Introduction
Nematodes are among the most successful group of organisms on Earth, in terms of both abundance and diversity (Dorris et al., Reference Dorris, De Ley and Blaxter1999), although an accurate estimate of the number of species is still lacking (Dobson et al., Reference Dobson2008). One of the reasons for this underestimation is the small body size, which together with imaging resolution limitations and the scarcity of taxonomic specialists restricts the choice and identification of appropriate morphological characters (De Ley et al., Reference De Ley2005). The implementation of molecular techniques has increased the number of new species described, frequently revealing the existence of cryptic species, undetected by traditional morphological approaches (Derycke et al., Reference Derycke2010; Nadler and Pérez-Ponce de León, Reference Nadler and Pérez-Ponce de León2011; Jorge et al., Reference Jorge2013; Ristau et al., Reference Ristau, Steinfartz and Traunspurger2013). When dealing with small organisms such as nematodes, molecular tools are not only useful but are now imperative to overcome taxonomic challenges.
The genus Parapharyngodon has been revised multiple times since its description by Chatterji in Reference Chatterji1933. This genus of oxyurid nematodes includes over 50 species distributed worldwide (Bursey and Goldberg, Reference Bursey and Goldberg2015; Pereira et al., Reference Pereira2017), many described very recently (e.g. Garduño-Montes de Oca et al., Reference Garduño-Montes de Oca, Mata-López and León-Règagnon2016; Pereira et al., Reference Pereira2017; Rizvi et al., Reference Rizvi, Maiti and Bursey2017). Still, the validity of the genus has been questioned repeatedly (Teixeira de Freitas, Reference Teixeira de Freitas1957). While many authors claim its generic status (e.g. Adamson, Reference Adamson1981; Roca, Reference Roca1985; Castaño-Fernández et al., Reference Castaño-Fernández, Zapatero-Ramos and Puertas1987; Hering-Hagenbeck et al., Reference Hering-Hagenbeck, Petter and Boomker2002), others consider it to be a subgenus of Thelandros Wedl, 1862 (e.g. Yamaguti, Reference Yamaguti1961) or a synonym (e.g. Baylis, Reference Baylis1936; Petter and Quentin, Reference Petter, Quentin, Anderson, Chabaud and Willmott2009; Dung et al., Reference Dung, Bursey and Goldberg2009 and references within). As currently considered, both genera exhibit distinct host preferences, with Parapharyngodon being typically found in insectivorous reptiles and amphibians, and Thelandros in herbivorous or omnivorous reptiles (Adamson, Reference Adamson1981). However, in several geographical regions both genera are found cohabiting the same host (e.g. Martin and Roca, Reference Martin and Roca2004, Reference Martin and Roca2005; Roca et al., Reference Roca2005; Carretero et al., Reference Carretero2006; Hassan, Reference Hassan2016). Morphologically, both genera exhibit differences in the posterior end of the body and egg shape upon egg deposition (Adamson, Reference Adamson1981; Bursey and Goldberg, Reference Bursey and Goldberg2005). Specifically, Parapharyngodon and Thelandros males differ in the shape and position of the lateral alae (generally absent or short in Thelandros and longer and wider in Parapharyngodon), presence of genital cone (absent in Parapharyngodon), presence and shape of the genital outgrowths (pedunculated in Thelandros, mammiliform in Parapharyngodon), number of caudal papillae (generally one in Thelandros, and two in Parapharyngodon), tail position and orientation (terminal and posteriorly directed in Thelandros; subterminal and dorsally oriented in Parapharyngodon), and presence/absence of caudal alae (Adamson and Nasher, Reference Adamson and Nasher1984, Astasio-Arbiza et al., Reference Astasio-Arbiza1988, Reference Astasio-Arbiza1989; Solera-Puertas et al., Reference Solera-Puertas1988; Dung et al., Reference Dung, Bursey and Goldberg2009). For females, together with egg development during posture, differences include traits such as the position of the mouth, location of the vulva, and size and shape of eggs (Adamson and Nasher, Reference Adamson and Nasher1984; Astasio-Arbiza et al., Reference Astasio-Arbiza1988, Reference Astasio-Arbiza1989; Solera-Puertas et al., Reference Solera-Puertas1988).
