Introduction
Root-knot nematodes (Meloidogyne spp.) are one of the most important pests, given their widespread distribution, host range, and potential damage to many crops (Hussey & Janssen, Reference Hussey, Janssen, Starr, Cook and Bridge2002). Wesemael et al. (Reference Wesemael, Viaene and Moens2011) reported that more than 90 species of root-knot nematodes had been described. They are significantly affecting agricultural products in the tropical, subtropical and temperate regions (Trudgill & Blok, Reference Trudgill and Blok2001; Hunt & Handoo, Reference Hunt, Handoo, Perry, Moens and Starr2009). The most common known species are M. incognita (Kofoid & White, Reference Kofoid and White1919) Chitwood, 1949; M. javanica (Treub, Reference Treub1885) Chitwood, 1949; M. arenaria (Neal, 1889) Chitwood, 1949; M. hapla Chitwood, 1949 and M. exigua (Göldi, Reference Göldi1887).
Twenty-one species of the Meloidogyne genus have been recorded on coffee (Souza, Reference Souza2008; Humphreys-Pereira et al., Reference Humphreys-Pereira, Flores-Chaves, Gómez, Salazar, Gómez-Alpízar and Elling2014). In Vietnam, only M. incognita, M. exigua and M. coffeicola have been recorded on coffee (Trinh et al., Reference Trinh, Pham and Nguyen2013), and M. incognita causes serious damage at almost all the coffee areas (Nguyen & Nguyen, Reference Nguyen and Nguyen2000, Reference Nguyen and Nguyen2001; Trinh et al., Reference Trinh, de la Peña, Nguyen, Nguyen and Moens2009).
Morphological analysis of perineal patterns in adult females of Meloidogyne species is often used as a rapid diagnostic tool. The basic morphology of the perineal pattern is shown as a collage of the alternative features throughout the genus. Jepson (Reference Jepson1987) proposed that the best way to examine the whole genus was to group species with similar morphology, and recognized six broad groups based on perineal pattern features.
However, this approach has some major limitations, including overlapping morphological characters, which can lead to misidentification (Souza, Reference Souza2008). Some coffee-parasitizing nematodes, such as M. izalcoensis, M. paranaensis and M. incognita, have overlapping distributions of characters that are related to the perineal patterns of females (Brito et al., Reference Brito, Powers, Mullin, Inserra and Dickson2004; Carneiro et al., Reference Carneiro, Randig, Almeida and Goncalves2005).
Ribosomal coding genes, such as the small subunit 18S internal transcribed spacer (ITS) and the large subunit 28S rRNA genes, evolve relatively slowly (Blaxter, Reference Blaxter, Kennedy and Harnett2001; De Ley et al., Reference De Ley, De Ley, Vierstraete, Karssen, Moens and Vanfleteren2002). These genes are useful for phylogenetic studies among groups of plant-parasitic, animal-parasitic, and free-living nematodes, or between the orders within the phylum Nematoda (Blaxter, Reference Blaxter, Kennedy and Harnett2001). Conversely, mitochondrial genes evolve more rapidly than ribosomal genes, which makes them useful for intraspecific and population genetic studies or intra-genus and intra-family analyses (Powers & Harris, Reference Powers and Harris1993; Blaxter, Reference Blaxter, Kennedy and Harnett2001; Plantard et al., Reference Plantard, Picard, Valette, Scurrah, Grenier and Mugniéry2008).
During our surveys in 2015 in the Western Highlands of Vietnam, we found root samples of coffee with untypical symptoms, including a high number of egg masses protruding outside the roots, and numerous small, white galls gathered around adventitious roots. These symptoms differ from those caused by other root-knot nematodes present in the region. Therefore, the objective of this study was to describe the new root-knot nematode using an integrated approach, including the diagnostics of morphometric, morphological and molecular characters.
Materials and methods
Nematode population
Nematodes were extracted from galled root and soil samples of Robusta coffee that were collected in Cu M'gar District, Dak Lak Province, Western Highlands, Vietnam.
Morphological characterization
Second-stage juveniles and males were extracted from soil samples using the modified Baermann tray method (Whitehead & Hemming, Reference Whitehead and Hemming1965). Measurements of heat-killed nematodes were made on permanent slides following transarticular screw fixation (TAF) and ethanol–glycerin dehydration, according to the method described by Seinhorst (Reference Seinhorst1962) and modified by De Grisse (Reference De Grisse1969).
Females were extracted from the root tissues under a stereomicroscope, using a scalpel. Perineal patterns of mature females were prepared following Hartman & Sasser (Reference Hartman, Sasser, Barker, Carter and Sasser1985) and mounted in lactophenol.
Light microscopy (LM)
All slides were examined under an Axio Lab.A1 light microscope (Carl Zeiss AG, Oberkochen, Germany). Measurements, line drawings and photographs were produced using ZEN lite software and a ZEISS Axiocam ERc 5s digital camera (Carl Zeiss AG, Oberkochen, Germany). Raw photographs were edited using Adobe Illustrator CS3 (Adobe Systems, San Jose, California, USA). Comparisons of morphological and morphometric characters were based on the diagnostic keys of Hewlett & Tarjan (Reference Hewlett and Tarjan1983), Eisenback (Reference Eisenback, Sasser and Carter1985), Jepson (Reference Jepson1987), Karssen (Reference Karssen2002) and Kazachenko & Mukhina (Reference Kazachenko and Mukhina2013).
Scanning electron microscopy (SEM)
Following examination and identification, some specimens in good condition were selected for observation under a scanning electron microscope, following Abolafia (Reference Abolafia2015). The nematode was hydrated in distilled water, dehydrated in a graded ethanol and acetone series, critical point dried, coated with gold, and examined under a Merlin scanning electron microscope (Carl Zeiss AG, Oberkochen, Germany).
