Introduction
Australia harbours a high animal diversity of over 600,000 animal species, with 80% of those endemic (Williams et al., Reference Williams, Read, Norton, Dovers, Burgman and Anderson2001; Pusey et al., Reference Pusey, Kennard and Arthington2004; Walker & Humphries, Reference Walker and Humphries2013). Despite Australia's incredible species richness, relatively few parasites have been recorded in teleost freshwater fishes in comparison with the North American and European faunas (Choudhury et al., Reference Choudhury, Aguirre-Macedo, Curran, de Nunez, Overstreet, de Leon and Santos2016; Poulin & Presswell, Reference Poulin and Presswell2016; Scholz et al., Reference Scholz, Besprozvannykh and Boutorina2016; Jorge & Poulin, Reference Jorge and Poulin2018). Most studies on fish parasites in Australia focused on marine and commercially valuable fishes, whereas freshwater fish parasites are generally less studied (e.g. Lymbery et al., Reference Lymbery, Hassan, Morgan, Beatty and Doupe2010), especially because the fauna of freshwater fish in this continent is depauperate (Allen et al., Reference Allen, Midgley and Allen2002).
Carp gudgeons (genus Hypseleotris Gill, 1863, family Eleotridae) are an important component of the freshwater fish biota of eastern Australia. They are widespread and frequently abundant, even in somewhat degraded habitats. Most species are small (<6 cm total length) and live for a year or two. They reproduce in spring, with males guarding a territory and providing protection for eggs until hatching (Unmack, Reference Unmack2000; Lintermans, Reference Lintermans2009). The taxonomy of carp gudgeons is complex, with multiple sympatric sexual species, most of which remain undescribed (Hoese et al., Reference Hoese, Larson, Llewellyn and McDowall1980; Thacker et al., Reference Thacker, Unmack, Matsui and Rifenbark2007; Unmack et al., Reference Unmack, Adams, Bylemans, Hardy, Hammer and Georges2019).
To date, three studies on the parasites of carp gudgeons have been published, with only two species of parasitic flatworms reported: the invasive bothriocephalidean cestode Schyzocotyle acheilognathi (Yamaguti, 1934) and trematode metacercariae of Apatemon hypseleotris Negm-Eldin & Davies, 2001 (Dove et al., Reference Dove, Cribb, Mockler and Lintermans1997; Dove & Fletcher, Reference Dove and Fletcher2000; Negm-Eldin & Davies, Reference Negm-Eldin and Davies2001). For almost 20 years, no parasitological studies have been carried out on these fishes, but sections of histological samples of carp gudgeons prepared recently at the Veterinary Diagnostic Laboratories, Charles Sturt University showed infection by other metazoan parasites (S. Raidal, pers. comm.).
We examined the parasite communities of three species of carp gudgeon in the Murrumbidgee River in New South Wales, Australia, where three species of Hypseleotris occur in sympatry (Unmack et al., Reference Unmack, Adams, Bylemans, Hardy, Hammer and Georges2019). Furthermore, we provide the first molecular data for most of the parasites found to aid their present and future identification, especially larval stages which can only currently be reliably identified by molecular data.
Materials and methods
Study area and fish sampling
Fish were collected from five sites associated with the Murrumbidgee River, four in the vicinity of Wagga Wagga and one near Narrandera. These sites were chosen due to the diversity of habitats and include an isolated floodplain billabong (−35.08753, 147.34909), a main river channel (−35.1042, 147.37527), an urban billabong (−35.1101, 147.36196), a large semi-urban lake (−35.17058, 147.36758) and a rearing/holding pond at the Narrandera Fisheries Research Centre (−35.14154, 147.44148). This area is inhabited by three different species from the carp gudgeon complex of species – the western carp gudgeon (Hypseleotris klunzingeri), Midgley's carp gudgeon (Hypseleotris sp. 4) and Lake's carp gudgeon (Hypseleotris sp. 5) – all of which can be visually differentiated (Unmack, Reference Unmack2000; Thacker & Unmack, Reference Thacker and Unmack2005; Thacker et al., Reference Thacker, Unmack, Matsui and Rifenbark2007).