Regarding molecular relationships, only four of the > 90 Parapharyngodon and Thelandros have been sequenced, representing < 5% of the described species. Genetic information, however, is inconclusive regarding the validity of the genera, since the two Thelandros species available (T. tinerfensis and T. scleratus, the latter formerly assigned to Parapharyngodon scleratus) are part of a clade including Parapharyngodon sequences (Chaudhary et al., Reference Chaudhary2015, Reference Chaudhary2017; Goswami et al., Reference Goswami2016). Therefore, there is a need for inclusion of other representatives to assess the evolutionary relationships and taxonomic validity of these two genera.
In the present study we combine available sequences and new genetic data from three markers (partial 18S rRNA, 28S rRNA and COI gene DNA sequences) from eight described species to investigate the relationships between Parapharyngodon and Thelandros genera. We use morphological data of two Parapharyngodon (P. echinatus and P. micipsae) and three Thelandros (T. filiformis, T. tinerfensis and T. galloti) species to identify distinctive morphological characters between genetic clades. Specifically, we aim to (1) determine the genetic distinctiveness of the two genera and, if distinct, (2) validate and identify those morphological characters that support discrimination of the main genetic clades.
Materials and methods
Nematode identification and DNA extraction
For this study a total of 34 pinworm specimens (25 males and 9 females) belonging to two Parapharyngodon (P. echinatus and P. micipsae) and three Thelandros species (T. filiformis, T. tinerfensis and T. galloti) were retrieved from 14 lizard hosts from Spain and Morocco (table 1). Nematodes were obtained from faecal pellets or intestines, stored in 96% ethanol, and then isolated, identified and counted using an Olympus SZX2-ILLT stereo-microscope (Olympus, Tokyo, Japan).
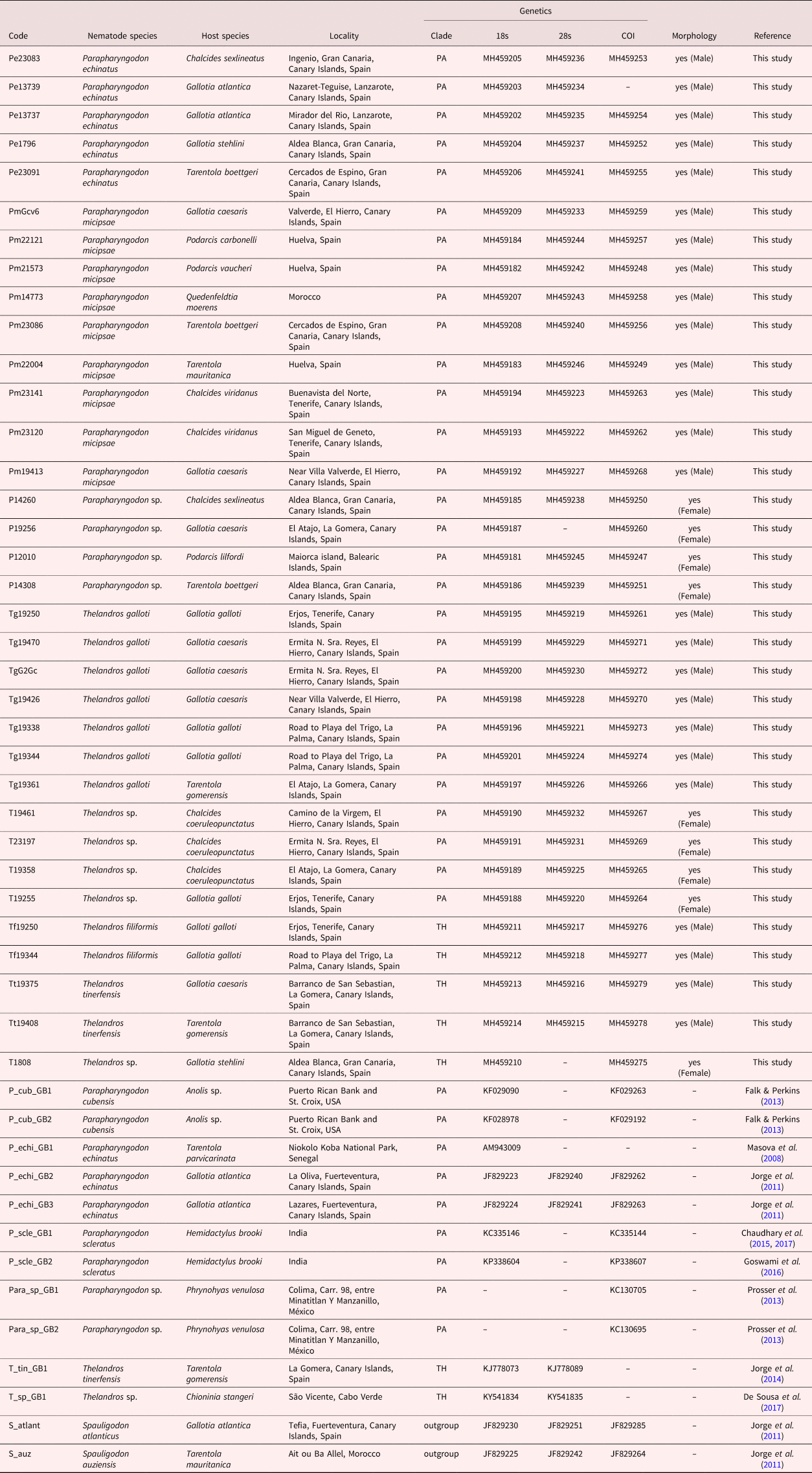
Table 1. List of the individuals included in the study. For each sample, species (identified by morphology), host, locality, country, genetic (clade, GenBank accession numbers) and morphological information (used in the morphological assessment, sex), and the reference are given.