Molecular characterization
DNA was extracted from individual second-stage juveniles using the Lysis buffer protocol as described by Holterman et al. (Reference Holterman, Oggenfuss, Kiewnick and Frey2009). The extracted crude DNA was kept at −70°C until it was used. The crude DNA of each specimen was amplified using the following sets of primers: for ITS rRNA genes using the reverse primer MelR5′–TCGTAACAAGGTAGCTGTAG–3′ and forward primer MelF5′–TGCTCTCGACTGAGTTCAG–3′, modified from Vrain et al. (Reference Vrain, Wakarchuk, Lévesque and Hamilton1992); for the mtDNA cytochrome c oxidase I (COI) genes using the Cox1 F/Cox1R primers (Cox1–5′–TGGTCATCCTGAAGTTTATG–3′/Cox1R–5′–CTACAACATAATAAGTATCATG–3′), modified from Kiewnick et al. (Reference Kiewnick, Holterman, van den Elsen, van Megen, Frey and Helder2014); for the mtDNA cytochrome c oxidase II (COII) genes using the primers Cox2–5′GTCAATGTTCAGAAATTTGTG–3′/16R–5′–TACCTTTGACCAATCACGCT–3′ (Powers & Harris, Reference Powers and Harris1993); and for the D2–D3 fragment of the 28S rRNA gene using the forward primer D2–5′–ACAAGTACCGTGAGGGAAAGTTG–3′/ and reverse primer D3–5′–TCCTCGGAAGGAACCAGCTACTA–3′ (De Ley et al., Reference De Ley, Felix, Frisse, Nadler, Sternberg and Thomas1999). All of the polymerase chain reactions (PCR) were performed in 25 μl Hot Start PCR Master Mix (Promega, Madison, Wisconsin, USA), 1 μM of each forward and reverse primer, 5 μl crude DNA and Milli-Q water (Merck Millipore, Burlington, Massachusetts, USA). After electrophoresis in 1% TAE-buffered agarose gel (1 h, 100 V) the PCR products were visualized under UV light, excised from the gel and purified using the Wizard SV Gel and PCR Clean-Up System (Promega, Madison, Wisconsin, USA). For direct sequencing of both DNA strands, 20 mg was used. The sequences were aligned using ClustalW in BioEdit (Hall, Reference Hall1999). The phylogenetic trees were generated based on Maximum Likelihood (ML) analysis in MEGA 6 (Tamura et al., Reference Tamura, Stecher, Peterson, Filipski and Kumar2013). The phylograms were bootstrapped 1000 times to assess the degree of support for the phylogenetic branching indicated by the optimal tree for each method.
Results
Systematics
Phylum: Nematoda Potts, 1932
