Introduction
The genus Thada was erected by Thorne (Reference Thorne1941) for two species, T. striata and T. cancellata, because of having a thick, deeply striated cuticle, valve-less median bulb, cap-like valvular apparatus at the pharyngo–intestinal junction and ovate cephalation of the spicules. Thorne & Malek (Reference Thorne and Malek1968) added a third species, T. tatra. The cuticle of the type species, T. striata, was marked only by transverse striae, while that of T. cancellata and T. tatra was marked by both transverse and longitudinal striae. Khan (Reference Khan1973) transferred T. cancellata and T. tatra to the genus Neothada Khan, 1973, because of the presence of longitudinal ridges, the action that was supported by Raski & Geraert (Reference Raski and Geraert1987). Geraert (Reference Geraert1974) urged that the genital system of T. striata (with elongate spermatheca filled with large sperms) is identical to the genital system in Ditylenchus Filipjev, 1936 and the other differences between T. striata and Ditylenchus species are small. Fortuner & Maggenti (Reference Fortuner and Maggenti1987) endorsed this view and accommodated Thada as a valid genus in the family Anguinidae Nicoll, 1935. Siddiqi (Reference Siddiqi2000) put Thada and Neothada in the subfamily Thadinae Siddiqi, 1986 within Tylenchidae Örley, 1880 and Geraert (Reference Geraert2008) considered Thada as a valid genus in the subfamily Boleodorinae Khan, 1964a within Tylenchidae, with T. striata as its type and the only species. It was found around roots of Atriplex confertifolia Torr. & Frém. and alfalfa crowns in Utah, USA (Thorne, Reference Thorne1941). Geraert (Reference Geraert1974) and Siddiqi (Reference Siddiqi1986, Reference Siddiqi2000) studied the type material and redescribed this species. Thada is marked by wide transverse annuli, flattened cephalic region, longitudinal amphidial slits that can continue on the lateral sides of the cephalic region, delicate spear without knobs but with a slight thickening at the base, pharynx with a slightly swollen median bulb without central sclerotized valve and lacking vulva membrane (Geraert, Reference Geraert2008).
De Man (Reference De Man1921) described Tylenchus eurycephalus based on a single male. Corbett (Reference Corbett1964) proposed the genus Ecphyadophoroides for this species and separated it from Ecphyadophora De Man, 1921 by the cephalic region strongly flattened dorso-ventrally (vs. rounded), the gradually narrowing body (vs. abruptly narrowed behind vulva and cloacal lips) and the shorter and stouter spicules of the male. Siddiqi (Reference Siddiqi1986) separated the genus Tenunemellus from Ecphyadophoroides by its longitudinal amphidial slit (vs. lacking) and having no clear incisures in the lateral field. According to Geraert (Reference Geraert2008), Tenunemellus has six valid species, including T. tenuis (Corbett, 1964) Siddiqi, 1986 as type species; T. graminis (Husain & Khan, 1968) Siddiqi, 1986; T. indicus (Verma, 1972) Siddiqi, 1986; T. leptocephalus (Raski et al., 1982) Siddiqi, 1986; T. microcephalus (Raski et al., 1982) Siddiqi, 1986; and T. sheri (Raski et al., 1982) Siddiqi, 1986. Tenunemellus eurycephalus (De Man, 1921) Siddiqi, 1986 has been considered as species inquirendae (Siddiqi, Reference Siddiqi2000; Geraert, Reference Geraert2008). The taxonomic studies on the members of the family Tylenchidae reported from Iran are well documented and discussed in Karegar (Reference Karegar, Ghaderi, Kashi and Karegar2018). Some other species of the genera Miculenchus Andrássy, 1959 (Panahandeh et al., 2019a), Coslenchus Siddiqi, 1978 (Hosseinvand et al., Reference Hosseinvand, Eskandari and Ghaderi2019), Labrys Qing & Bert, 2018 (Panahandeh et al., Reference Panahandeh, Abolafia, Pourjam, Jahanshahi Afshar, Giblin-Davis and Pedram2018, Reference Panahandeh, Pourjam, Abolafia, Roshan-bakhsh, Mojerlou, Jahanshahi Afshar and Pedram2019b), Sakia Khan, 1964b (Panahandeh et al., Reference Panahandeh, Atighi, De Ley, Pourjam, Mundo-Ocampo, Abolafia, Koolivand, Jahanshahi Afshar and Pedram2019c), Basiria Siddiqi, 1959 (Eisvand et al., Reference Eisvand, Farrokhi Nejad and Azimi2019) and Malenchus Andrássy, 1959 (Jalalinasab et al., Reference Jalalinasab, Adeldoost, Abolafia and Heydari2019) have since been reported or described from Iran through morphological and molecular studies. The present study aims to describe the newly recovered species, named Thada populus n. sp., and to characterize a known species of Tenunemellus (T. indicus) using morphological and molecular approaches and, moreover, discussing their phylogenetic relationships with other species. This is the first report of the genera Thada and Tenunemellus from Iran.