Detailed morphological examination of the specimens was conducted by mounting semi-permanent slides prepared using a glycerol–water solution (1:1) as described by Borges et al. (Reference Borges2012), and observed under a light microscope (Olympus CX41, Olympus Australia Pty Ltd). Species identification was based on previously published traits (Roca, Reference Roca1985; Moravec et al., Reference Moravec, Barus and Rysavy1987; Astasio-Arbiza et al., Reference Astasio-Arbiza1988, Reference Astasio-Arbiza1989; Solera-Puertas et al., Reference Solera-Puertas1988; Mašová et al., Reference Mašová2008, Reference Mašová2009; see table 2). For all individuals, the following body measurements were taken: body length (BL) and width at mid-body region (BW), tail length (TL), distance from nerve ring to mouth (NR), oesophageal bulb length (OBL) and width (OBW), and oesophagus length (OL) and width (OW). In addition, in males we also measured lateral alae length (LAL) and width (LAW), tail width above the tail papillae pair (TW1) and below the tail papillae pair (TW2), spicule length (SL) and spicule width (SW). In females, vagina length (VL), vulva position (Vu), and average egg length (ELA) and width (EWA, both calculated from three eggs) were also measured. Details of the characters measured are in supplementary fig. S1. We also recorded the following categorical variables for males: spicule shape (SS, blunt, semi-sharp or sharp), number of cloacal papillae (GP, either four or six), number of caudal papillae (CP, one or two), and presence of caudal alae (CA, present or absent). These characters were chosen given their importance in the diagnosis of these two genera. Photographs were taken using an Olympus DP25 digital camera (Olympus, Tokyo, Japan), and saved and edited using Cell^B v.3.4 (Olympus Soft Imaging Solutions). Linear measurements were taken using ImageJ software version 1.48 (Wayne Rasband, National Institute of Health, USA) and were recorded by the same person (AS).
Table 2. Description of the main characters used to identify morphologically the species of Parapharyngodon and Thelandros included in the study.