Class: Chromadorea Inglis, 1983
Order: Rhabditida Chitwood, 1933
Suborder: Tylenchina Thorne, 1949
Infraorder: Tylenchomorpha De Ley & Blaxter, 2002
Superfamily: Tylenchoidea Örley, 1880
Family: Meloidogynidae Skarbilovich, 1959
Genus: Meloidogyne Göldi, 1887
Meloidogyne daklakensis n. sp.
Description
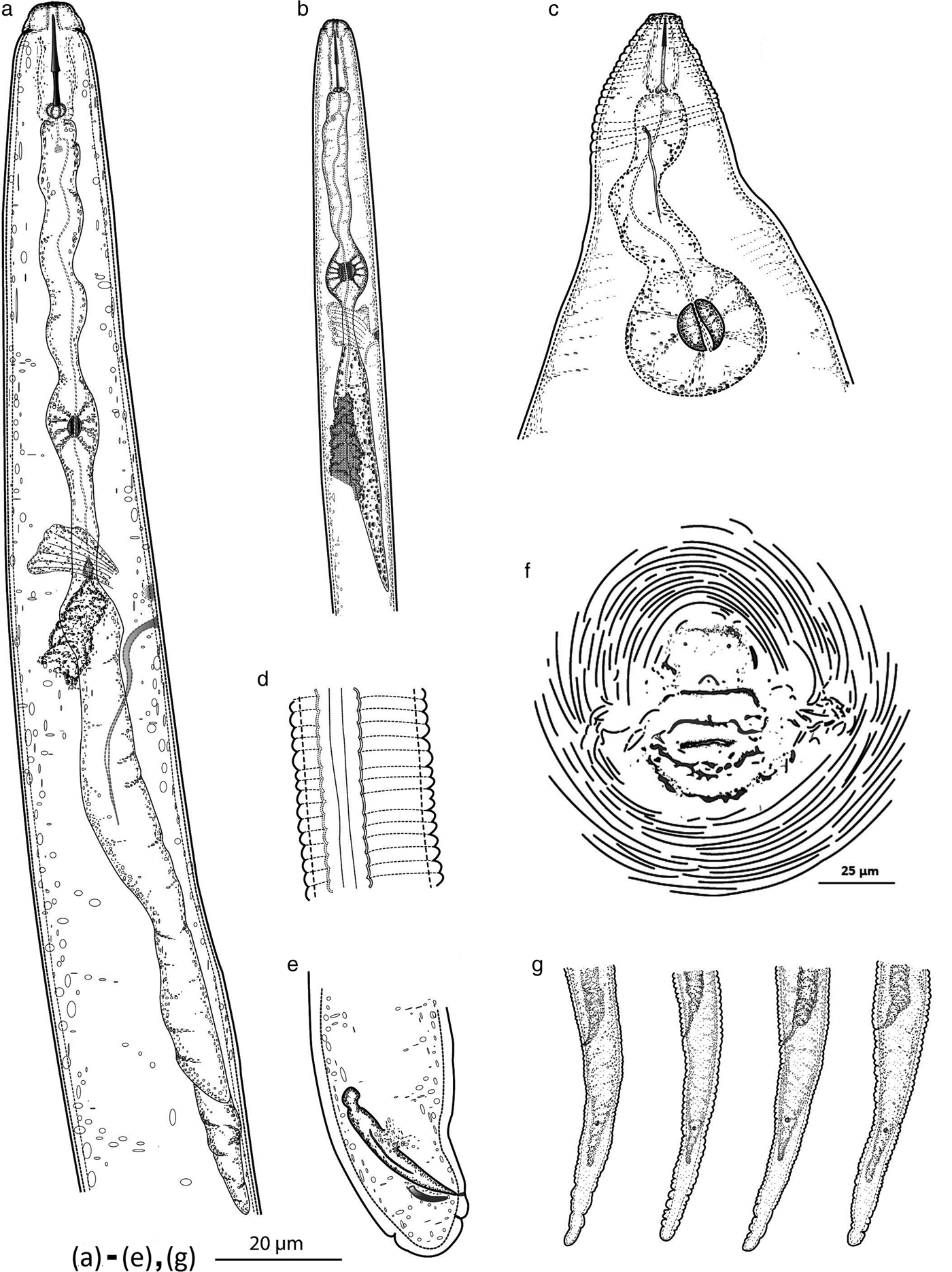
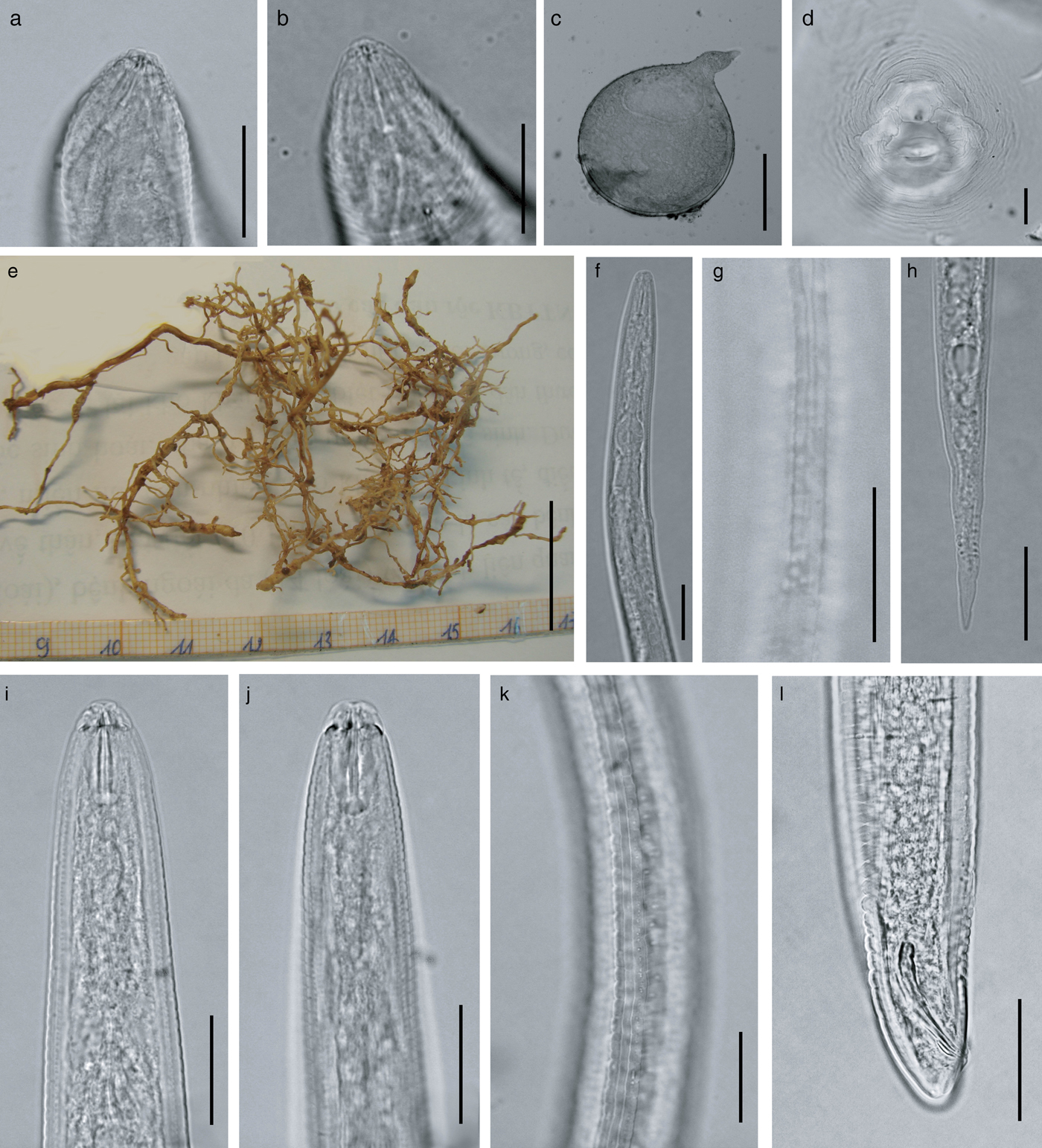
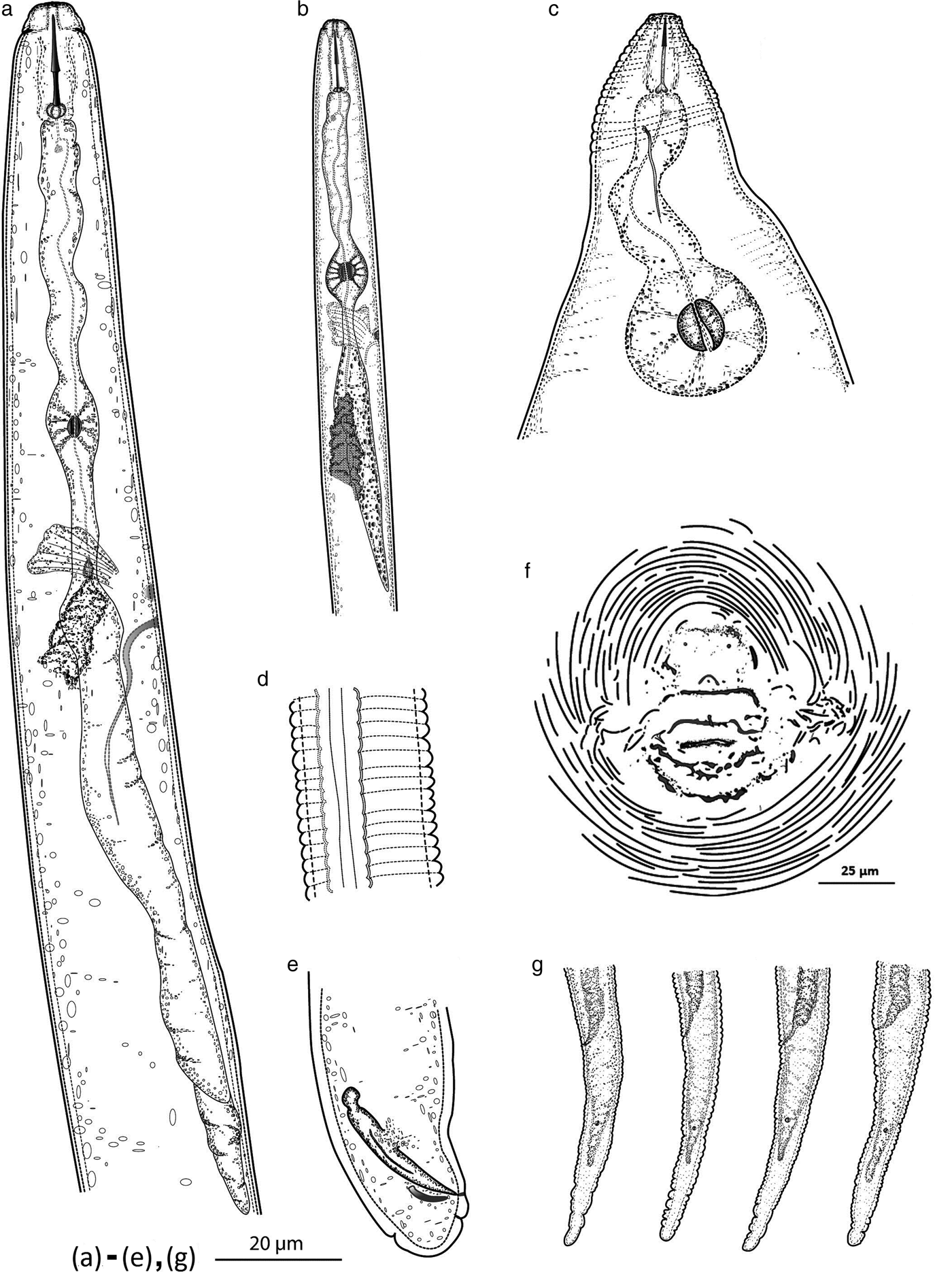
Females. Body pearly white, pear-shaped, and anterior body portion commonly off-centre from a median plane (fig. 2c). Anterior end pointed; posterior end varying from rounded to slightly flattened. Neck usually short, not curved (bent). Stoma slit-like; lip region sclerotized and slightly offset from rest of body, bearing one annulus; labial disc not fused with medial lips, elevated; lateral sectors enlarged, extension prominent; amphidial openings elongated (fig. 3a–e). Stylet short, cone base triangular and wider than shaft. Stylet tip normally straight (sometimes curved dorsally), shaft cylindrical, same diameter throughout its length (figs 1c and 2a, b). Three stylet knobs, oval and sloping posteriorly (figs 1c and 2a, b). Distance from base of stylet to dorsal pharyngeal gland orifice (DGO) c. 5 ± 0.2 μm. Pharyngeal lumen of procorpus wide, often showing rounded protuberances. Excretory pore situated far behind stylet knobs, within range of level of procorpus to metacorpus (figs 1c and 3b). Metacorpus large and rounded, valve apparatus oval-shaped and sclerotized. Perineal patterns of females rounded to oval; striae smooth, regular and continuous; lateral lines reduced, resulting in appearance of thick prominent lateral part in SEM; dorsal arch low, rounded and encloses distinct vulva, anus and tail tip. Phasmids indistinct. Vulva slit centrally located in the unstriated area, and longer than the distance between vulva and anus (figs 1f, 2d and 3f).