Specimens (41 western carp gudgeon, 39 Midgley's carp gudgeon and 57 Lake's carp gudgeon; table 1) were collected at each locality using a seine net, euthanized using an overdose of clove oil and dissected shortly after. Fish were identified and measured (total length) prior to parasitological examination. Collecting and handling of specimens was conducted under approval number 20180442 from the University of Canberra ethics committee.
Table 1. Sample sizes and total lengths of carp gudgeons (Hypseleotris spp.) from five sites in the study area from the Murrumbidgee River, Australia.
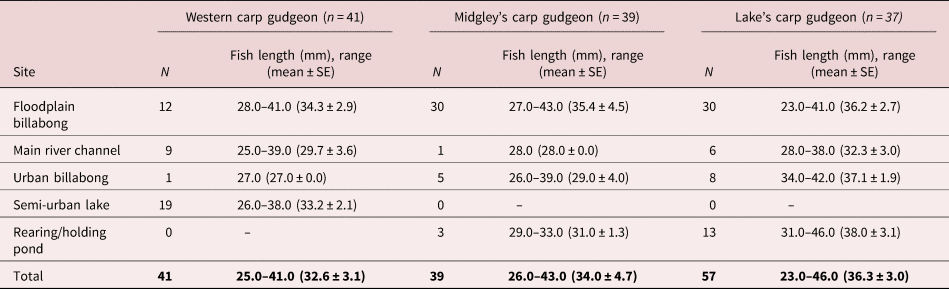
SE, standard error.
Parasite collection and identification
Fish were dissected under a stereomicroscope and all organs and tissues were examined for metazoan parasites. Parasites were preliminary sorted by groups under the stereomicroscope and since most of them were larval stages, they were fixed in absolute ethanol (99%) for molecular analyses.
The parasite identification, mainly based on molecular data, was refined with morphological assessment when sufficient samples were available and to the best of our possibilities since most specimens were larval stages. Specimens of larval cestodes and trematodes were prepared as whole-mounts, as described by Justine et al. (Reference Justine, Briand and Bray2012), and examined under the light microscope (Zeiss Axioplan, Göttingen, Germany). Ethanol-fixed gryporhynchid larvae were identified to the genus or species level by flattening in order to better observe their rostellar hooks (Bona, Reference Bona1975). All voucher specimens are deposited in the Platyhelminthes collection of the Natural History Museum of Geneva, Switzerland (accession numbers MHNG-PLAT-135210–MHNG-PLAT-135224).
DNA was extracted from 54 specimens of Pseudodactylogyrus sp., Gyrodactylus sp., Apatemon cf. hypseleotris, Clinostomum sp., S. acheilognathi, Paradilepis sp., Dendrouterina sp., Parvitaenia sp., Cyclophyllidea gen. sp., Procamallanus sp. and larvae of spirurine nematode using Chelex® (Bio-Rad, Cressier, Switzerland) in deionized water containing 0.1 mg/ml proteinase K. A partial fragment of the large or small ribosomal subunit was amplified. The large ribosomal subunit (28S rDNA) was chosen as it is a common marker used to identify parasitic flatworms (Blasco-Costa et al., Reference Blasco-Costa, Cutmore, Miller and Nolan2016). The following primers were used for trematodes: U178 [5′-GCA CCC GCT GAA YTT AAG-3′] and L1642R [5′-CCA GCG CCA TCC ATT TTC A-3′] (Lockyer et al., Reference Lockyer, Olson and Littlewood2003); and for cestodes: LSU5 F [5′-TAG GTC GAC CCG CTG AAY TTA AGC-3′] and 1500R [5′-GCT ATC CTG AGG GAA ACT TCG-3′] (Littlewood et al., Reference Littlewood, Curini-Galletti and Herniou2000; Olson et al., Reference Olson, Cribb, Tkach, Bray and Littlewood2003). In addition, a partial fragment of the small ribosomal subunit (18S rDNA) was amplified for nematodes, as it is the most common marker used for this group, with the primers PhilonemaF [5′-GCC TAT AAT GGT GAA ACC GCG AAC-3′] and PhilPCRr0 [5′-CCG TT CAA GCCACT GC ATT A-3′] (Černotíková et al., Reference Černotíková, Horák and Moravec2011). Polymerase chain reaction (PCR) amplifications were performed using a total volume of 20 μl. The enzyme MyFi™ Mix (Bioline, Paris, France) was used as follows: 4 μl of DNA template, 2× MyFi™ Mix (containing DNA polymerase, dNTPs, magnesium chloride and enhancers at optimal