After morphological identification, DNA extractions were performed on individual specimens using DNeasy® Blood & Tissue Kit (QIAGEN) according to the manufacturer's protocol. Two partial nuclear genes – 28S ribosomal RNA (28S rRNA) and 18S ribosomal RNA (18S rRNA) – and the mitochondrial gene cytochrome oxidase c subunit 1 (COI) were amplified using the polymerase chain reaction (PCR) method. As currently available COI markers failed to amplify most of the samples, three new sets of primers were designed, targeting Thelandros and Parapharyngodon (table 3). Amplified 28S rRNA products were sequenced for both strands, whereas in most cases for 18S rRNA and COI products only the forward strand was sequenced. PCR product purification and sequencing was performed by a commercial facility (Beckman Coulter Genomics, UK).
Table 3. Primers and PCR conditions used to amplify the gene fragments targeted in this study.

An initial denaturation step at 95°C for 3 minutes and a final elongation step at 72°C for 10 minutes were performed.
Phylogenetic analysis
Sequences obtained were compared against GenBank using BLAST to confirm the identity of the amplified products, and imported into Geneious Pro version 4.8.5 (http://www.geneious.com, Kearse et al., Reference Kearse2012) for analysis. In addition, other sequences of Parapharyngodon (seven sequences for 18S rRNA, four for 28S rRNA and six for COI) and Thelandros (two sequences for 18S rRNA and two for 28S rRNA) available from GenBank were also included (table 1). Spauligodon atlanticus and Spauligodon auziensis (Jorge et al., Reference Jorge2011) were used as outgroups. Alignments were performed in the online version of MAFFT v.7 (Katoh et al., Reference Katoh2002, Reference Katoh, Rozewicki and Yamada2017; Katoh and Standley, Reference Katoh and Standley2013; available at http://mafft.cbrc.jp/alignment/server/) using the “auto” strategy for the COI dataset, and the “Q-INS-I” strategy, which considers secondary RNA structure, for the 18S and 28S rRNA datasets. Gblocks software was used to eliminate poorly aligned positions using default parameters (Castresana, Reference Castresana2000; Talavera and Castresana, Reference Talavera and Castresana2007). COI sequences were translated into amino acids using the invertebrate mitochondrial DNA genetic code to confirm the absence of stop codons and assess the reading frame. Substitution saturation in the third codon position was assessed by plotting the uncorrected pairwise genetic distances against model corrected genetic distances (K81) in R (Ape package; Paradis et al., Reference Paradis, Claude and Strimmer2004). As the relationship between distances was approximately linear, the third positions were considered to be unsaturated and were kept in the analysis.
Phylogenetic relationships were investigated using Bayesian inference (BI) approaches for each single gene and for the concatenated dataset. Most appropriate evolutionary models and partition schemes for the COI and concatenated datasets were inferred following the Bayesian information criterion (BIC) using PartitionFinder2 (Lanfear et al., Reference Lanfear2012) and implemented in the phylogenetic analyses. Bayesian (BI) analyses were performed using MrBayes version 3.2.6 (Ronquist et al., Reference Ronquist2012), run on XSEDE in the CIPRES Science Gateway v. 3.3. The analysis was run for 100 million generations, with random starting trees, employing a Markov Chain Monte Carlo (MCMC) approach for sampling the joint posterior probability distribution saved every 1000 generations. Four chains and two independent runs were performed to ensure consistent results. Convergence of the runs was monitored using the average standard deviation of split frequencies (< 0.01), potential scale reduction factor (PSRF, close to 1.0; Gelman and Rubin, Reference Gelman and Rubin1992) and estimated sample size (ESS, values above 100), all provided in MrBayes output. Considering this, the first 2500 trees (25%) were discarded as burn-in. Resulting trees were summarized in a 50% majority-rule consensus tree with clade credibility support values (Bayesian posterior probability, BPP) and branch length information. BI outputs were imported to FigTree v. 1.4.2 (Rambaut, Reference Rambaut2014) for graphical visualization and editing.
Average uncorrected genetic distances (p-distance) between and within main genetic groups were calculated in MEGA v. 6 (Tamura et al., Reference Tamura2013) using the pairwise-deletion option.