Males. Body vermiform, anterior end tapering and posterior region bluntly rounded. Lip region slightly offset from body, with a high lip region cap consisting of a large labial annulus. Medial lips and labial disc are bow-tie shaped. Lateral lips large and triangular, slightly lower than labial disc and medial lips. Amphidial openings appear as long, wide slits located between labial disc and lateral lips. Post-labial annulus divided into four lobes by longitudinal slits (figs 1a, 2i, 2k and 3g–k). Stylet robust and straight, one lateral knob projected, two others sloping posteriorly (figs 1a and 2i, l). Distance from base of stylet to DGO = 5 ± 1 μm. Procorpus distinctly outlined, three times longer than metacorpus. Metacorpus ovoid, with a strong valve apparatus. Excretory duct curved. Lateral field with four incisures forming three bands, areolation of whole body (figs 2k and 3k). One testis, occupying 58% of the body cavity. Spicules slightly curved ventrally, with bluntly rounded terminus. Gubernaculum short and crescent-shaped (figs 1e and 2l).
Second-stage juveniles (J2). Body slender, tapering to an elongated tail. Lip region narrower than body, weak and slightly offset from body; labial disc offset; stoma slit-like; medial lips and labial disc bow-tie shaped; labial disc rounded; amphidial openings enlarged, covered lateral lips (figs 1b and 3m–q). Stylet slender; cone weakly expanded at junction with shaft; knobs rounded (figs 1b and 2f). Distance from stylet knobs to dorsal gland orifice c. 4 ± 0.8 μm. Metacorpus broadly oval, valve large and heavily sclerotized. Pharyngeal–intestinal junction at level of nerve ring. Excretory pore slightly behind nerve ring, opening just behind hemizonid. Lateral field with four incisures, not areolated or prominent (figs 2g and 3o). Phasmids small, distinct. Tail elongate–conoid (figs 1g, 2h and 3q).

Fig. 1. Meloidogyne daklakensis n. sp. (a) Anterior of the male, showing pharynx and excretory canal; (b) anterior of the second-state juvenile, showing pharynx and excretory canal; (c) female anterior end; (d) lateral field of male; (e) tail region; (f) perineal pattern; (g) tail variation of juvenile.

Fig. 2. Light micrographs of M. daklakensis n. sp. Female: (a, b) anterior end; (c) body; (d) perineal pattern; (e) coffee root. Second-stage juvenile: (f) anterior end; (g) lateral fields; (h) tail region. Male: (i, j, k) anterior end; (l) lateral fields; (m) tail region. (Scale bar: (a, b, d, f, g, h, i, k, l, m): 25 μm; (c): 200 μm; (e): 2000 μm).

