmonic mean estimation (HME, Newton & Raftery, Reference Newton and Raftery1994) and a posterior simulation-based analogue of the Akaike information criterion through Markov Chain Monte Carlo (MCMC) analysis (AICM, Raftery et al., Reference Raftery, Newton, Satagopan, Krivitsky and Bernardo2007) implemented in Tracer v1.6 (Rambaut et al., Reference Rambaut, Suchard, Xie and Drummond2014). Bayesian reconstructions were conducted under a Yule process for species-level phylogenies as a tree prior, using BEAST v. 1.8.3 (Drummond et al., Reference Drummond, Suchard, Xie and Rambaut2012) with a MCMC simulation for 60 million generations for the concatenated dataset, sampling trees every 1000 generations with a burn-in of 10%. Convergence diagnostics were conducted in Tracer and reliable ESS values (>200) were ensured. Then, the maximum clade credibility tree was generated from the combined trees in TreeAnnotator v 1.6.1 (Drummond et al., Reference Drummond, Suchard, Xie and Rambaut2012). Finally, the editing of the trees was carried out in Figtree v 1.4 (Morariu et al., Reference Morariu, Srinivasan, Raykar, Duraiswami, Davis, Koller, Schuurmans, Bengio and Bottou2008).
RESULTS
Phylogenetic analysis
The observed saturation indexes were significantly different and lower than the expected indexes, Iss < Iss.c (0.139 < 0.369–0.6756; P < 0.05) for the 12S rRNA and Iss < Iss.c (0.115 < 0.3847–0.6912; P < 0.05) for the 16S rRNA, suggesting that our mitochondrial data show little saturation. The partition homogeneity test was not significant (P = 0.01) suggesting that there is no conflict between partitions, so all genes were combined in the subsequent analyses.