Materials and methods
Nematode sampling and morphological identification
Soil samples were collected from the rhizosphere of different plants in Khuzestan province, south-west Iran. Nematodes were extracted by the tray method (Whitehead & Hemming, Reference Whitehead and Hemming1965), killed and fixed by hot FPG (4:1:1, formaldehyde:propionic acid:glycerol), processed to anhydrous glycerol (De Grisse, Reference De Grisse1969), mounted in glycerol on permanent slides using paraffin wax and studied using a light microscope, equipped with a Dino-Eye microscope eye-piece camera and its software, Dino Capture version 2.0 (https://www.dinolite.us/dinocapture). Specimens were identified at species level using the available identification keys (Geraert, Reference Geraert2008).
DNA extraction, polymerase chain reaction (PCR) and sequencing
Following morphological confirmation on a temporary slide, nematode DNA was extracted from single individuals of each species, as described by Tanha Maafi et al. (Reference Tanha Maafi, Subbotin and Moens2003). Two overlapping fragments of 18S ribosomal DNA (rDNA) were amplified from each specimen using primer sets 1096F (5′-GGTAATTCTGGAGCTAATAC-3′) and 1912R (5′-TTTACGGTYAGAACTAGGG-3′) for the first fragment, and 1813F (5′-CKGCGYKAGAGGTGAAAT-3′) and 2646R (5′- GCTACCTTGTTACGACTTTT-3′) for the second fragment (Holterman et al., Reference Holterman, van der Wurff, van den Elsen, van Megen, Bongers, Holovachov, Bakker and Helder2006). The D2–D3 expansion segments of 28S rDNA were amplified using the forward D2A (5′-ACAAGTACCGTGAGGGAAAGT-3′) and reverse D3B (5′-TCGGAAGGAACCAGCTACTA-3′) primers (Subbotin et al., Reference Subbotin, Sturhan, Chizhov, Vovlas and Baldwin2006). The 30 μl PCR contained 15 μl 2 × Taq DNA polymerase mix (Ampliqon, Denmark), 1 μl (10 pmol μl−1) each of forward and reverse primers, 2 μl of DNA template and 11 μl deionized water. This mixture was placed into a Hybaid Express thermal cycler (Hybaid, Ashford, Middlesex, UK). The thermal cycling program was denaturation at 95°C for 4 min, then 33 cycles of denaturation at 94°C for 30 s, annealing at 54°C for 30 s (18S rDNA) and 57°C for 30 s (28S rDNA), and extension at 72°C for 90 s. A final extension was performed at 72°C for 10 min. The quality of the PCR products was checked by electrophoresis of 4 μl of each PCR product in 1% agarose gel containing ethidium bromide under ultraviolet light. The length and concentration of each PCR product was measured by comparison with a low DNA mass ladder (Invitrogen, Carlsbad, California, USA). The PCR product was purified and sequenced directly for both strands using the same primers with an ABI 3730XL sequencer (Bioneer, Seoul, South Korea). The newly obtained sequences were submitted to the GenBank database under accession numbers MN557353 for the 18S and MN535624 for the 28S regions.