concentrations) and 4 μM of each forward and reverse primers. The PCR amplification protocol for the 28S followed Blasco-Costa et al. (Reference Blasco-Costa, Balbuena, Kostadinova and Olson2009) and for the 18S of nematodes followed Černotíková et al. (Reference Černotíková, Horák and Moravec2011). After verification of single-band PCR products via electrophoresis, amplicons were purified with a mix of exonuclease I and thermosensitive alkaline phosphatase enzymes (Werle et al., Reference Werle, Schneider, Renner, Volker and Fiehn1994). Purified amplicons were sent to Macrogen Europe (Amsterdam, the Netherlands) for sequencing from both strands using the same PCR forward and reverse primers used for amplification and an additional internal primer for the 28S fragments, L1200R [5′–GCA TAG TTC ACC ATC TTT CGG–3′] (Littlewood et al., Reference Littlewood, Curini-Galletti and Herniou2000).
Phylogenetic analyses
Sequences were assembled and inspected for errors in Geneious 8.1.9 (Drummond et al., Reference Drummond, Ashton and Buxton2010; Kearse et al., Reference Kearse, Moir and Wilson2012). Available sequences for taxa belonging to the same family (species included are listed in the supplementary table S1) as the suspected taxa were extracted from GenBank and aligned together with our 54 newly obtained sequences using MAFFT (Katoh et al., Reference Katoh, Kuma, Toh and Miyata2005) implemented in Geneious to confirm species or generic identification or refine the preliminary identification. As outgroup, a sister taxon outside the family was included to each alignment based on previous studies (e.g. Olson et al., Reference Olson, Cribb, Tkach, Bray and Littlewood2003; Černotíková et al., Reference Černotíková, Horák and Moravec2011; Hernández-Mena et al., Reference Hernández-Mena, García-Varela and Pérez-Ponce de León2017; Ortega-Olivares & García-Varela, Reference Ortega-Olivares and García-Varela2018). The alignments were checked by eye and the uneven ends of the sequences were trimmed to match the shortest. Phylogenetic reconstructions were used to confirm the affiliation of our sequenced specimens to a genus or species whenever possible.
We ran phylogenetic analyses under maximum likelihood (ML) and Bayesian inference (BI) criteria. The model of nucleotide evolution GTR (general time-reversible model) with gamma-distribution among site rate variation was applied to all analyses. The model was selected using JModelTest 2.1.7 with Akaike Information Criterion (Darriba et al., Reference Darriba, Taboada, Doallo and Posada2012). ML analyses were conducted using RAxML 8 (Stamatakis, Reference Stamatakis2006). All model parameters and bootstrap nodal support values (1000 repetitions) were estimated using RAxML. BI trees were constructed using MrBayes 3.2.6 (Ronquist et al., Reference Ronquist, Teslenko and van der Mark2012), running two independent Markov chain Monte Carlo runs of four chains for ten million generations and sampling tree topologies every 1000th generations. Burn-in periods were automatically set to 25% of generations. RAxML and MrBayes analyses were carried out for each individual dataset on the public computational resource CIPRES (Miller et al., Reference Miller, Pfeiffer and Schwartz2010).
Infection parameters
Infection descriptors, which include parasite prevalence (%), mean intensity and mean abundance, were calculated as described by Bush et al. (Reference Bush, Lafferty, Lotz and Shostak1997).