The MP analysis resulted in four equally parsimonious trees with 2441 steps for the concatenated dataset. The General Time Reversible models (GTR + I + G) were selected for ML estimation based on the AIC criterion. In the case of Bayesian inference, Bayes factors based on HME and AICM favoured the GTR + I + G and the strict clock models over the HKY + I + G and the relaxed clock models for the concatenated dataset (Supplementary material SM2).
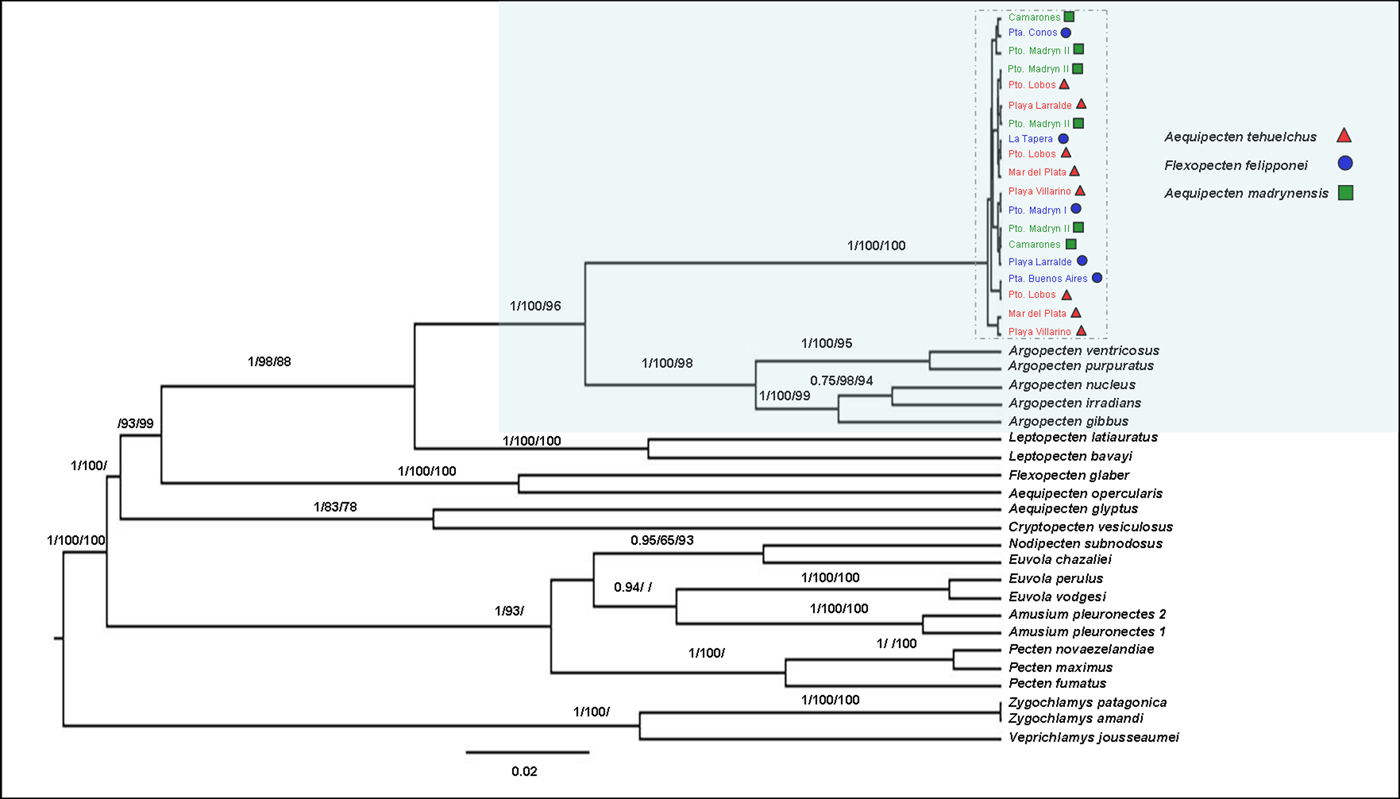
The phylogenetic reconstruction based on maximum parsimony, maximum likelihood and Bayesian analyses of the concatenated dataset (Figure 3) and of separate genes (Supplementary material SM1) supports the hypothesis that the two putative subspecies of the Tehuelche scallop, A. t. tehuelchus and A.t. madrynensys, and F. felipponei form a monophyletic clade, without differentiating at the specific level. The genetic distances between and within the 16S rRNA sequences from the three forms were 0.1%. This monophyletic clade is more closely related to the genus Argopecten than to the other members of the genus Aequipecten. Argopecten and Lectopecten are monophyletic, while Aequipecten is resolved as polyphyletic. From a phylogenetic point of view, Aequipecten is an artificial group, therefore does not reflect the phylogeny.
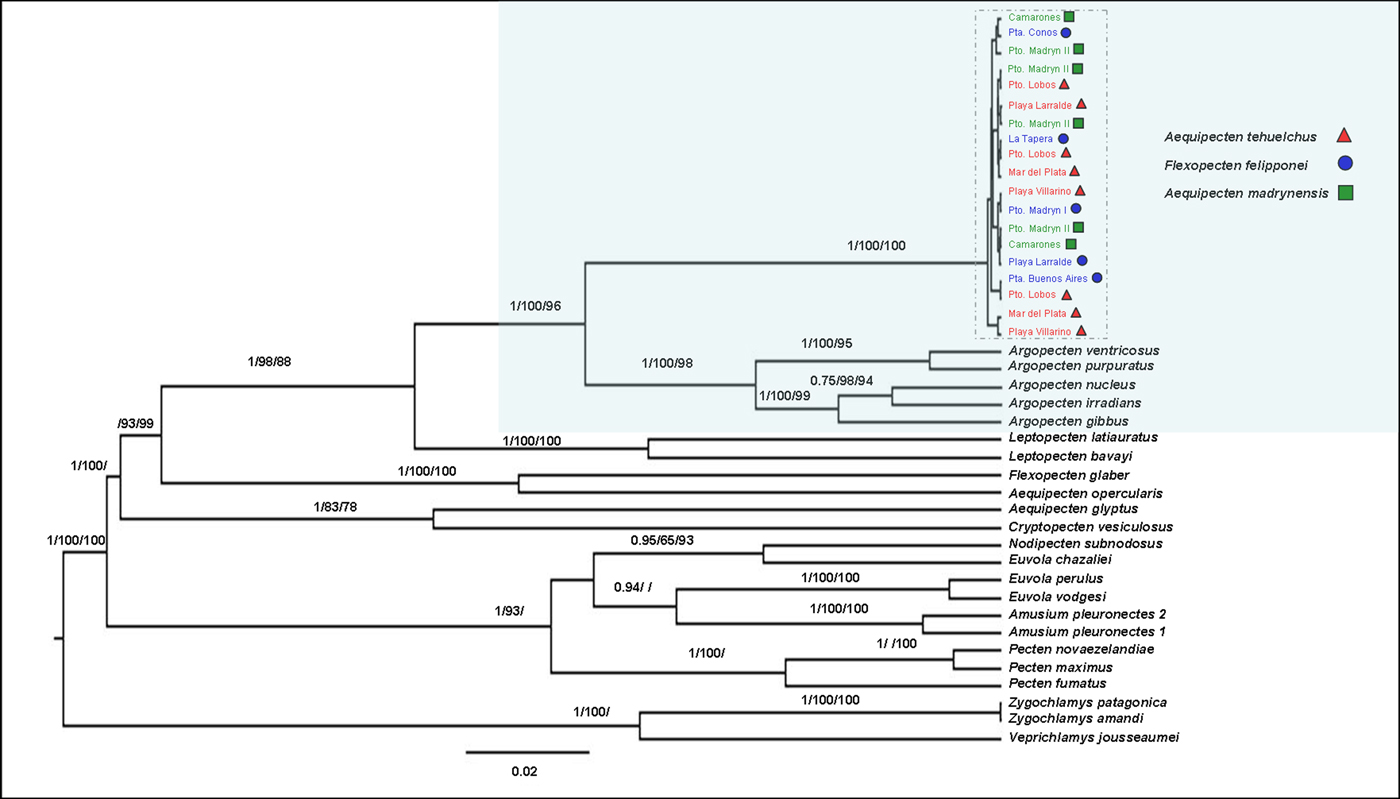
Fig. 3. Bayesian tree for the Tehuelche scallop based on the concatenated dataset of 12S rRNA −16S rRNA −28S rRNA -H3. Numbers above the branches represent the Maximum parsimony and Maximum likelihood bootstrap values and Bayesian posterior probabilities (>60 only) for the supported nodes.