Morphological analysis
We performed morphological analyses to identify putative phenotypic differences between the two main groups retrieved in the phylogenetic analysis (Parapharyngodon vs Thelandros genetic clades). Given sexual dimorphism, analyses were performed separately for each sex. In cases in which some linear measurements could not be properly recorded, missing data were substituted by the average of the group (species and sex). Before analysis, linear measurements were log-transformed and checked for homoscedasticity and normality. As some of the variables did not meet the assumptions, correlation between body length (BL) and other linear measurements was tested using a Spearman test (function rcorr, R package Hmisc, Harrel et al., Reference Harrell2017). When variables were correlated to BL, body length was included as covariate in the corresponding variance analysis.
To assess the general pattern of morphological variation of the nematodes analysed and to identify morphological similarities without a priori information on genetic assignation, we performed a principal component analysis (PCA) on the linear measurements (R function prcomp; R Core Team, 2016). As only a single individual of Thelandros was available in the female dataset, univariate analyses were performed only in the male dataset. Differences in male linear measurements between Parapharyngodon and Thelandros genetic clades were tested using permutational analyses of variance (permANOVA, Geomorph R package; Adams and Otárola-Castillo, Reference Adams and Otárola-Castillo2013). In body length correlated variables, BL was included as covariate in the model. Differences in categorical variables between Thelandros and Parapharyngodon male genetic clades were analysed using the Fisher exact test (fisher.test; R Core Team, 2016).
All statistical analyses were performed in Rstudio v. 0.98.1103 (R Core Team, 2016; RStudio Team, 2015).
Results
Phylogenetic relationships
Single-gene aligned datasets included 45 sequences for 18S rRNA (706 bp), 38 sequences for 28S rRNA (830 bp) and 43 sequences for COI (503 bp), including outgroups. Following PartitionFinder, the most appropriate substitution models were K80 + I for the 18S dataset and HKY + G for 28S. For COI, PartitionFinder chose three partitions corresponding to the first (with HKY + I+G as best model), second (F81 + I) and third (GTR + G) codon positions. For the concatenated dataset (2039 bp), the best-fit partitioning schemes included all the separate gene fragments and gene codon positions (for COI) evaluated separately implementing the same models of evolution as described above.
Phylogenetic analyses identified two main clades (fig. 1 & supplementary fig. S2a,b,c) well supported in all single-gene and concatenated datasets (in all cases BPP = 1; fig. 1 & supplementary fig. S2). Uncorrected p-distances among main lineages for each individual gene are detailed in supplementary table S1. The first one, hereafter called Parapharyngodon clade, includes all individuals morphologically identified as Parapharyngodon spp., plus all T. galloti males and several Thelandros sp. females. The second clade, referred to as Thelandros clade, comprises individuals morphologically identified as T. tinerfensis, T. filiformis and several Thelandros spp.

Fig. 1. Phylogenetic tree depicting the relationships between Parapharyngodon and Thelandros samples using the 18S + 28S + COI concatenated dataset. The topology of the tree corresponds to a Bayesian inference (BI) 50% majority-rule consensus tree, and support values for the main lineages represent Bayesian posterior probabilities above 0.70. Sequences generated in this study are highlighted in bold. Additional information regarding sample codes is in table 1.
Within the Parapharyngodon clade, five lineages arise (fig. 1 & supplementary fig. S2). The first group (P. cubensis lineage) includes P. cubensis infecting reptiles from the Caribbean region, and the second (Parapharyngodon sp. lineage) clusters two sequences of Parapharyngodon sp. from Mexico. A third lineage includes published sequences of P. scleratus from India plus a sequence identified as P. echinatus from Senegal (AM943009). All the other P. echinatus samples clustered into a fourth group, named P. echinatus lineage. Individuals morphologically identified as T. galloti are found in a fifth group (hereafter T. galloti lineage), sister to P. echinatus lineage. Interestingly, individuals morphologically identified as P. micipsae are scattered throughout the T. galloti and P. echinatus lineages. Lineages within the Parapharyngodon clade are, in general, congruent across datasets, although relationships between current species can only be partially inferred due to the incomplete data (i.e. sequences missing in single gene inferred trees). All three genes support the clustering of all T. galloti individuals into a distinct lineage, sister group to P. echinatus lineage. However, relationships within P. echinatus are not clear, as the phylogram estimated from 18S does not support the monophyly of the P. echinatus lineage as inferred in the other analyses. The phylogram estimated from the COI sequences shows P. scleratus as sister group to T. galloti/P. echinatus, and P. cubensis as the most basal lineage within Parapharyngodon clade. This pattern, however, is not supported in the inference tree derived from 18S (supplementary fig. S2). Unfortunately, no sequences of any of these species are available for 28S.