Phylogenetic analyses
The newly obtained sequences were edited and aligned with other relevant sequences of 18S and 28S rDNA sequences available in GenBank using the MUSCLE alignment tool implemented in MEGA7 (Kumar et al., Reference Kumar, Stecher and Tamura2016). The ambiguously aligned parts and divergent regions were removed using the online version of Gblocks 0.91b (Castresana, Reference Castresana2000). The best-fitting model of nucleotide substitution used for the phylogenetic analysis was statistically selected using jModelTest 2.1.10 (Darriba et al., Reference Darriba, Taboada, Doallo and Posada2012). Phylogenetic trees were generated with the Bayesian inference (BI) method using MrBayes 3.2.2 (Huelsenbeck & Ronquist, Reference Huelsenbeck and Ronquist2001; Ronquist et al., Reference Ronquist, Teslenko and van der Mark2012). Aphelenchus avenae Bastian, 1865 was chosen as the outgroup for both 18S and 28S trees, respectively (accession numbers in trees). The analysis under the GTR + I + G model was initiated with a random starting tree and run for 1 × 106 generations. The Markov chain Monte Carlo method within a Bayesian framework was used to estimate the posterior probability values of the clades. The trees were visualized and saved with FigTree 1.4.3 (Rambaut, Reference Rambaut2016) and edited with Adobe® Acrobat® XI Pro 11.0.1.
Results
Thada populus n. sp.
Description
Measurements. See table 1.
Table 1. Morphometrics of Thada populus n. sp. and Tenunemellus indicus (Verma, 1972) Siddiqi, 1986 from Iran.

All measurements are in μm and in the form: mean ± standard deviation (range) where appropriate. CV, coefficient of variation.
Female (figs 1 and 2). Body short, straight or slightly curved ventrally upon fixation. Cuticle annulated, transverse annuli 1.0 (0.9–1.2) μm wide. Lateral field with four lines, 4.5 (3.6–5.6) μm wide and 30% (22–35%) of body diameter. Cephalic region low and truncate, continuous with adjacent part of body, 7.7 (7.1–8.5) μm wide and 2.5 (2.0–3.0) μm high, with one or two annuli. Amphidial aperture sigmoid, bearing near oral aperture and extending as lateral longitudinal slits. Cephalic framework weakly sclerotized. Stylet delicate, conus 3.3 (3.0–3.6) μm or 36% (33–40%) of total stylet length, basal knobs distinct, posteriorly directed, 1.9 (1.2–2.3) μm wide. Dorsal pharyngeal gland opening (DGO) 1.2 (1.0–1.5) μm posterior to the stylet knobs. Procorpus cylindrical, median bulb weakly developed, non-muscular and without valve, 5.5 (4.5–6.5) μm wide or 40% (34–45%) of the corresponding body diameter, isthmus slender, basal pharyngeal bulb pyriform, 8.5 (7–11) μm wide and 18.5 (16.7–24.0) μm long, cardia large and rounded. Deirids 62 (59–65) μm from the anterior end, at level of secretory–excretory pore or anterior to it. Secretory–excretory pore at the anterior level of basal pharyngeal bulb. Reproductive system monodelphic–prodelphic, outstretched, oocytes mostly in single row, oviduct not well discernible, spermatheca almost oval, 9.4 (7.5–10.0) μm wide and 15.1 (13.8–16.0) μm long, along the reproductive system and without sperm. Crustaformeria quadricolumellate. Vagina with thin wall, slightly anteriorly directed, 4.0 (3.3–5.0) μm long or 29% (23–33%) vulval body diameter, vulva without lateral membranes. Post-vulval uterine sac (PUS) short, 4.7 (3.4–6.8) μm long or 3.5 (0.2–0.5) times the vulval body diameter. Tail conical with finely to broadly rounded terminus.