Fig. 3. Scanning electron micrographs of M. daklakensis n. sp. Female: (a) neck region; (b) anterior end, lateral view; (c) anterior end, ventral view; (d) en face view; (e) lip region, lateral view; (f) perineal pattern. Male: (g) anterior end, ventral view; (h) anterior end, lateral view; (i) en face view; (j) lip region, lateral view; (k) lateral fields; (l) tail region. Second-stage juvenile: (m) anterior end, ventral view; (n) anterior end, subdorsolateral view; (o) lateral field; (p) en face view; (q) posterior end. (Scale bar: (a–f): 10 μm; (g–l, q, m): 5 μm; (l, p): 1 μm, (o): 2 μm).
Taxonomic summary
Type host. Robusta coffee (Coffea canephora Pierre ex A. Froehner).
Type locality. Western Highlands, Dak Lak Province (12°47′12′'N, 108°05′43″E).
Etymology. The species epithet refers to the Dak Lak Province, Western Highlands, Vietnam, where this new species was found.
Type material. Holotype (females) and paratypes that were isolated from the roots of the type host are deposited in the nematode collection of the Nematology Department at the Institute of Ecology and Biological Resources (IEBR), Vietnam Academy of Science and Technology (VAST). Five female paratypes are deposited in the nematode collection of the Zoology Museum, Ghent University, K.L. Ledeganckstraat 35, Ghent, Belgium. The ITS, D2D3, COI and COII sequences are deposited in GenBank under the accession numbers (KU243339, MG266058), (KU243338, MG266060), (MG431339, MG341340) and (KU243337, MG431341), respectively.
Pathology. The coffee roots infected with M. daklakensis sp. n. had relatively small galls (1–2 mm diameter). Each gall contained a single female, with the exposure of an egg mass outside of the gall (fig. 2e).
Differential diagnosis
Meloidogyne daklakensis n. sp. is closest to M. marylandi Jepson & Golden, 1987, M. naasi Franklin, 1965, M. ovalis Riffle, 1963, M. panyuensis Liao, Yang, Feng & Karssen, 2005, M. lopezi Humphreys-Pereira, Flores Chaves, Gomez, Salazar, Gomez-Alpizar & Elling, 2014, M. mali Itoh, Ohshima & Ichinohe, 1969 and M. baetica Castillo, Vovlas, Subbotin & Troccoli, 2003 (the perineal patterns are characterized by a round/oval-shaped, moderately high dorsal arch, and smooth, regular and continuous striae). However, the species can be distinguished from each other by certain features (tables 1 and 2).
Table 1. Morphometric indices of the females, males and second-stage juveniles (J2) of M. daklakensis n. sp. All measurements are in μm and in the form: mean ± SD (range).

Table 2. Morphological differences between M. daklakensis n. sp. and some closely related Meloidogyne species. All measurements are in μm and in the form: mean ± SD (range).
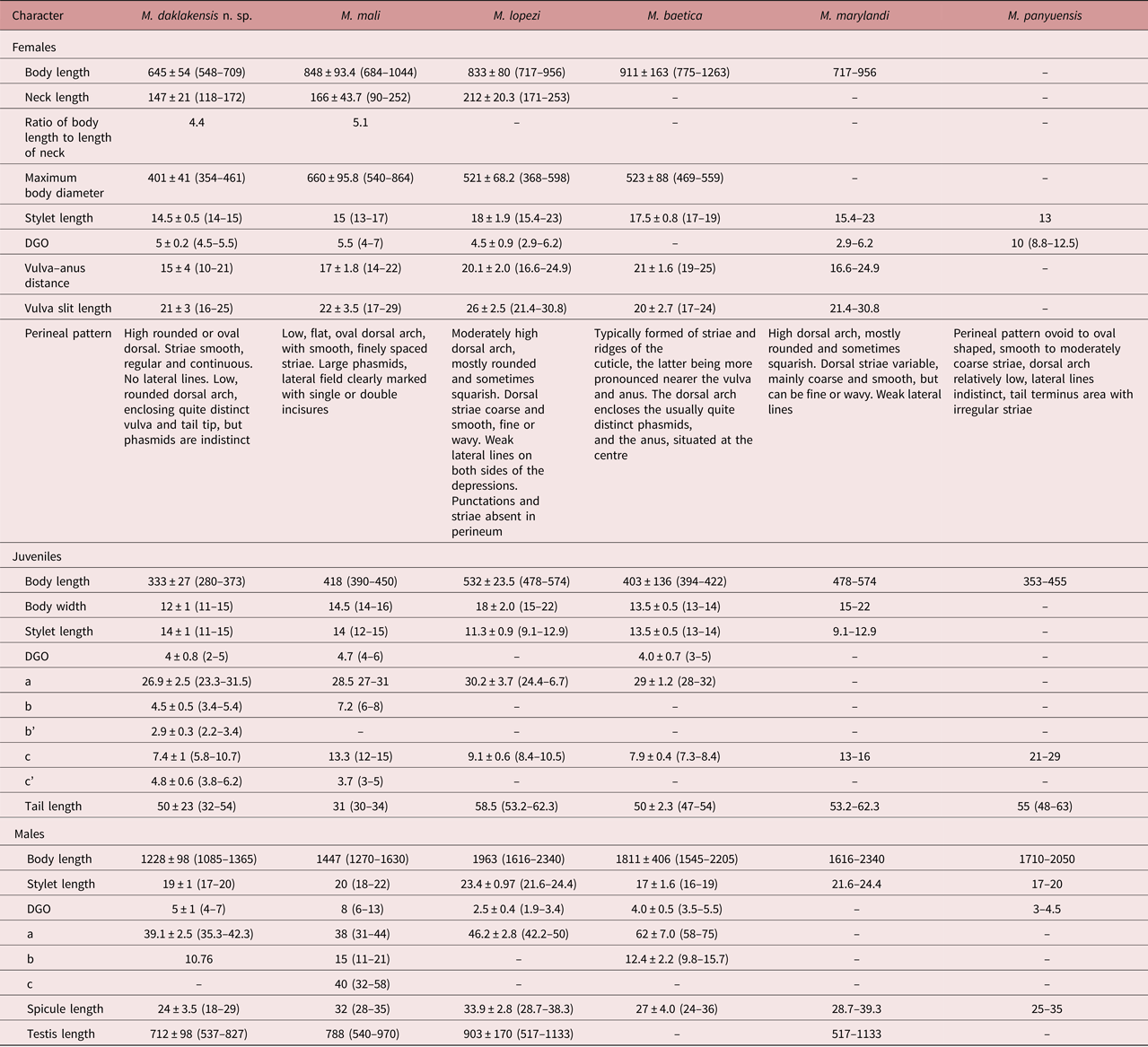
Meloidogyne daklakensis n. sp. differs from M. marylandi Jepson & Golden, 1987 in that females have shorter body length (548–709 vs 717–956 μm) and stylet length (14–15 vs 15.4–23 μm); males have shorter body length (1085–1365 vs 1616–2340 μm) and spicule length (18–29 vs 28.7–39.3 μm); and second-stage juveniles have shorter body length (280–373 vs 478–574 μm), smaller body diameter (11–16 vs 15–22 μm) and shorter tail length (32–54 vs 53.2–62.3 μm).