Regarding the Thelandros clade, relationships between T. filiformis and T. tinerfensis samples are not resolved. Although the estimate of relationships from 28S supports the genetic differentiation between T. filiformis and T. tinerfensis, phylogenetic trees derived from 18S and COI do not indicate the same genetic clusters (supplementary fig. S2).
Morphological analysis
To identify consistent morphological differences between Thelandros and Parapharyngodon genetic clades, individuals were grouped according to the clades retrieved in the phylogenetic analysis (fig. 1). Therefore, T. tinerfensis and T. filiformis were assigned to the Thelandros clade (TH), and P. echinatus, P. micipsae and T. galloti were considered part of the Parapharyngodon clade (PA) (see table 1 for details on individual morphological and genetic assignation).
Principal component analysis based on linear measurements showed that variation across the first three axes is lower in males than in females (fig. 2; supplementary table S2). In males, the first three axes explained only 62.8% of total variation, whereas in females the same axes explained 96.0%. In males, the main contributors (individual loading values higher than 0.65) to the variation across PC1 (34% variation) were BL, BW, OBL and OBW; LAL and TL explained variation across PC2 (16% variation), and NR across PC3 (12%; supplementary table S2). In females, the main source of variation was body length (all variables had high loadings), which accounts for 86.6% of the variation across PC1. Variation across PC2 (5.2%) was explained mainly by NR and OBL, and by NR across PC3 (4.1%; supplementary table S2). For females, PCA plot dots highlighted by genetic lineage showed that the single Thelandros female analysed was morphologically distinct from the Parapharyngodon females (fig. 2). However, in males, morphological differentiation was not so evident (fig. 2). Interestingly, T. galloti individuals exhibited higher morphological diversity, and many showed morphological characteristics intermediate between Thelandros and Parapharyngodon (fig. 2).

Fig. 2. Principal component analysis (PCA) plots for the morphological variation in Parapharyngodon and Thelandros males (left) and females (right). Values in parentheses in the axis labels correspond to the eigenvalues and to the amount of variation (%) explained for each principal component. Individuals are colour-coded by genetic clade (Parapharyngodon clade in grey, Thelandros clade in black), and different lineages are shape-coded: P. echinatus (grey square), T. galloti (grey circle), T. filiformis (black square) and T. tinerfensis (black circle). Given that P. micipsae individuals were scattered across both P. echinatus and T. galloti lineages, they are also highlighted in a different shape (grey triangle).
Permutational ANOVA/ANCOVA analyses showed that males of the Thelandros and Parapharyngodon clades have similar body length (BL), but differ in NR, LAL and TL (table 4). Males of the Thelandros clade have longer tails, their nerve ring is located further away from the mouth structure, and they have shorter lateral alae than males of the Parapharyngodon clade (table 4; supplementary fig. S1). Given that only a single female was available for the Thelandros clade, this analysis was not performed on the female dataset.
Table 4. Results of the permANOVA/permANCOVA analyses on male body measurements. Significant p-values are in bold. Asterisk indicates the variables correlated to body length and for which ANCOVA analyses were performed. BL, body length; BW, width at mid-body region; LAL, lateral alae length; LAW, lateral alae width; NR, nerve ring distance to the mouth; OBL, oesophageal bulb length; OBW, oesophageal bulb width; OL, oesophagus length; OW, oesophagus width; SPI, spicule length; SW, spicule width; TL, tail length; TW1, tail width above the tail papillae pair; TW2, tail width below the tail papillae pair.