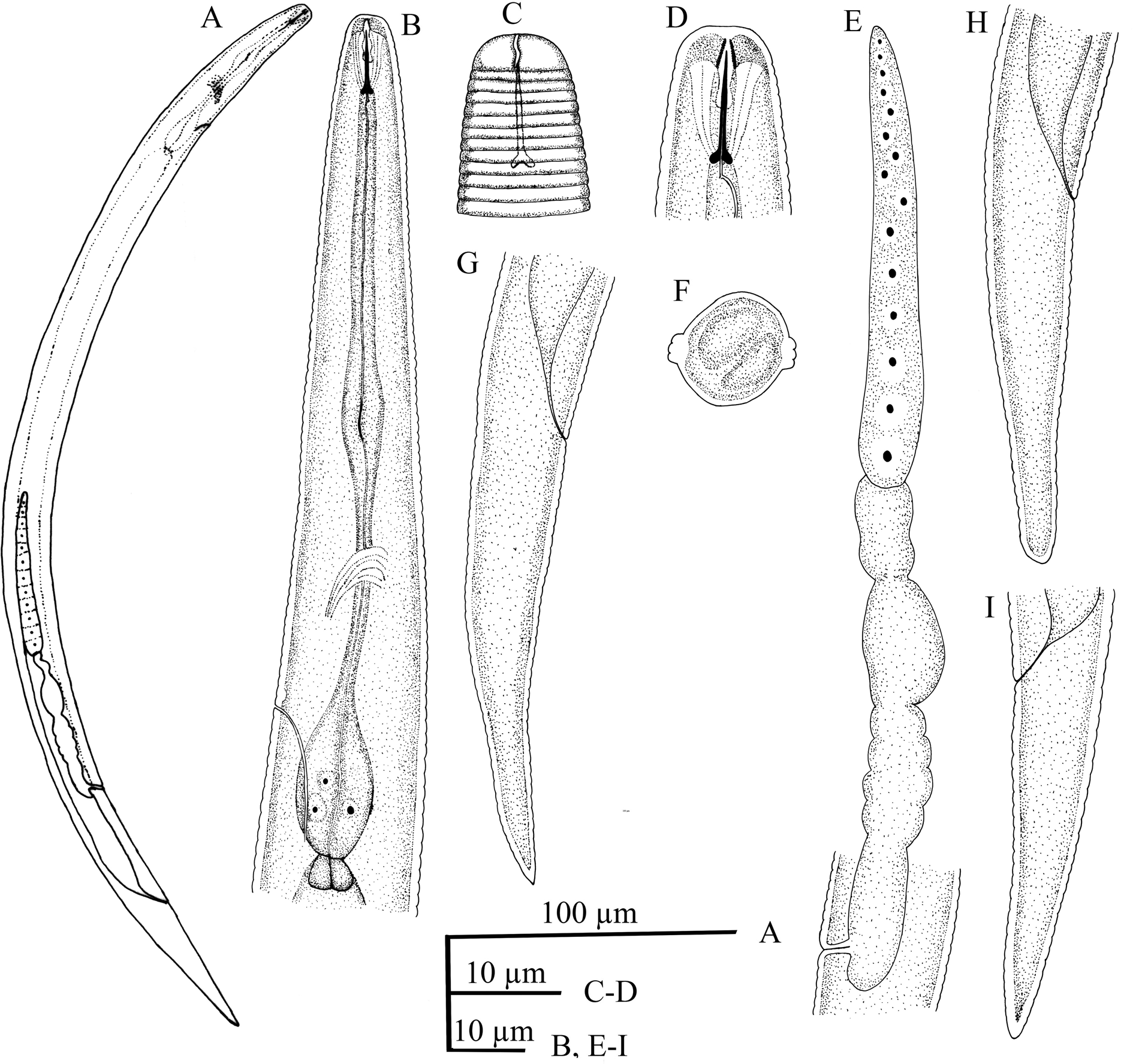
Fig. 1. Thada populus n. sp. female (line drawing): (A) entire body; (B) anterior end and pharynx; (C) amphidial slit; (D) cephalic region and stylet; (E) reproductive system; (F) cross section of body; (G–I) tail.

