INTRODUCTION
It is widely known that many marine annelids utilize hard calcareous substrates such as corals, coralline algae and mollusc shells as habitats (Blake & Evans, Reference Blake and Evans1973; Martin & Britayev, Reference Martin, Britayev and Ansell1998). Some species of Polydora and related genera, i.e. the so-called polydorid spionid annelids, are well known for inhabiting burrows excavated in mollusc shells or mud tubes in crevices on the surfaces of the shells (Blake, Reference Blake, Blake, Hilbig and Scott1996; Sato-Okoshi, Reference Sato-Okoshi1999, Reference Sato-Okoshi2000; Simon & Sato-Okoshi, Reference Simon and Sato-Okoshi2015). Among the species which utilize commercially important mollusc shells as a habitat, many are increasingly regarded as harmful invaders from the perspective of aquaculture as they often reduce the commercial value of the molluscs by damaging their shells, decreasing their growth rate and meat yield, and causing heavy mortality (Mori et al., Reference Mori, Sato, Nomura and Imajima1985; Okoshi & Sato-Okoshi, Reference Okoshi and Sato-Okoshi1996; Sato-Okoshi & Takatsuka, Reference Sato-Okoshi and Takatsuka2001; Lleonart et al., Reference Lleonart, Handlinger and Powell2003; Simon et al., Reference Simon, Ludford and Wynne2006; Sato-Okoshi & Abe, Reference Sato-Okoshi and Abe2012; Sato-Okoshi et al., Reference Sato-Okoshi, Abe, Okoshi, Teramoto, Shaw, Koh, Kim, Hong, Li and Ceccaldi2015). These species have therefore become targets for monitoring and tracking in aquaculture towards reducing infestation.
An expansion of mollusc aquaculture has resulted in the global distribution of certain commercially important molluscs including oysters and abalone (e.g. Cohen & Carlton, Reference Cohen and Carlton1998; Simon & Sato-Okoshi, Reference Simon and Sato-Okoshi2015). This intentional movement of molluscs by humans has been accompanied by the inter- and intraregional spread of the polydorid species which associate with them (e.g. Bailey-Brock, Reference Bailey-Brock2000; Radashevsky & Olivares, Reference Radashevsky and Olivares2005; Simon et al., Reference Simon, Thornhill, Oyarzun and Halanych2009; Simon, Reference Simon2011; Simon & Sato-Okoshi, Reference Simon and Sato-Okoshi2015; Williams, Reference Williams2015). These organisms are not only a source of concern economically but may also pose a threat ecologically since they can infest indigenous species in their invasive ranges should they escape from aquaculture facilities (Cohen & Carlton, Reference Cohen and Carlton1998; Mack et al., Reference Mack, Simberloff, Lonsdale, Evans, Clout and Bazzaz2000; Moreno et al., Reference Moreno, Neill and Rozbaczylo2006; Miura, Reference Miura2007). However, the effective management of these invasions relies on accurate identification of species.
The taxonomy of spionids, particularly that of polydorids, can be complicated by several factors. For example, polydorids include several complexes of species that are morphologically indistinguishable from each other but actually represent genetically distinct species (Mustaquim, Reference Mustaquim1988; Radashevsky & Pankova, Reference Radashevsky and Pankova2006; Rice et al., Reference Rice, Karl and Rice2008). On the other hand, some species possess a high degree of intraspecific morphological variation, particularly with respect to pigmentation patterns (Read, Reference Read2010; Sato-Okoshi & Abe, Reference Sato-Okoshi and Abe2013; Teramoto et al., Reference Teramoto, Sato-Okoshi, Abe, Nishitani and Endo2013). This may lead to either the under- or overestimation of polydorid diversity, respectively. Adding to these difficulties is the frequent absence of type material combined with simple or inadequate original species descriptions which can hinder confirmation of species identification. Consequently polydorid species identification is often very complex and requires detailed descriptions to avoid misidentification and confusion (see, for example, Read, Reference Read2010). It is therefore not surprising that recent investigations of polydorid species from different regions have highlighted the possibility of confusion in the identifications of widespread pests, based on morphological similarities, which need to be clarified (Walker, Reference Walker2014; Simon & Sato-Okoshi, Reference Simon and Sato-Okoshi2015; Williams, Reference Williams2015).