DISCUSSION
We have placed the Tehuelche scallop within a partial phylogenetic framework of the family Pectinidae outlined by Alejandrino et al. (Reference Alejandrino, Puslednik and Serb2011). The hypothesis that Aequipecten tehuelchus, A. madrynensys and Flexopecten felipponei correspond to different phenotypes of the same species was supported by the results of this study. The estimated interpopulational genetic distances obtained for the Tehuelche scallop were similar to the intraspecific variation expected for Pectinids (0.1% average, 1% threshold, Feng et al., Reference Feng, Li, Kong and Zheng2011). Our results are consistent with those of Real et al. (Reference Real, Julio, Gardenal and Ciocco2004), who found no evidence of interspecific genetic differentiation using allozymes. Of their 10 loci, five had significant FST values, averaging F ST = 0.032, suggesting some genetic differentiation along the Patagonian coast. However, the level of genetic differentiation was not sufficient to justify the classification of the phenotypes ‘tehuelchus’ and ‘madrynensis’ as different species or subspecies, as had been proposed by Castellanos (Reference Castellanos1971) and Orensanz et al. (Reference Orensanz, Pascual, Fernandez and Shumway1991). Consistently, Huber (Reference Huber2010), using morphological characters (number of ribs, strength of sculpture on the main ribs and colour), concluded that there was only one highly variable species of Aequipecten from southern Brazil to southern Argentina. The current evidence suggests the occurrence of high phenotypic plasticity, resulting in three different phenotypes which are genetically similar.
Phenotypic plasticity, i.e. the capacity of a genotype to produce different phenotypes under varying environmental conditions (Via, Reference Via and Real1994), is a common trait in marine molluscs (e.g. Vermeij, Reference Vermeij1973; Melatunan et al., Reference Melatunan, Calosi, Rundle, Widdicombe and Moody2013) and has been used to explain differences in shell shapes in several bivalve populations (Krapivka et al., Reference Krapivka, Toro, Alcapán, Astorga, Presa, Pérez and Guiñez2007; Leyva-Valencia et al., Reference Leyva-Valencia, Álvarez-Castañeda, Lluch-Cota, González-Pelez, Pérez-Valencia, Vadopalas, Ramírez-Pérez and Cruz-Hernández2012). The occurrence of morphological adaptation of shell shapes to different local environmental conditions appears likely in the Tehuelche scallop given the geographic segregation of the two phenotypes (Figure 1). This, however, cannot be the case of the smooth phenotype F. felipponei, which has a sympatric distribution with the other two phenotypes. An alternative explanation that we cannot entirely discard is that the molecular markers used in this study may not be sufficiently variable to resolve the status of these three forms. Future studies should include more variable markers such as the cytochrome c oxidase subunit I (COI), which we failed to amplify.
With regard to within-population differences, Real et al. (Reference Real, Julio, Gardenal and Ciocco2004) found that the San Jose Gulf population of Tehuelche scallops was the most distant genetically, which they attributed to different hydrodynamic conditions of the study areas. According to Orensanz (Reference Orensanz1986), the Tehuelche scallops from San José Gulf have an outline that is more typical of sedentary species, characterized by higher valves with more asymmetric auricles than those from San Matías Gulf. Such sedentary outline might improve scallop attachment to the bottom and thus be favoured by selective pressures in high-current environments (Stanley, Reference Stanley1970, Reference Stanley1972; Orensanz et al., Reference Orensanz, Pascual, Fernandez and Shumway1991). Further efforts to understand the effect of the environment on the genetic structure of the Tehuelche scallop would require the use of microsatellites. Although some advances have been made in the development of microsatellites in this species (Domínguez-Contreras et al., Reference Domínguez-Contreras, Munguía-Vega, Getino-Mamet, Soria and Parma2017), the genetic structure of the population has not been studied to date. Based on a model of water circulation in the south-western Atlantic Ocean proposed by Tonini & Palma (Reference Tonini and Palma2017), we hypothesized that the northward coastal current described could connect the scallop populations along the Argentinean coast. Amoroso et al. (Reference Amoroso, Parma, Orensanz and Gagliardini2011), postulated a physical mechanism capable of dispersing larvae over long distances from San José Gulf into the adjacent San Matias Gulf. It would be interesting to investigate, using microsatellites, if there are intraspecific genetic differentiations among the populations in relation with these patterns of circulation.