Fig. 2. Thada populus n. sp. female (light microscope): (A, B) entire body; (C, E) anterior end and stylet; (D) en face view of cephalic region; (F) median pharyngeal bulb; (G) basal pharyngeal bulb and secretory–excretory pore; (H) amphidial slit; (I) cross section of body; (J) vulval region; (K) lateral field; (L–N) tail. Scale bars: (A, B) 50 μm; (C–N) 5 μm.
Male. Not found.
Diagnosis and relationships
Thada populus n. sp. is characterized by its 365–453 μm body length, cuticle with transverse annuli, annulus 1.0 (0.9–1.2) μm wide, lateral fields with four lines, low lip region 7.0–8.5 μm wide at base, with one or two annuli, longitudinal or slightly sigmoid amphidial slit, delicate stylet 8.4–9.8 μm long with weak posteriorly directed knobs, DGO at 1.0–1.5 μm posterior to the stylet knobs, non-muscular and valve-less median pharyngeal bulb, pyriform and offset basal pharyngeal bulb, almost oval spermatheca, posterior position of vulva (V = 75.4–78.7%) without lateral membrane, short PUS and conical tail with finely to broadly rounded terminus. It differs from the type and the only species of the genus, T. striata, by having a shorter body (365–453 vs. 570–590 μm), shorter pharynx (70–87 vs. 104 μm) and thinner cuticular annuli (0.9–1.2 vs. 2.5–3.0 μm), presence of stylet knobs (vs. slight basal thickening present), width of cephalic region (7.1–8.5 vs. 5.5 μm), position of DGO (1.0–1.5 vs. 4.0–4.5 μm), shape of spermatheca (slightly oval vs. very long and sac-like) and absence of males (vs. present).
Taxonomic summary
Type material. Holotype and 11 paratype females were deposited in the collection of the Department of Plant Protection, College of Agriculture, University of Zanjan, Zanjan, Iran. Five paratype females were deposited in the collection of the Department of Plant Protection, School of Agriculture, University of Shiraz, Shiraz, Iran.
Type locality and habitat. Thada populus n. sp. was found in the rhizosphere of Populus euphratica Oliv. in a forest in the Hamidabad region (south of Dezful), Khuzestan province, south-west Iran, in March 2018 (GPS coordinates: 32°07.38′N, 48°25.10′E).
Etymology. The species epithet refers to the type-related plant, P. euphratica, in which the nematode was recovered from its rhizosphere.
Iranian population of Tenunemellus indicus (Verma, 1972) Siddiqi, 1986
Description
Measurements. See table 1.
Female (figs 3 and 4). Body straight or curved ventrally upon fixation. Cuticle finely annulated, annulus 0.7 (0.5–1.0) μm wide. No lateral fields visible in cross section at mid-body. Cephalic region continuous with the body contour, smooth, anteriorly flattened, long and elevated, 5.1 (4.5–5.4) μm wide and 5.0 (4.4–5.5) μm high, with weak framework. Amphidial aperture with long straight longitudinal slit. Stylet delicate, conus 3.7 (3.3–4.1) μm or 38% (35–41%) of its total length, knobs rounded, slightly posteriorly directed, 1.2 (1.1–1.5) μm in width. Dorsal pharyngeal gland orifice 1.1 (0.7–1.5) μm posterior to stylet knobs. Median bulb fusiform, slightly developed with small distinct valve. Isthmus slender. Nerve ring at the middle of isthmus, 65 (59–72) μm from anterior end. Secretory–excretory pore at level of anterior part of pharyngeal bulb, hemizonid just anterior to the pore. Basal bulb elongated, cylindroid and set off from intestine. Cardia small. Reproductive system monodelphic–prodelphic, composed of an outstretched ovary with oocytes mostly arranged in single row. Spermatheca set off, filled with spheroid sperm. Crustaformeria quadricolumellate, uterus simple. Vulva a transverse slit lacking flap or epiptygma. Vagina varying from perpendicular to body axis to slightly anteriorly directed, occupying 42% (37–54%) of the corresponding body diameter. PUS 15 (14–17) μm, 1.7 (1.5–1.9) times longer than body diameter at vulval region. Tail elongate, gradually tapering to a finely rounded terminus.