Results
Sixteen species of Digenea, Cestoda, Nematoda and Arthropoda were distinguished based on morphological and molecular characteristics (table 2). Eleven species were larval stages (fig. 1) and five species were found as adults (fig. 2 and table 3). Based on molecular data and morphological (fig. 3) examination, only four species could be identified to the species level, namely the invasive Asian fish tapeworm S. acheilognathi, Paradilepis patriciae Baer & Bona, 1960, P. cf. kempi (Southwell, 1921) and A. cf. hypseleotris. Some of the parasites may represent species new to science, but their identification to the lower taxonomic level was not possible. An ectoparasitic copepod of the genus Lernaea Linnaeus, 1758, another invasive parasite, as well as monogeneans belonging to Pseudodactylogyrus Gusev, 1965 and Gyrodactylus von Nordmann, 1832 found on the gills of Lake's carp gudgeon and Midgley's carp gudgeon could not be identified to the species level. Among the juvenile stages, we recognized metacercariae of A. cf. hypseleotris (encysted in the abdominal cavity and muscles) and Clinostomum sp. (encysted in the abdominal cavity), and merocerci of four distinct sequences of Paradilepis spp. (genetic distance between 0.8 and 2.7%) encysted in the liver; two morphotypes were identified on the basis of number and shape of the hooks (Paradilepis patriciae and P. cf. kempi, the two morphotype are represented in fig. 3) and Dendrouterina sp. (free in the gall bladder and morphologically rostellar hooks were similar to those of Dendrouterina herodiae Fuhrmann, 1912) and Parvitaenia sp. and two lineages of Cyclophyllidea gen. sp. genetically distinct from each other by 1.8% (in the intestinal lumen), and unidentified larvae of spirurine nematodes (see table 2). Specifically, the cestode identification was mainly based on the molecular data, but the study of rostellar hooks was used to further refine the identification.

Fig. 1. Maximum likelihood (ML) phylograms based on 28S rRNA gene sequences of our larval specimens and selected sequences from GenBank: (a) diplostomatoidean trematodes, with two sequences of taxa belonging to the Brachylaimoidea as outgroup; (b) cyclophylidean cestodes, with sequences of nippotaeniids serving as outgroup. Bootstrap values are followed by Bayesian posterior probabilities (BI) above the branches. Newly acquired sequences are marked in bold. Full circles illustrate high support (ML > 90, BI = 1) and empty circles illustrate moderate support (ML = 70–90, BI = 0.90–0.99). Scale bar indicates the number of substitutions per site. The asterisks represent the partial sequences.

Fig. 2. Maximum likelihood (ML) phylograms based on 28S rRNA gene sequences of adult specimens and selected sequences from GenBank: (a) bothriocephalidean cestodes, with three trypanorhynch sequences included as outgroup; (b) Monopisthocotylea, with a sequence of the catenulid Stenostomum leucops as outgroup; and (c) 18S rRNA gene sequences of camallanid nematodes, with the philometrid Philonema oncorhynchi as outgroup. Bootstrap values are followed by Bayesian posterior probabilities (BI) above the branches. Newly acquired sequences are marked in bold. Full circles illustrate high support (ML > 90, BI = 1) and empty circles illustrate moderate support (ML = 70–90, BI = 0.90–0.99). Scale bar indicates the number of substitutions per site.

Fig. 3. Illustration of the rostellar hooks of specimens of Paradilepis spp.: (a) Paradilepis cf. kempi; (b) Paradilepis patriciae.
Table 2. Infection parameters for the parasite community of carp gudgeons from the Murrumbidgee River, Australia, expressed as prevalence of infection (PI), mean intensity of infection (MI) and mean abundance (MA).

a Values for encysted metacestodes of gryporhynchids were pooled because individual larvae were not identified to the genus/species level.
b Invasive parasites.
L, larval stages (metacercariae, merocercoids or larval nematodes).
Table 3. Stage, life cycle, site of infection and record of carp gudgeon parasites from Eastern Australia.