Phylogenetic relationships
The non-commercial Flexopecten felipponei formed a monophyletic group with the two subspecies of Aequipecten tehuelchus. Waller (Reference Waller and Shumway1991, Reference Waller, Shumway and Parsons2006) concluded that Flexopecten s.s. would be related to Aequipecten and restricted to the Mediterranean and adjacent eastern Atlantic with a single ‘outlier’ species, F. felipponei, occurring in the western South Atlantic. Orensanz et al. (Reference Orensanz, Pascual, Fernandez and Shumway1991), on the examination of soft tissue and simultaneous hermaphroditism of numerous individuals with intermediate characters, proposed that ‘F.’ felipponei would be only a phenotypic variant of A. tehuelchus, a hypothesis supported by our results. We found that Flexopecten glaber is grouped together with Aequipecten opercularis within the Pectinidae, both being phylogenetically distant from F. felipponei. This result is in agreement with those of Canapa et al. (Reference Canapa, Barucca, Marinelli and Olmo2000), but in contrast with the results of Waller (Reference Waller and Shumway1991) who placed Flexopecten as a member of the Decatopecten group, distant from Aequipecten.
Our phylogenetic reconstruction suggests that the clade formed by the three morphotypes of the Tehuelche scallop distributed along the south-western Atlantic Ocean is more closely related to the genus Argopecten from the Eastern Pacific and North and Central-Western Atlantic than to the other members of the genus Aequipecten, including the type species of the genus, Aequipecten opercularis (Linnaeus 1758), a species distributed in the Norwegian and Mediterranean seas (Huber, Reference Huber2010). Aequipecten tehuelchus was originally classified in Chlamys (d'Orbigny 1842) and considered a species of this genus until Waller (Reference Waller and Shumway1991) and Del Río (Reference Del Río1992) transferred it to Aequipecten, even though Aequipecten is primarily distributed to the North Atlantic. A more comprehensive phylogenetic reconstruction of the family Pectinidae (Alejandrino et al., Reference Alejandrino, Puslednik and Serb2011; but see also: Saavedra & Peña, Reference Saavedra and Peña2006; Puslednik & Serb, Reference Puslednik and Serb2008; Feng et al., Reference Feng, Li, Kong and Zheng2011) showed that the monophyletic genus Chlamys, including the type species Chlamys islandica (O. F. Müller, 1776), is very distant to both genera Argopecten and Aequipecten. In our phylogeny, the Tehuelche scallop is located in a basal position within the clade formed by the species belonging to the genus Argopecten. Therefore, we recommend transfer of the species including the forms: A. t. tehuelchus, A. t. madrynensis and Flexopecten felipponei, to the genus Argopecten Monterosato, 1889.
Future directions to test species origins
Two alternative hypotheses about the origin of the Tehuelche scallop are implicit in the literature. The extant species ‘Aequipecten’ tehuelchus (d'Orbigny 1846) originated in the south-western Atlantic from an endemic stock of ‘Aequipecten’ paranensis tehuelchus (d'Orbigny 1842), a species from the Miocene, which was classified as a subspecies of A. tehuelchus by del Rio (1992). Alternatively, A. tehuelchus is an austral vicariant of a genus primarily distributed in the North Atlantic (Waller, Reference Waller and Shumway1991). Our phylogenetic results give support to a third hypothesis: the Tehuelche scallop is part of the genus Argopecten, which is primarily distributed in the eastern Pacific and North and Central western Atlantic. The evaluation of these hypotheses requires the use of palaeontological and morphological data, as well as a molecular clock to estimate the separation time of ‘Aequipecten’ tehuelchus from the other species of the genus. These would allow formulation of possible scenarios for the Tehuelche scallop evolutionary history, including potential causes of speciation such as geological or historical events that may have intervened (e.g. formation of the Isthmus of Panama).
SUPPLEMENTARY MATERIAL
The supplementary material for this article can be found at https://doi.org/10.1017/S0025315418000292
ACKNOWLEDGEMENTS
We dedicate this paper to our co-author and friend José M. (Lobo) Orensanz (JMO) who passed away in January 2015. We are grateful to Alejo Irigoyen, Pedro Fiorda, David Galván, Gastón Trobbiani, Enrique Morsán and Emilio Ocampo for kindly assisting with collection of samples.
FINANCIAL SUPPORT
This study was funded by Project PICT/ Raices 57, FONCYT, Argentina.