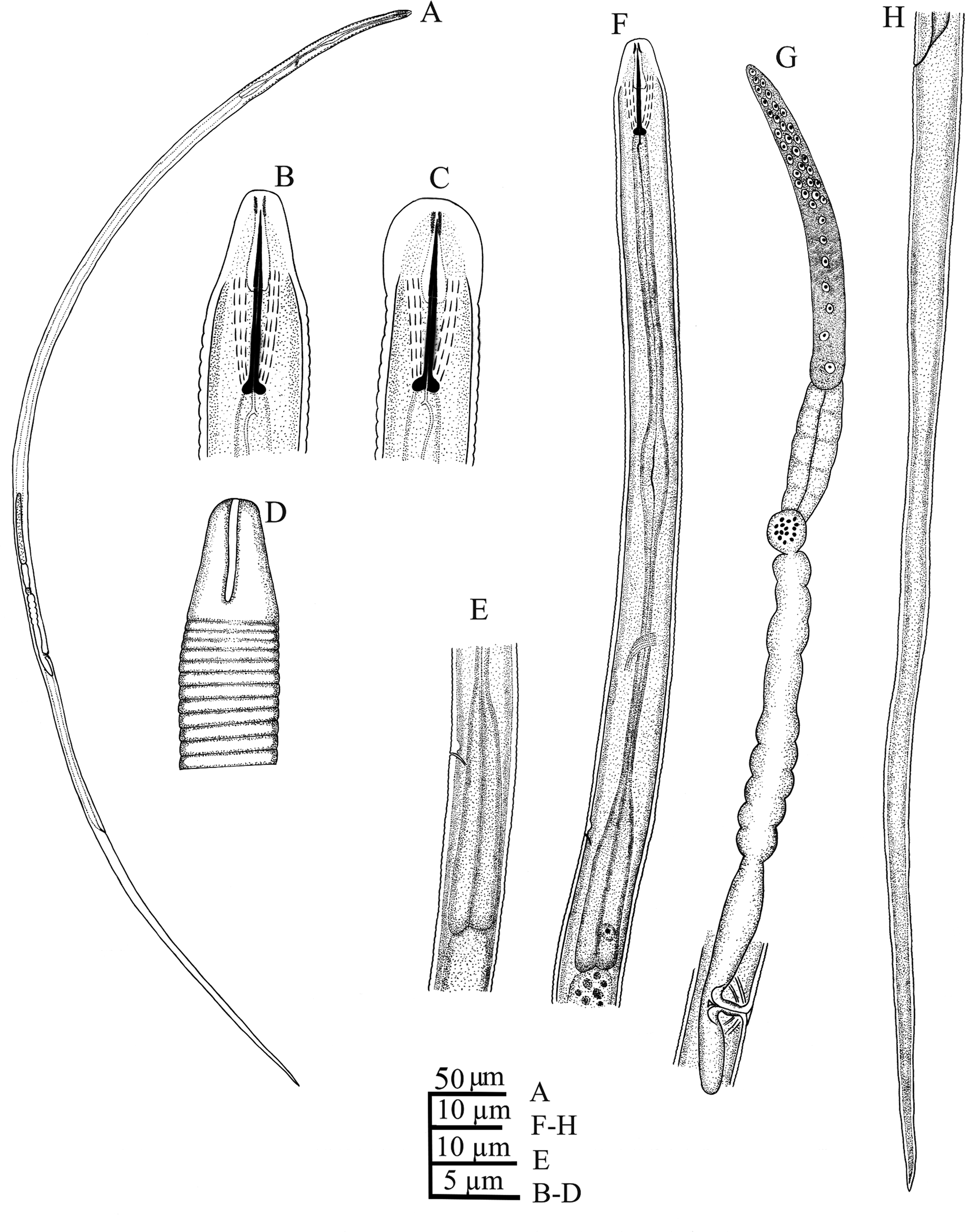
Fig. 3. Iranian population of Tenunemellus indicus (Verma, 1972) Siddiqi, 1986 female (line drawing): (A) entire body; (B) lateral view of cephalic region; (C) dorso-ventral view of cephalic region; (D) amphidial slit; (E) basal pharyngeal bulb; (F) anterior end and pharynx; (G) reproductive system; (H) tail.