Polydora uncinata Sato-Okoshi, Reference Sato-Okoshi1998 and Polydora hoplura Claparède, Reference Claparède1870 resemble each other very closely in terms of morphology, reproductive strategies and habit; both belong to the Polydora ciliata/websteri group (Blake, Reference Blake, Blake, Hilbig and Scott1996), are associated with mollusc shells and have recently been identified as two of the most common and harmful pests of mollusc aquaculture (Simon & Sato-Okoshi, Reference Simon and Sato-Okoshi2015). Each species shows wide distributions overlapping in Australia: P. uncinata from Japan, Korea, Australia and Chile (Sato-Okoshi, Reference Sato-Okoshi1998; Sato-Okoshi et al., Reference Sato-Okoshi, Okoshi and Shaw2008, Reference Sato-Okoshi, Okoshi, Koh, Kim and Hong2012; Radashevsky & Olivares, Reference Radashevsky and Olivares2005) and P. hoplura from the Gulf of Naples, England, Spain, Australia, New Zealand and South Africa (Fauvel, Reference Fauvel1927; Wilson, Reference Wilson1928; Claparède, Reference Claparède1868; Read, Reference Read1975; Blake & Kudenov, Reference Blake and Kudenov1978; Hutchings & Turvey, Reference Hutchings and Turvey1984; Carazzi, Reference Carazzi1893; Simon et al., Reference Simon, Ludford and Wynne2006; Bilbao et al., Reference Bilbao, Núñez, Viera, Sosa, Fernández-Palacios and Hernández-Cruz2011; Walker, Reference Walker2011).
Adelphophagy has been widely reported as the larval developmental mode for both species, but more recently, planktotrophy was also reported for P. hoplura from populations in South Africa (David et al., Reference David, Matthee and Simon2014; Simon, Reference Simon2015). However, Walker (Reference Walker2014) and Williams (Reference Williams2015) recently provided strong morphological and molecular evidence to suggest that these two species may actually represent a single species. Here we clarify the issue by not only investigating the morphological characteristics originally used to distinguish between them but also use molecular analyses to compare P. uncinata from Japan and Australia and P. hoplura from South Africa, to determine whether they represent independent or a single species.
MATERIALS AND METHODS
Morphological observation
Individuals of P. uncinata and P. hoplura were collected from the shells of cultured molluscs: oyster Crassostrea gigas, abalone Haliotis discus discus and Haliotis laevigata from Japan, Australia and South Africa (Table 1, Figure 1). After the worms were extracted by fracturing the shells with cutting pliers and a hammer, their morphological characteristics were observed under a stereomicroscope (Olympus SZX16) in live animals and/or after being fixed in 10% neutral formalin in seawater. Pictures of both species were taken under the same condition of the specimens preserved in 80–95% ethanol by a camera (Olympus DP73) fixed to a stereomicroscope (Olympus SZX16) and an optical microscope (Olympus BX53). Specimens are deposited in the National Museum of Nature and Science, Tokyo (NSMT), Tohoku University, and the Iziko Museum, Cape Town, South Africa.

Fig. 1. Maps showing the locations of the sampling sites. (A) The locality of Saldanha Bay, South Africa and Albany, Australia. (B) The locality of sampling sites in Japan.
Table 1. Polydora uncinata and P. hoplura specimens applied for morphological observation and molecular analysis collected from mollusc shells in Japan, Australia, and South Africa with their GenBank accession numbers. The number of individuals used for molecular analysis is indicated in parentheses after each accession number.

a Previously reported in Sato-Okoshi & Abe (Reference Sato-Okoshi and Abe2012).
Phylogenetic analysis
Nuclear 18S rRNA and 28S rRNA, and mitochondrial 16S rRNA and cytochrome b (cyt b) gene analyses were performed on P. uncinata and P. hoplura collected from Japan, Australia and South Africa (Table 1). Genomic DNA was extracted from live or ethanol-preserved tissues and nuclear 18S rRNA gene analysis was performed according to the methods described by Sato-Okoshi & Abe (Reference Sato-Okoshi and Abe2012, Reference Sato-Okoshi, Okoshi, Abe and Li2013) and Teramoto et al. (Reference Teramoto, Sato-Okoshi, Abe, Nishitani and Endo2013). For nuclear 28S rRNA, mitochondrial 16S rRNA, and cyt b gene analysis, three primer pairs D1R/D2C (forward: ACCCGCTGAATTTAAGCATA, reverse: CCTTGGTCCGTGTTTCAAGA) (Scholin et al., Reference Scholin, Herzog, Sogin and Anderson1994), 16Sar/16Sbr (forward: CGCCTGTTTATCAAAAACAT, reverse: CCGGTCTGAACTCAGATCACGT) (Palumbi et al., Reference Palumbi, Martin, Romano, McMillan, Stice and Grabowski1991), and cytbPuh10F/cytbPuh382R (forward: AGCAATTCCYTACTTTGGCGA, reverse: AGGAAGTATCATTCAGGTTGAATATG) (this study) were used, respectively. PCR cycling conditions were 94°C for 2 min followed by 36 cycles of denaturation for 10 s at 98°C, annealing for 30 s at 52°C, and extension for 1 min at 68°C for nuclear 28S rRNA gene, 94°C for 2 min followed by 36 cycles of denaturation for 10 s at 98°C, annealing for 30 s at 56°C, and extension for 45 s at 68°C for mitochondrial 16S rRNA gene, and 94°C for 2 min followed by 34 cycles of denaturation for 10 s at 98°C, annealing for 30 s at 58°C, and extension for 30 s at 68°C for mitochondrial cyt b gene. PCR reaction mixtures and procedures following PCR cycling were the same as that for 18S rRNA gene analysis. The forward and reverse complementary sequences of the PCR products were assembled and aligned using GeneStudio Pro v.2.2.0.0 software (GeneStudio Inc., GA, USA) and BioEdit sequence alignment editor v7.2.5 (Ibis Biosciences, CA, USA). Neighbour-joining trees were generated using Molecular Evolutionary Genetics Analysis (MEGA) software v 4.0 (Tamura et al., Reference Tamura, Dudley, Nei and Kumar2007) with the default setting and bootstrapping with 1000 replications used to estimate the reliability of the phylogenetic trees.