Meloidogyne daklakensis n. sp. differs from M. naasi Franklin, 1965 in that females have a perineal pattern with indistinct phasmids vs large phasmids, and second-stage juveniles have shorter body length (280–373 vs 418–465 μm); b = 3.4–5.4 vs 7–8; c = 7.4 vs 6.2.
Meloidogyne daklakensis n. sp. differs from M. ovalis Riffle, 1963 in that females have indistinct phasmids vs large phasmids; males have shorter body length (1085–1365 vs 1300–1800 μm) and spicule length (18–29 vs 31–38 μm); and second-stage juveniles have shorter body length (280–373 vs 350–430 μm); a = 27 vs 22; c = 7.4 vs 8.
Meloidogyne daklakensis n. sp. differs from M. panyuensis Liao et al., 2005 in that in females the distance from anterior end to excretory pore is shorter (18–21 vs 32.5–37.5 μm) and DGO = 4.5–5.5 vs 8.8–12.5 μm; males have shorter body length (1085–1365 vs 1710–2050 μm), DGO = 4–7 vs 3–4.5 μm, and shorter spicule length (18–29 vs 25–35 μm); and second-stage juveniles have shorter body length (280–373 vs 353–455 μm), c =5.8–10.8 vs 21–29, shorter distance from anus to posterior end (32–54 vs 48–63 μm), and longer hyaline part (9–17 vs 7.5–10 μm).
Meloidogyne daklakensis n. sp. differs from M. lopezi Humphreys-Pereira et al., 2014 in that females have shorter body length (548–709 vs 717–956 μm), stylet length (14–15 vs 15.4–23 μm) and distance from anterior end to excretory pore (18–21 vs 22–59 μm), and the perineal pattern has moderately smooth, regular and continuous dorsal striae and reduced lateral lines vs coarse, fine or wavy striae and weak lateral lines; males have shorter body length (1085–1365 vs 1616–2340 μm), smaller body diameter (27–34 vs 37–47 μm), shorter spicule length (18–29 vs 28.7–38.3 μm), DGO = 4–7 vs 1.9–3.4 μm), lip region offset from body, with a high lip region cap consisting of a large labial annulus and a prominent post-labial annulus vs lacking annulation; and second-stage juveniles have shorter body length (280–373 vs 478–574 μm), smaller body diameter (11–15 vs 15–22 μm), longer stylet length (11–16 vs 9.1–12.9 μm), shorter distance from anterior end to excretory pore (55–78 vs 95–109 μm), and shorter tail length (32–54 vs 53.2–62.3 μm).
Meloidogyne daklakensis n. sp. differs from M. mali Itoh et al., 1969 in that females have smaller maximum body diameter (548–709 vs 684–1044 μm), reduced lateral lines vs clearly visible lateral lines, and indistinct phasmids vs large phasmids; males have shorter body length (1085–1365 vs 1270–1630 μm) and spicule length (18–29 vs 28–35 μm); and second-stage juveniles have shorter body length (280–373 vs 390–450 μm), smaller maximum body diameter (11–15 vs 14–16 μm) and longer tail length (32–54 vs 30–34 μm).
Meloidogyne daklakensis n. sp. differs from M. baetica Castillo et al., 2003 in that females have the excretory pore situated within the range from level of procorpus to level of metacorpus vs constantly anterior to the level of stylet knob, indistinct vs quite distinct phasmids, shorter body length (548–709 vs 755–1263 μm) and stylet length (14–15 vs 17–19 μm), smaller maximum body diameter (322–462 vs 469–559 μm), and smaller distance from vulva to anus (10–21 vs 19–25 μm); males have shorter body length (1085–1365 vs 1545–2205 μm); and second-stage juveniles have shorter body length (280–373 vs 394–422 μm) and shorter length from anterior end to excretory pore (55–78 vs 85–100 μm).
Molecular characterization
Amplification of the ITS rDNA region of M. daklakensis n. sp. yielded a single fragment approximately 600 bp in length (gene sequence was 516 bp). The ITS sequences of the new species are most similar to those of M. panyuensis, M. mali and M. marylandi, by 82%, 81% and 79%, respectively. Phylogenetic trees generated based on the Maximum Likelihood (ML) method show that M. daklakensis n. sp. is closest to M. panyuensis with high bootstrap (89%), and separated from other Meloidogyne species (fig. 4).