Fig. 4. Iranian population of Tenunemellus indicus (Verma, 1972) Siddiqi, 1986 female (light microscope): (A) lateral view of cephalic region and stylet; (B) dorso-ventral view of cephalic region; (C) amphidial slit; (D) body annuli at mid-body; (E) posterior end of tail; (F) entire body; (G) vulval region; (H) tail. Scale bars: (A–E) 5 μm; (F) 100 μm; (G) 10 μm; (H) 15 μm.
Male. Not found.
Voucher specimens
Ten females were deposited in the collection of the Department of Plant Protection, College of Agriculture, University of Zanjan, Zanjan, Iran. Five females were deposited in the collection of the Department of Plant Protection, School of Agriculture, University of Shiraz, Shiraz, Iran.
Locality and habitat
The species was recovered from a soil sample collected from the rhizosphere of P. euphratica Oliv. in a forest in the Hosseinabad region (west of Shush), Khuzestan province, south-western Iran, in March 2018 (GPS coordinates: 32°02.53′N, 48°14.04′E).
Morphological remarks
The morphological characters of the Iranian population of T. indicus is in agreement with the those of the type population of the species; and minor (usually overlapping) differences in some measurements like body length (806–949 vs. 910–1010 μm), stylet length (9.0–10.8 vs. 11–12 μm), pharynx length (99–120 vs. 166 μm), the distance of secretory–excretory pore from anterior end (80–98 vs. 96–105 μm), annulus width (0.5–1.0 vs. 0.4–0.5 μm) and PUS length (1.5–1.9 vs. 1.0–1.5 times vulval body diameter) were observed, which may be attributed to the intraspecies variations.
Molecular profiles and phylogenetic relationships
The PCR amplifications yielded a single fragment of 1603 bp (partial 18S rDNA) for T. populus n. sp. and a single fragment of 645 bp (partial 28S rDNA D2–D3) for T. indicus. The newly obtained sequences were deposited in the GenBank database under accession numbers MN557353 for the 18S and MN535624 for the 28S regions.
The 18S rDNA dataset contained 53 sequences, including A. avenae (AB368918) as outgroup and the sequence of the new species. The 50% majority-rule consensus phylogenetic tree inferred using the abovementioned dataset by BI analysis under the GTR + I + G model is presented in fig. 5. In this tree, the new species is in sister relation to a clade containing Malenchus spp., Filenchus facultativus (Szczygiel, 1970) Raski & Geraert, 1987 (KJ869310), F. fungivorus Bert et al., 2010 (FJ949564) and Filenchus sp. (FJ949565). The 18S rDNA sequence of T. populus n. sp. differed by 63 bp (3.2%) and 65 bp (3.3%) from those of Malenchus ovalis (Siddiqi, 1979) Andrássy, 1981 (KX156297 and KX156298, respectively), 171 bp (5.3%) from F. fungivorus, 184 bp (6.6%) from Filenchus sp. and 184 (6.8%) from F. facultativus.

Fig. 5. Bayesian inference tree from the known and newly sequenced Thada populus n. sp. based on sequences of the partial 18S rDNA region. Bayesian posterior probabilities (50%) are given for each clade. Scale bar shows the number of substitutions per site.