RESULTS
Phylogenetic analysis
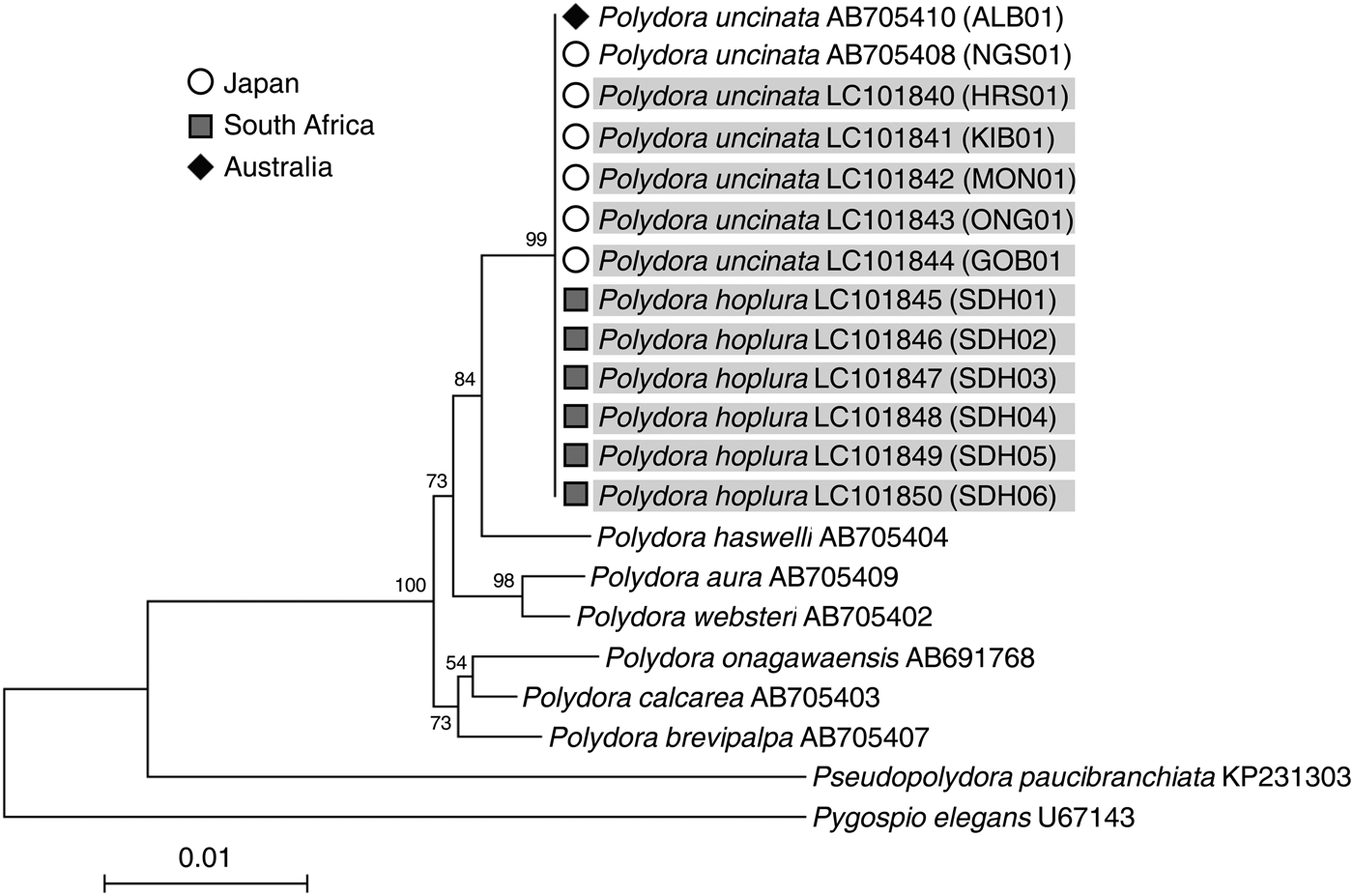
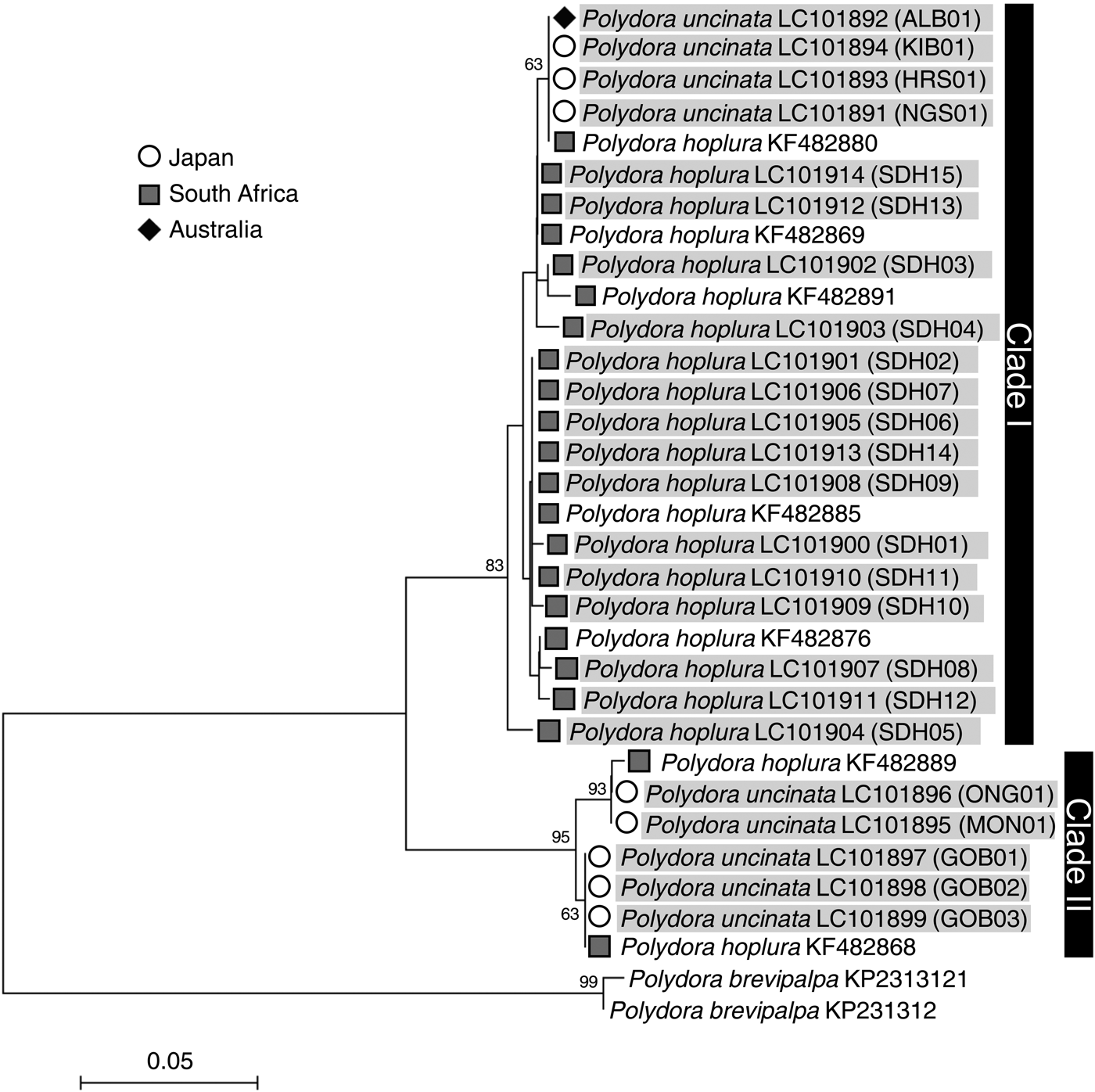
Genetic analyses revealed that the nucleotide sequences of both the 18S rRNA and 28S rRNA genes were identical between P. uncinata and P. hoplura (Figures 2 & 3) indicating that the two species represent a single species. The mitochondrial 16S rRNA and cyt b gene analysis showed intraspecific variations, separating the species into two major clades (Figure 4). The mitochondrial cyt b gene analysis, including previously analysed individuals of P. hoplura from South Africa (David et al., Reference David, Matthee and Simon2014), showed the distribution of both species across two major clades which each included specimens of both putative species (Figure 5). Additionally, there were two shared haplotypes, one including specimens from Australia, Japan and South Africa and the other including specimens from Japan and South Africa. These results suggested that there were no genetic differences between the two species. There were also no genetic differences among individuals collected from different host species (Crassostrea gigas, Haliotis discus discus and Haliotis laevigata).