Regarding categorical variables, males of the two clades showed differences in the shape of the spicule, number of caudal papillae and presence of caudal alae (Fisher exact test, spicule shape: P = 0.009; number of caudal papillae P < 0.001; caudal alae P = 0.014; fig. 3), but not in the number of cloacal papillae (P = 0.231). However, after excluding P. micipsae from the analyses, differences in spicule shape between clades disappeared (P = 0.118). The only diagnostic character that was found to discriminate between the Parapharyngodon and Thelandros genetic clades was the number of caudal papillae. All the individuals from the Parapharyngodon clade had two papillae, whereas all individuals belonging to the Thelandros clade had only one (fig. 3 & supplementary fig. S1). Regarding the presence of caudal alae, all Parapharyngodon individuals lacked them, whereas they were present in all Thelandros, with one exception (fig. 3 & supplementary fig. S1).

Fig. 3. Frequency plots (number of individuals) of male categorical variables between the main lineages of Parapharyngodon and Thelandros.
Discussion
Nematode identification has traditionally relied on morphological characters, but molecular approaches are crucial to detect cryptic and sibling species and to clarify taxonomic relationships (Nadler and Pérez-Ponce de León, Reference Nadler and Pérez-Ponce de León2011; Jorge et al., Reference Jorge2014; Ott et al., Reference Ott, Leisch and Gruber-Vodicka2014). Despite this, the use of genetic tools in oxyurids is still not routinely implemented in most current taxonomic descriptions. Until this study, from over 50 species of Parapharyngodon (Bursey and Goldberg Reference Bursey and Goldberg2015; Pereira et al., Reference Pereira2017) and over 40 of Thelandros (Rizvi et al., Reference Rizvi, Maiti and Bursey2017) described, only four (P. cubensis, P. echinatus, P. scleratus and T. tinerfensis) have been sequenced.
Our genetic results confirm the presence of two main clades, mostly congruent with Parapharyngodon and Thelandros genera (e.g. Adamson, Reference Adamson1981; Roca, Reference Roca1985; Castaño-Fernández et al., Reference Castaño-Fernández, Zapatero-Ramos and Puertas1987), but reveal several incongruences regarding currently described species. Whereas all the Parapharyngodon species included in the analysis are part of the Parapharyngodon clade, Thelandros seems to be polyphyletic. Two of the three Thelandros species analysed (T. tinerfensis and T. filiformis) are placed in the Thelandros clade, but the third one (T. galloti) falls within the Parapharyngodon clade. When described, T. galloti was assigned to the genus Thelandros based on the existence of a prominent genital cone with a V-shape sclerotized structure supporting the posterior cloacal lip (Astasio-Arbiza et al., Reference Astasio-Arbiza1988). However, contrary to what is common in other Thelandros, this species lacks caudal alae and exhibits long and wide lateral alae, both characteristics usually associated with Parapharyngodon (Astasio-Arbiza et al., Reference Astasio-Arbiza1988). Such intermediate morphological traits were corroborated by our results. Univariate preliminary analysis with our dataset (results not shown) confirmed that T. galloti males have wider alae and longer oesophagus than Parapharyngodon representatives. Despite these intermediate morphological characteristics, genetic data support T. galloti as part of the Parapharyngodon clade, and as such the taxonomic status should be reviewed and the species reassigned as Parapharyngodon galloti. Regarding Parapharyngodon, three of the species analysed (P. echinatus, P. scleratus and P. cubensis) appear as distinct groups (with the exception of a single sequence of P. echinatus from a gecko from Senegal, which clusters with P. scleratus), supporting their taxonomic validity. However, individuals morphologically identified as P. micipsae are scattered across the P. echinatus and T. galloti lineages. This suggests that P. micipsae may represent an alternative morphotype. Existence of alternative male morphotypes in oxyurid nematodes has been confirmed genetically in Spauligodon and Skrjabinodon (Ainsworth, Reference Ainsworth1990; Jorge et al., Reference Jorge2014) and may arise in response to alternative reproductive tactics (Jorge et al., Reference Jorge2014). Alternatively, P. micipsae could be a morphological misidentification due to poor conservation of the specimens (Chabaud and Golvan, Reference Chabaud and Golvan1957). These two cases of incongruence between molecular and morphological evidence indicate that at least some of the diagnostic characters currently in use may not be appropriate to identify species of these two genera.