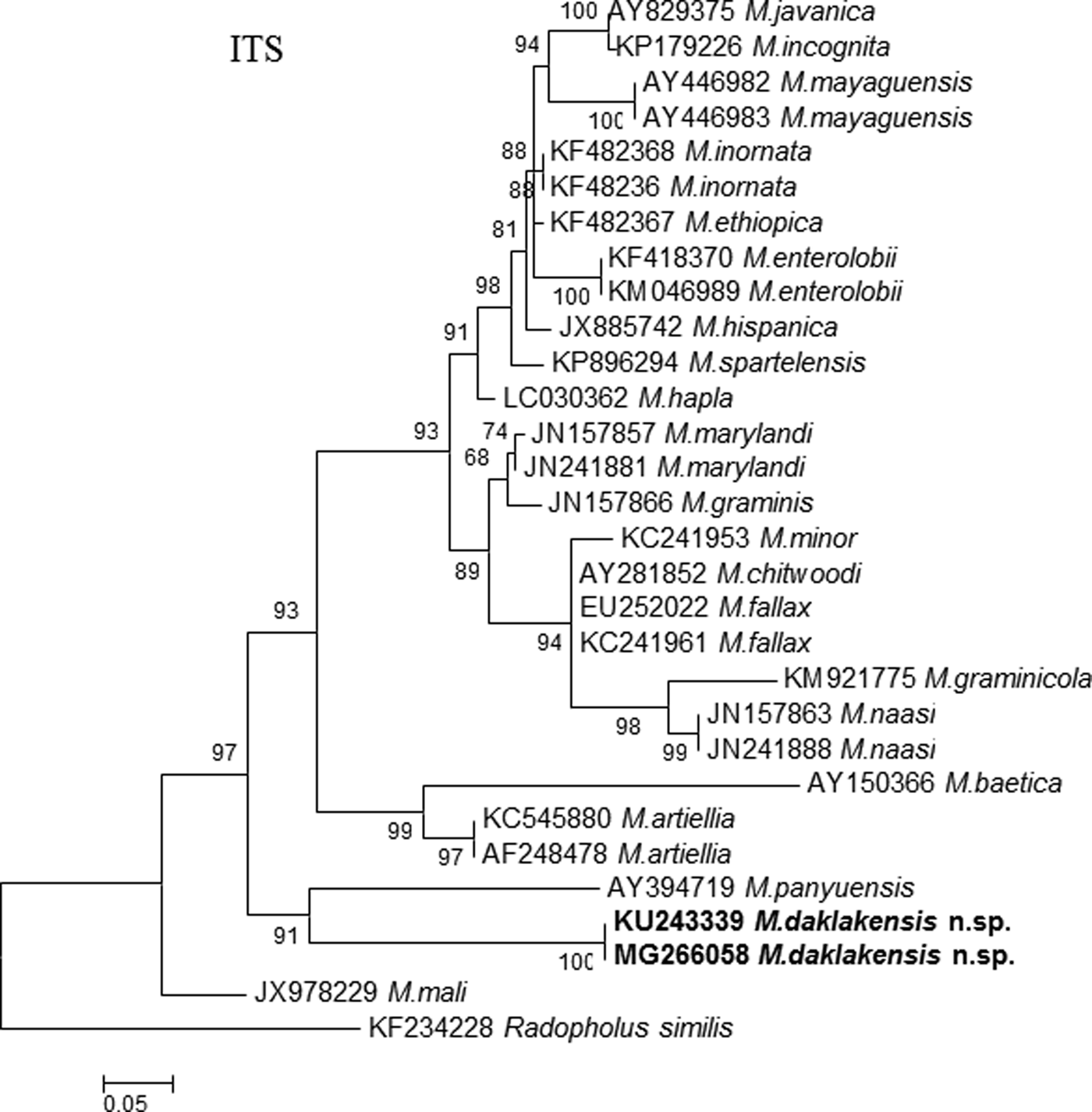
Fig. 4. Phylogenetic relationships of M. daklakensis n. sp. with other root-knot nematodes, from the analysis of the ITS sequences. Numbers to the left of the branches are bootstrap values for 1000 replications.
The D2D3 aligned sequence of M. daklakensis n. sp. was 727 bp. The D2D3 sequence of M. daklakensis n. sp. is similar to those of M. ichinohei, M. camellia and M. mali by 86%, 84% and 83%, respectively. The phylogenetic relationships between M. daklakensis n. sp. and other Meloidogyne species based on the 28S ribosomal RNA sequences indicated that M. daklakensis n. sp. was grouped with M. ichinohei and M. camellia with moderate bootstrap value (84%). Meloidogyne daklakensis n. sp. was clearly separated from other related Meloidogyne species.
On the other hand, the COI sequence length of M. daklakensis n. sp. was 364 bp. The Basic Local Alignment Search Tool (BLAST) search for the COI sequence of M. daklakensis n. sp. revealed the highest identical match with the sequences of M. ichinohei and M. mali, by 92% and 86%, respectively. The phylogenetic tree of COI sequences placed M. daklakensis n. sp. in a clade together with M. ichinohei (bootstrap value = 93%), and they were also a sister group of M. mali with 99% bootstrap (fig. 7). The COII–16S sequence length of M. daklakensis n. sp. was 526 bp, which was most similar to M. mali and M. marylandi, by 77% and 75%, respectively. The COII–16S phylogenetic tree showed that M. daklakensis n. sp. belonged to a clade together with M. mali and M. marylandi (99% support). The new species was clearly separated from other root-knot nematodes (fig. 7).

Fig. 5. Phylogenetic relationships of M. daklakensis n. sp. with other root-knot nematodes, from the analysis of the D2D3 (28S–rRNA) sequences. Numbers to the left of the branches are bootstrap values for 1000 replications.

Fig. 6. Phylogenetic relationships of M. daklakensis n. sp. with other root-knot nematodes, from the analysis of the COI–rRNA sequences. Numbers to the left of the branches are bootstrap values for 1000 replications.

Fig. 7. Phylogenetic relationships of M. daklakensis n. sp. with other root-knot nematodes, from the analysis of the alignment of the COII–16S–rRNA sequences. Numbers to the left of the branches are bootstrap values for 1000 replications.