Fig. 2. Neighbour-joining trees inferred from nuclear 18S rRNA gene sequences of Polydora uncinata (Japan and Australia) and Polydora hoplura (South Africa). Gene sequences obtained in this study are shaded. Bootstrap values of >50 as a percentage of 1000 bootstrap replicates are given at the respective nodes. The scale bar represents the number of substitutions per site. Each individual ID (Table 1) is indicated in parentheses.

Fig. 3. Neighbour-joining trees inferred from nuclear 28S rRNA gene sequences of Polydora uncinata (Japan and Australia) and Polydora hoplura (South Africa). Gene sequences obtained in this study are shaded. Bootstrap values of >50 as a percentage of 1000 bootstrap replicates are given at the respective nodes. The scale bar represents the number of substitutions per site. Each individual ID (Table 1) is indicated in parentheses.

Fig. 4. Neighbour-joining trees inferred from mitochondrial 16S rRNA gene sequences of Polydora uncinata (Japan and Australia) and Polydora hoplura (South Africa). Gene sequences obtained in this study are shaded. Bootstrap values of >50 as a percentage of 1000 bootstrap replicates are given at the respective nodes. The scale bar represents the number of substitutions per site. Each individual ID (Table 1) is indicated in parentheses.

Fig. 5. Neighbour-joining trees inferred from mitochondrial cyt b gene sequences of Polydora uncinata (Japan and Australia) and Polydora hoplura (South Africa). Gene sequences obtained in this study are shaded. Bootstrap values of >50 as a percentage of 1000 bootstrap replicates are given at the respective nodes. The scale bar represents the number of substitutions per site. Each individual ID (Table 1) is indicated in parentheses.
Comparison of morphological characteristics
The two species, Polydora uncinata (Figure 6) and P. hoplura (Figure 7) showed high, and overlapping, intraspecific variation in morphology, suggesting that the two species should be synonymized.

Fig. 6. Polydora uncinata. Preserved specimens in ethanol collected from Kitaibaraki, Japan. (A) Anterior region in dorsal view showing black pigmentation on prostomium, part of peristomium and chaetigers 1 & 2. (B) Anterior region in ventral view showing black pigmentation on anterior end of prostomium and chaetigers 1 & 2. (C) Modified falcate spines with lateral flange in chaetiger 5. (D) Modified falcate spines with lateral flange in chaetiger 5 in opposite side of C. (E) Worn modified falcate spine in chaetiger 5. (F) Posterior chaetigers and pygidium with special recurved hooks. (G) Magnified recurved hooks. Bars (A & B) 500 µm; (C–E) 100 µm; (F) 200 µm; (G) 100 µm.

Fig. 7. Polydora hoplura. Preserved specimens in ethanol collected from Saldanha Bay, South Africa. (A) Anterior region in dorsal view showing black pigmentation on palps, prostomium, caruncle, peristomium and chaetigers 1–3. (B) Anterior region in ventral view showing black pigmentation on peristomium and chaetigers 1 & 2. (C) Modified falcate spines with lateral flange in chaetiger 5. (D) Modified falcate spines with lateral flange in chaetiger 5 in opposite side of C. (E) Special posterior recurved hooks. Bars (A & B) 500 µm; (C–E) 100 µm.
Polydora Bosc, 1802
Polydora hoplura Claparède, Reference Claparède1868
Polydora hoplura Claparède, Reference Claparède1868: pp. 318–319, pl. XXII, figure 2; Reference Claparède1870: pp. 58–59, pl. XXII, figure 2. Carazzi, Reference Carazzi1893: pp. 20–21, pl. 2, figures 6, 7, 13, 16, 18. Fauvel, Reference Fauvel1927: figures 17A–F. Wilson, Reference Wilson1928: pp. 578–585, figure 2, pls. V–VII. Read, Reference Read1975: pp. 411–412, figure 6. Blake & Kudenov, Reference Blake and Kudenov1978: p. 264, figure 47. Lleonart, Reference Lleonart2001: figures 4–6, 9, 26–32. David et al., Reference David, Matthee and Simon2014: figures 4–6
Polydora hoplura hoplura: Day, Reference Day1967: p. 468, figure 18.2.