Our morphological analysis identified some discriminatory characteristics between Thelandros and Parapharyngodon genetic samples (Chatterji, Reference Chatterji1933; Yamaguti, Reference Yamaguti1961; Adamson, Reference Adamson1981; Roca, Reference Roca1985; Castaño-Fernández et al., Reference Castaño-Fernández, Zapatero-Ramos and Puertas1987), at least in males. Individuals identified genetically as part of the Thelandros clade have longer tails, the nerve ring is located further away from the anterior end and they have shorter lateral alae than the ones ascribed to Parapharyngodon. Also, all individuals from the Thelandros clade analysed have a single caudal papilla and all but one exhibit caudal alae. Although some of the variables detected here (lateral alae shape, tail length, number of caudal papillae and presence or not of caudal alae) were already reported to discriminate between Parapharyngodon and Thelandros in previous literature (Solera-Puertas et al., Reference Solera-Puertas1988; Astasio-Arbiza et al., Reference Astasio-Arbiza1989), we confirm that the location of the nerve ring is also a useful trait to distinguish between Parapharyngodon and Thelandros genetic clades. On the other hand, the spicule shape and the number of cloacal papillae were not reliable to discriminate between both clades, according to our data. Although the presence of genital cone, presence and shape of outgrowths and position of excretory pore have been described as useful characters to distinguish between both Parapharyngodon and Thelandros genera, we were not able to collect enough data to perform well-supported analyses. Concerning females, the only Thelandros individual analysed seemed to have a distinctive morphology. A broader molecular and morphological assessment including a larger number of taxa and additional characters (such as cephalic structures, egg development during posture and position of the operculum in the egg) are needed to confirm reliable diagnostic characters and to describe the diversity and evolutionary relationships within these groups. Unfortunately, neither of the type species of the two genera (Thelandros alatus Wedl, 1862 and Parapharyngodon maplestoni Chatterji, 1933) have been sequenced to date, preventing us from making further decisions regarding the taxonomy of the two genera. Further studies including representatives of these and other species are also needed to assess the evolutionary and morphological relationships within this group.
In conclusion, this study highlights the urgent need for incorporating genetic information in taxonomic studies of nematodes. Only with molecular data will it be possible to detect polyphyly, alternative morphotypes and cryptic diversity, as well as identifying those morphological characters with more phylogenetic signal and less homoplasy, all of which are fundamental to understand the evolutionary and taxonomic relationships within this group.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/S0022149X1800069X
Acknowledgements
We thank the local Environmental Authorities from the Canary Islands (Cabildos Insulares), Spain (Junta de Andalucia) and Morocco (Haut Commisariat aux Eaux et Fôrets et à la Lutte Contre la Désertification) for the lizard collecting permits. We thank C. Rato, A. Kaliontzopoulou, J.A. Mateo, M. López-Darias and B. Fariña for their help in the field.
Financial support
This work was supported by the Fundação para a Ciência e Tecnologia (AP, contract number IF/01257/2012; DJH, contract number IF/01627/2014) under the Programa Operacional Potencial Humano – Quadro de Referência Estratégico Nacional from the European Social Fund and Portuguese Ministério da Educação e Ciência, and by the Norte Portugal Regional Operational Programme (NORTE2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (MAC, project AGRIGEN: NORTE-01-0145-FEDER-000007). This study was funded by the Fundação para a Ciência e Tecnologia and Compete program (project number IF/01257/2012/CP0159/CT0005 to AP and PTDC/BIA-BDE/67678/2006, FCOMP-01-0124-FEDER-007062 to MAC).
Conflict of interest
None.