Discussion
Molecular markers are an important tool for identification of nematodes and remain essential for accurate diagnosis of root-knot nematodes (Perry & Moens, Reference Perry and Moens2013). Furthermore, DNA-based methods can be applied to various stages of the nematodes to distinguish individual species from mixed populations (Onkendi & Moleleki, Reference Onkendi and Moleleki2013). The 28S region is relatively conserved with high similarity among the three most common tropical root-knot nematode species, M. incognita, M. arenaria and M. javanica. Therefore, conserved ribosomal DNA is not useful for identifying these species (Ye et al., Reference Ye, Zeng and Kerns2015; Janssen et al., Reference Janssen, Karssen, Topalović, Coyne and Bert2017). However, it can be a useful tool for identification of M. marylandi, M. graminis and M. naasi, or M. enterolobii, M. hapla, M. chitwoodi and M. fallax (Onkendi & Moleleki, Reference Onkendi and Moleleki2013; Ye et al., Reference Ye, Zeng and Kerns2015). In order to confirm identification of Meloidogyne at species level, mitochondrial DNA is usually used (Tigano et al., Reference Tigano, Carneiro, Jeyaprakash, Dickson and Adams2005; Landa et al., Reference Landa, Rius, Vovlas, Carneiro, Maleita, Abrantes and Castillo2008). The combined analyses of 18S and 28S nuclear rDNA sequences showed the haplotypes of Meloidogyne species on coffee (Herrera et al., Reference Herrera, Bryngelsson, Monzón and Geleta2011). The mitochondrial DNA has a faster rate of evolution than the corresponding nuclear genes, creating sufficient nucleotide variation for species-level analyses (Hugall et al., Reference Hugall, Moritz, Stanton and Wolstenholme1994; Blok & Powers, Reference Blok, Powers, Perry, Moens and Starr2009). Extensive geographical sampling of the COI can be conducted to assess interspecific variability (Kiewnick et al., Reference Kiewnick, Holterman, van den Elsen, van Megen, Frey and Helder2014; Janssen et al., Reference Janssen, Karssen, Verhaeven, Coyne and Bert2016). The mitochondrial haplotypes are strongly linked and consistent with traditional esterase isozyme patterns, which suggests that different parthenogenetic lineages can be identified reliably using mitochondrial haplotypes (Janssen et al., Reference Janssen, Karssen, Verhaeven, Coyne and Bert2016). The COII is a highly conserved region, and therefore it was easy to distinguish different Meloidogyne species (Blouin, Reference Blouin2002; Tigano et al., Reference Tigano, Carneiro, Jeyaprakash, Dickson and Adams2005; Onkendi & Moleleki, Reference Onkendi and Moleleki2013). As an alternative to the morphological identification of Meloidogyne spp., esterase patterning has been used for diagnosing Meloidogyne spp. from a wide range of samples and has been proved to be species-specific for a number of species (Carneiro & Almeida, Reference Carneiro and Almeida2001; Carneiro & Cofcewics, Reference Carneiro, Cofcewics and Souza2008). Isozyme electrophoresis patterning has discriminated between all of these cryptic species, but this technique is restricted to females (Carneiro & Cofcewics, Reference Carneiro, Cofcewics and Souza2008). Isozyme electrophoretic profiles, often using esterase (EST) and malate dehydrogenase (MDH), have been established for a number of species and can provide a useful routine diagnostic test, particularly for morphologically variable species (Blok & Powers, Reference Blok, Powers, Perry, Moens and Starr2009). Certain limitations have also been associated with biochemical analysis, such as the absence of enzymatic patterns for all the species of Meloidogyne and the dependence on the nematode development phase (Blok & Powers, Reference Blaxter, Kennedy and Harnett2009). Other limitations can include the occurrence of intraspecific variations and the difficulty in resolving the same esterase phenotype between species. In addition, weak bands on the polyacrylamide gel may require a larger number of females per well to resolve (Carneiro et al., Reference Carneiro, Almeida and Quénéhervé2000). Furthermore, some Meloidogyne species have been misidentified based on the esterase and malate dehydrogenase isozyme phenotypes, such as M. arenaria and M. morocciensis (Carneiro & Cofcewics, Reference Carneiro, Cofcewics and Souza2008), and M. ulmi and M. mali (Ahmed et al., Reference Ahmed, van de Vossenberg, Cornelisse and Karssen2013).
The mtDNA variation, although limited, is strongly structured, with as much divergence between two lineages of M. arenaria as between either of them and M. javanica (Pagan et al., Reference Pagan, Coyne, Carneiro, Kariuki, Luambano, Affokpon and Williamson2015). The low diversity of mtDNA suggests that these parthenogenetic lineages arose from distinct but closely related sexually mature females, a pattern observed in other parthenogenetic complexes (Hugall et al., Reference Hugall, Moritz, Stanton and Wolstenholme1994). The molecular analysis based on amplification of specific genome regions stands out among the tested methods because of its speed and accuracy, particularly in the case of identifying morphologically similar nematode species or a mixture of species in the samples. The molecular method is also very useful for routine analysis in nematology laboratories, facilitating the processing of a large number of samples simultaneously (Oliveira et al., Reference Oliveira, Monteiro and Blok2011; Ahmed et al., Reference Ahmed, van de Vossenberg, Cornelisse and Karssen2013; Baidoo et al., Reference Baidoo, Joseph, Mengistu, Brito, McSorley, Stamps and Crow2016).
In this study, at the molecular level, phylogenetic trees based on the ITS, 28S rDNA genes and mtDNA (COI and COII) of M. daklakensis n. sp. indicated that the new species is closest to M. marylandi, M. naasi, M. panyuensis, M. mali and M. ichinohei, but they are clearly separated, with high genetic distance. Molecular and morphological characters strongly support the case that M. daklakensis. n. sp. is a new species that is clearly distinct from other Meloidogyne species.
Acknowledgments
We thank Prof. Dr Maurice Moens for his constructive advice. We are also grateful to the reviewers for their valuable comments.
Financial support
This research was supported by the National Foundation for Science & Technology Development (NAFOSTED) of Vietnam (Number code: 106–NN.03–2013.56) and the World Academy of Sciences (TWAS) (Code: 14–246 RGA).
Conflict of interest
None
Ethical standards
Specimens were collected under the license for the collection of biological material (4555) granted by the Department of Nematology, Institute of Ecology and Biological Resources (IEBR), Vietnam Academy of Science and Technology (VAST).