Polydora uncinata Sato-Okoshi, Reference Sato-Okoshi1998: pp. 278–280, figure 1; Reference Sato-Okoshi1999: p. 835. Radashevsky & Olivares, Reference Radashevsky and Olivares2005: pp. 491–494, figures 2–4. Sato-Okoshi et al., Reference Sato-Okoshi, Okoshi and Shaw2008: pp. 493–495, figures 2 & 3; Reference Sato-Okoshi, Okoshi, Koh, Kim and Hong2012: p. 87, figure 4. Sato-Okoshi & Abe, Reference Sato-Okoshi and Abe2012: pp. 43–44, figure 3. Sato-Okoshi et al., Reference Sato-Okoshi, Abe, Okoshi, Teramoto, Shaw, Koh, Kim, Hong, Li and Ceccaldi2015: p. 36, figure 1C, D. New synonymy
New materials examined
Individuals collected from shells of Crassostrea gigas from Moune, Onagawa, Gobuura, Kitaibaraki, Hiroshima, Japan, coll. Sato-Okoshi W.; individuals collected from shells of Haliotis discus discus from Nagasaki, Japan, coll. Sato-Okoshi W.; individuals collected from shells of Haliotis laevigata from Albany, Australia, coll. Sato-Okoshi W., Okoshi K. & Shaw J. (Table 1, Figure 1). Individuals collected from shells of Crassostrea gigas from Saldanha Bay, South Africa, coll. Williams, L.-G. (Table 1, Figure 1).
MORPHOLOGY
Specimens measuring up to 35 mm long, 1.5 mm wide at chaetiger 5, for 180 chaetigers from Japan (>100 inds) and Australia (30 inds), and up to 28 mm long, 1.3 mm wide at chaetiger 5, for 150 chaetigers (30 inds) from South Africa were examined. Prostomium weakly incised or rounded anteriorly, caruncle extending to chaetiger 2 or end of 3; short or inconspicuous occipital tentacle present; nuchal organs ciliated grooves along margin of caruncle; up to 4 eyes present, if present arranged in trapezoid. A pair of palps may lack pigmentation completely, be crossed by brown to black bands (Figure 7A), have dark pigmentation along the edge, or dense to weak dark pigmentation on whole or part of palps. Varying degrees of dark pigmentation on anterior of prostomium; on and along caruncle, peristomium, and between chaetigers 1–4 on both dorsal and ventral sides (Figures 6A, B & 7A, B). Some specimens have a conspicuous black spot on dorsal right and left of peristomium. Some without conspicuous pigmentation. Branchiae present from chaetiger 7, fairly long, overlapping in mid-dorsum, continuing almost to end of body. Chaetiger 1 without notochaetae, only winged capillary neurochaetae. Chaetigers 2–4, 6 with winged capillary neuro- and notochaetae; only winged capillary notochaetae from chaetiger 7 onwards. Bidentate hooded hooks replacing winged capillary neurochaetae from chaetiger 7, up to 15 hooks per fascicle, decreasing to one posteriorly. Hooks with constriction on shaft and main fang at right angle to shaft, acute angle between main fang and apical tooth. One or two recurved hooks present on posterior notopodia for about last 10% of the total chaetigers (Figures 6F, G & 7E). Hook slightly bent at tip, broadening at its middle, then tapering to base (Figures 6G & 7E).
Modified spines of modified chaetiger 5 falcate with lateral flange (Figures 6C, D & 7C, D), but some worn spines distally rounded and flange appears as a small lateral tooth (Figure 6E) or absent; major spines alternating with pennoned companion chaetae; short dorsal and ventral unilimbate chaetae present.
Pygidium a small to flaring disc with dorsal notch (Figure 6F). Some with dense pigmentation on and along the edge of pygidium.
MORPHOLOGICAL VARIATION
A wide range of pigmentation patterns were observed among specimens from different locations: variable black pigmentation on chaetigers 1–4 dorsally (Albany, Australia); dark pigmentation in anterior region both ventrally and dorsally (Figures 6A, B) (Kitaibaraki, Japan); no to slight dark pigmentation along the caruncle (Hiroshima, Japan); zero to up to 14 black bands on palps, slight to complete black pigmentation on prostomium, peristomium, chaetigers 1–4, and pygidium (Nagasaki, Japan); partially to completely black on palps with intermittent lines along the palps but never bands, dense black pigmentation on anterior end and pygidium (Moune, Japan); transparent to black bands on palps, dark pigmentation on peristomium, prostomium, along caruncle and chaetigers 1–4 or no pigmentation (Gobuura, Japan). A wide range of pigmentation was also observed from Saldanha Bay, South Africa. Especially strong black pigmentation was observed in peristomium, anterior end of prostomium, chaetigers 1 & 2 both dorsally and ventrally (Figure 7A, B). No particular differences in pigmentation patterns were observed under different fixation and preservation conditions.