The 28S rDNA dataset contained 34 sequences, including A. avenae (MF125328) as outgroup and sequence of the Iranian isolate of T. indicus. The 50% majority-rule consensus phylogenetic tree generated from the 28S dataset by BI analysis under the GTR + I + G model is presented in fig. 6. The 28S rDNA sequence of the Iranian isolate differed by 97 bp (4.7%) from that of Lelenchus leptosoma (De Man, 1880) Andrássy, 1954 (KX156322), 121 and 153 bp (6.4 and 6.5%) from that of Labrys fuzhouensis Qiao et al., 2019 (MK039727 and MK039728, respectively) and 199 bp (32.5%) and 214 bp (32.7%) from that of Tenunemellus sheri (MG994966 and MG994967, respectively). In the 28S rDNA tree, the two currently sequenced species of Tenunemellus have occupied separate placements. Tenunemellus sheri (MG994967 and MG994966), L. leptosoma (KX156335 and KP730042) and Lelenchus brevislitus Soleymanzadeh et al., 2016 (KU234169) have fallen into the clade A. The Iranian isolate of T. indicus has fallen into the clade B, also including L. fuzhouensis (MK039727 and MK039728), L. leptosoma from the Netherlands (KX156322), Malenchus spp. and three isolates of Filenchus discrepans (Andrássy, 1954) Andrássy, 1972.

Fig. 6. Bayesian inference tree from the known and newly sequenced Tenunemellus indicus based on sequences of the D2–D3 expansion fragments of the 28S rDNA region. Bayesian posterior probabilities (50%) are given for each clade. Scale bar shows the number of substitutions per site.
Discussion
The present study enriched the Tylenchidae genera occurring in Iran. The two genera Thada and Tenunemellus were reported from the country for the first time. Of the resolved clades in the present 18S tree, six clades are considered (fig. 5). Group I includes species and genera with amphidial slits in front of the cephalic region that are not elongated or are confined to the first annulus after labial plate viz. Ecphyadophora, Labrys, Filenchus misellus (Andrássy, 1958) Raski & Geraert, 1987 and F. chilensis Raski & Geraert, 1987. Group II includes those species and genera with sigmoid amphidial slit, beginning near oral aperture and extending as lateral longitudinal slits viz. Malenchus, Ottolenchus Husain & Khan, 1967 and T. populus n. sp. Group III includes representatives of the genera Basiria, Boleodorus Thome, 1941 and Neopsilenchus Thome & Malek, 1968, with a transverse amphidial slit. Group IV includes the taxa with straight elongate amphidial slit, beginning at edge or near labial plate, extending laterally through three or four cephalic region annuli viz. Filenchus Andrássy, 1954 species with four lateral lines and the genus Tylenchus Bastian, 1865. Group V includes the genus Lelenchus Andrássy, 1954, characterized by long amphidial slits, beginning near oral aperture. And group VI includes the taxa having small amphidial slits that are confined to the oral plate, including the genera Coslenchus and Cephalenchus Goodey, 1962. The current result concurs with previous studies (Geraert & Raski, Reference Geraert and Raski1987; Qing & Bert, Reference Qing and Bert2018). The first phylogenetic study of Thada using small subunit rDNA data, although using one representative, revealed that its placement under Boleodorinae (Geraert, Reference Geraert2008) is a superficial grouping, and its placement under the subfamily Thadinae according to Decraemer & Hunt (Reference Decraemer, Hunt, Perry and Moens2013) seems more logical.
According to the 28S tree, and based upon currently available data, it may be concluded that Tenunemellus is not monophyletic, although further data of other species and other genomic markers need to be examined.
Acknowledgements
The authors thank anonymous reviewers and the editor of this manuscript for their valuable suggestions to improve the paper.
Financial support
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
Conflicts of interest
None.
Ethical standards
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional guides on the care and use of laboratory animals.