REMARKS
Sato-Okoshi (Reference Sato-Okoshi1998) described the new species P. uncinata from Japan. Although she acknowledged the resemblance to P. hoplura, she noted several differences which she deemed sufficient to warrant the erection of a new species. These included its smaller size, the presence of an occipital tentacle, a proportionately longer caruncle and morphology of the modified posterior curved spine compared with P. hoplura. After examining individuals of both species in this study, in combination with published records, it is clear that these differences fall within the range of variation which occurs within the species (Read, Reference Read1975; Blake & Kudenov, Reference Blake and Kudenov1978; Sato-Okoshi, Reference Sato-Okoshi1998; Van Niekerk, Reference Van Niekerk2014; Walker, Reference Walker2014) or may be the consequence of inaccurate illustrations. The specimens examined by Sato-Okoshi (Reference Sato-Okoshi1998) were just over half the length of those described by Read (Reference Read1975) and Blake & Kudenov (Reference Blake and Kudenov1978), but ~3 mm shorter than those described by Van Niekerk (Reference Van Niekerk2014) and Walker (Reference Walker2014). This indicates a great range in body size, which is confounded by the inconsistent relationship between the number of chaetigers and length which may be a consequence of inconsistent shrinkage after fixation. For example, while Sato-Okoshi (Reference Sato-Okoshi1998) and Blake & Kudenov (Reference Blake and Kudenov1978) recorded up to 160 chaetigers for worms that were 25 and 40 mm long, respectively, Van Niekerk (Reference Van Niekerk2014) and Walker (Reference Walker2014) recorded 140 and 87 chaetigers, respectively, for worms of up to 28 mm long. The length of the caruncle probably depends on the number of chaetigers; the specimens with the most chaetigers (180) had the longest caruncles (to the start of chaetiger 4) (Read, Reference Read1975) while the shortest specimens (up to 87 chaetigers) had the shortest caruncles (to the end of chaetiger 2 or anterior of chaetiger 3) (Walker, Reference Walker2014). The inconspicuous nature of the occipital tentacle might have led to it being overlooked and therefore not reported in earlier descriptions of European and South African material (Fauvel, Reference Fauvel1927; Day, Reference Day1967; Carazzi, Reference Carazzi1893). This was, however, later described in material from New Zealand, Australia and South Africa (Read, Reference Read1975; Blake & Kudenov, Reference Blake and Kudenov1978; Van Niekerk, Reference Van Niekerk2014; Walker, Reference Walker2014). The posterior spine illustrated for P. hoplura by Day (Reference Day1967) was clearly sickle shaped. However, Van Niekerk (Reference Van Niekerk2014) suggested that this figure was mislabelled, and probably represented the posterior modified hooks of Polydora colonia since the shape of the spines of the P. hoplura specimens collected by John Day conformed to the shape as described by Sato-Okoshi (Reference Sato-Okoshi1998) and Radashevsky & Olivares (Reference Radashevsky and Olivares2005). Similarly, in the illustrations of P. hoplura from New Zealand and Australia (Read, Reference Read1975; Blake & Kudenov, Reference Blake and Kudenov1978), the shaft of the hook appears to be of a similar diameter along the length, with an elongated hook, whereas the shaft of specimens from Japan and Chile (Sato-Okoshi, Reference Sato-Okoshi1998; Radashevsky & Olivares, Reference Radashevsky and Olivares2005) and South Africa (Van Niekerk, Reference Van Niekerk2014) is broader in the middle before forming a shorter, more pronounced curve. Alternatively, the shape as illustrated by Read (Reference Read1975) and Blake & Kudenov (Reference Blake and Kudenov1978) was accurately represented, and represents phenotypic variation in this character. Here we also describe high variability in pigmentation patterns, ranging from absent to conspicuous black pigmentation on palps, anterior chaetigers, prostomium, peristomium and pygidium (see also Van Niekerk, Reference Van Niekerk2014). Thus the variably developed brown dusky pigments limited to the prostomium anterior to the palps and on anterior peristomium described for P. hoplura by Read (Reference Read1975) fall within the range of variation described for this species.
DISCUSSION
Here we provide conclusive evidence that Polydora hoplura and Polydora uncinata represent a single species making P. uncinata the junior synonym. No genetic differences were found between P. uncinata and P. hoplura collected from Japan, Australia and South Africa in this study; a single sequence type was found for each of the nuclear genes tested and although the mitochondrial analyses showed intraspecific variation within both putative species separating the specimens into two major clades, these were shared across P. uncinata and P. hoplura (including individuals analysed by David et al., Reference David, Matthee and Simon2014). The morphological examination of P. uncinata and P. hoplura from Japan, Australia, and South Africa also revealed that the variation in morphology of the two species largely overlapped and there were no distinct differences.
The mitochondrial 16S rRNA and cyt b gene analyses reflect the unintentional, artificial, transportation of the species. Although individuals from Japan, Australia and South Africa formed two major clades for both mitochondrial genes, these did not reflect strict geographic differences. Furthermore, the haplotypes that were common to South Africa, Australia and Japan, and Japan and South Africa, respectively, suggest that worms had either been transported between these sites, or arose from a common source population (cf. Simon et al., Reference Simon, Thornhill, Oyarzun and Halanych2009).
Polydora is the most speciose genus among alien polychaetes (Çinar, Reference Çinar2013). More specifically, Polydora hoplura is considered introduced to South Africa, Australia and New Zealand, while ‘Polydora uncinata’ is also reported to have been accidentally transported from Japan to Chile with abalone broodstock, and now also occurs in Korea (Radashevsky & Olivares, Reference Radashevsky and Olivares2005; Sato-Okoshi et al., Reference Sato-Okoshi, Okoshi, Koh, Kim and Hong2012; Çinar, Reference Çinar2013; Simon & Sato-Okoshi, Reference Simon and Sato-Okoshi2015). The synonymization of these two species means that P. hoplura is now the most widespread pest of aquaculture, infesting the widest range of pests (Simon & Sato-Okoshi, Reference Simon and Sato-Okoshi2015). The spread of this species was probably facilitated by the transportation of infested cultured molluscs (Sato-Okoshi et al., Reference Sato-Okoshi, Okoshi and Shaw2008; Simon & Sato-Okoshi, Reference Simon and Sato-Okoshi2015). However, polydorid species may also be transported by ships’ ballast water and hull fouling (Carlton & Geller, Reference Carlton and Geller1993; Çinar, Reference Çinar2013), and indeed, this has been suggested as the original vector of transportation for P. hoplura to South Africa and Australia (Walker, Reference Walker2014; Williams, Reference Williams2015). Further analyses of population gene structure for P. hoplura from its entire distribution range will shed light on its origin and global transportation pathways.
In spite of widespread distribution and observations of adelphophagic larval development (Wilson, Reference Wilson1928; Radashevsky & Olivares, Reference Radashevsky and Olivares2005; Sato-Okoshi et al., Reference Sato-Okoshi, Okoshi and Shaw2008, Reference Sato-Okoshi, Okoshi, Koh, Kim and Hong2012), it is interesting to see poecilogony only from South Africa (David et al., Reference David, Matthee and Simon2014). Furthermore, Simon (Reference Simon2015) revealed planktotrophy in off-shore culture systems and adelphophagy in on-shore systems. There is, however, increasing evidence demonstrating the flexibility in larval developmental mode in polydorids. For example, an increase in nurse egg production accompanied by a decrease in numbers of adelphophagic larvae per brood occurred in response to sperm limitation in Polydora cornuta (Rice & Rice, Reference Rice and Rice2009) or an increase in latitude for Boccardia proboscidea (Oyarzun et al., Reference Oyarzun, Mahon, Swalla and Halanych2011) while the opposite effect occurred when female B. proboscidea were exposed to bisphenol A (Hart et al., in Gibson et al., Reference Gibson, Hart, Coulter and Xu2012). It is therefore possible that the poecilogony demonstrated by David et al. (Reference David, Matthee and Simon2014) and Simon (Reference Simon2015) was induced by environmental conditions unique to South Africa. Further studies of populations around the world may increase our understanding of larval developmental mode of this species.
The present study emphasizes the usefulness of combining morphological and gene sequence analyses to effectively resolve taxonomic confusion within the polydorids. We have demonstrated conclusively that specimens identified as P. uncinata from Japan and Australia and P. hoplura from South Africa represent a single species. However, without specimens from the type locality we cannot confirm, with 100% certainty, that this species is in fact P. hoplura; it could well represent a different, widespread pest of aquaculture. This issue can only effectively be resolved with the inclusion of specimens from the type locality and the designation of a lectotype, but this was beyond the scope of our study. In the meantime we feel that to avoid causing unnecessary confusion, the most parsimonious approach would be to continue referring to this species as P. hoplura until such time that a comparison with specimens from the type locality proves otherwise.
Accurate species identification would be the first and most important step towards ensuring safe and sustainable transportation of commercially important molluscs by reducing the risk of transporting harmful species outside of their native ranges and causing economic and ecological damage in the invaded region. Moreover, accurate species identification can contribute to the implementation of effective measures for preventing or controlling infestation by harmful species by considering their biological characteristics, i.e. timing of settlement and reproduction and lifespan (Sato-Okoshi et al., Reference Sato-Okoshi, Sugawara and Nomura1990; Sato-Okoshi, Reference Sato-Okoshi1994; Simon & Booth, Reference Simon and Booth2007). It is important to recognize the contribution which aquaculture activities have made to the unintentional movement of many species (Miura, Reference Miura2007; Haupt et al., Reference Haupt, Griffiths, Robinson and Tonin2010; Mead et al., Reference Mead, Carlton, Griffiths and Rius2011), the confusion which may occur when species are not identified correctly, and the influence which these species may have on the environment.
FINANCIAL SUPPORT
C.A.S.'s visit to Japan was funded by the HB & MJ Thom and Oppenheimer Memorial Trust travel grants and research funds were provided by the National Research Foundation (Thuthuka Programme) and Stellenbosch University. This study was partially funded by a research grant from the Ministry of Education, Science, Sports and Culture of Japan to W.S.-O. (grant number 15K07540).