Western carp gudgeon harboured the lowest parasite richness and overall parasite prevalence (only three taxa and 39% of fish were infected), whereas Lake's carp gudgeon and Midgley's carp gudgeon had more than twice the parasite richness (11 and eight taxa, and prevalence of infection of 93 and 95%, respectively; see table 2). The most prevalent parasites were metacercariae of A. cf. hypseleotris present in 60% of fish, followed by metacestodes of gryporhynchid cestodes encysted in the liver and gall bladder, which were present in 29% of individuals examined – that is, mixed infection with two Paradilepis spp., P. patriciae, P. cf. kempi and Dendrouterina sp., and two unidentified lineages of cyclophyllidean cestodes typified by the presence of numerous small hooks attached to the lumen of the intestine. Metacercariae of Clinostomum sp. and unidentified larvae of a spirurine nematode were found in 10 and 6% of individuals, respectively. Adult parasites were represented by S. acheilognathi (prevalence of 2% in western carp gudgeon), the nematode Procamallanus and the ectoparasitic copepod Lernaea sp. Monogeneans were rare, with overall prevalence of only 10%.
Discussion
We found the species diversity in the parasite community of carp gudgeons occurring in the Murrumbidgee River six times higher than previously reported. Out of 16 parasite taxa distinguished, all but two represent new host records for carp gudgeons. Most of these parasites were found at the larval stage, which often hampered their accurate identification. Since most parasites could not be identified to the species level, we were unable to reliably assess the number of putative new species. At the very least, our study considerably enriched the existing database of molecular data on Australian freshwater fish parasites with the generation of 54 sequences of parasites of carp gudgeons. The only metazoan parasite found in carp gudgeon for which molecular data were already available is the invasive S. acheilognathi (see Kuchta et al., Reference Kuchta, Choudhury and Scholz2018).
The considerable increase in the number of parasites detected in this fish group can be explained by several factors, including: (1) the large number of individuals of three different species examined; (2) the geographic span of our study including five different water bodies; (3) the parasitological examination focused also on larval stages occurring in parenteral sites – that is, outside of the fish intestine; (4) the DNA sequencing of morphologically similar larvae such as metacestodes of gryporhynchids; or (5) simply because this study represents the first survey focused on the parasite fauna of carp gudgeons. We suggest the use of this comparative approach of different geographic locations and closely related fish species, and combined examination of larval and adult parasites with morphological and molecular approaches as much as possible, to reveal the diversity of poorly known freshwater parasite fauna of fishes in Australia and elsewhere.
There were clear differences between the parasite communities of different species of carp gudgeons, despite the host fish species being closely related and often ecologically sympatric. Especially noteworthy is the depauperate fauna of metazoan parasites in the western carp gudgeon compared to the two other congeners. These differences are difficult to explain based on the available data, but they may reflect slightly different ecology, feeding habits, behaviour, immune response to parasite infection and other factors yet unknown that may differ between these three species of carp gudgeons studied.
Our knowledge of the life cycles of freshwater fish parasites in Australia is still very fragmentary because life-history studies have not been the main focus of parasitologists in Australia for decades, despite their importance for understanding host–parasite interactions (Cribb, Reference Cribb1998; Thomas et al., Reference Thomas, Guegan, Michalakis and Renaud2000; Blasco-Costa & Poulin, Reference Blasco-Costa and Poulin2017). Therefore, the present study helps us to better understand the role of parasites of carp gudgeons in ecosystems. Numerous larval stages of parasites, and high prevalence and intensity of infection of some of them, highlight the key role of carp gudgeons as intermediate hosts for multiple parasites with complex life cycles.
The common occurrence of parasites (trematode and cestode larvae), which mature in fish-eating birds, indicates that these fishes serve as a prey for birds such as Platibis flavipes (Gould, 1838), species of Phalacrocorax Brisson, 1760 or Egretta novaehollandiae (Latham, 1790) for the larval cestodes (Paradilepis, Dendrouterina and Parvitaenia) and Columba livia (Gmelin, 1789) or Nycticorax caledonicus (Gmelin, 1789) for the two trematodes A. cf. hypseleotris and Clinostomum sp. (see Bona (Reference Bona1975), Matthews & Cribb (Reference Matthews and Cribb1998) or Negm-Eldin & Davies (Reference Negm-Eldin and Davies2001) for data on the definitive hosts of gryporhynchid cestodes and trematodes). Metacercariae of Clinostomum sp. appeared closely related to C. complanatum (Rudolphi, 1814) in the phylogenetic tree, but they diverged genetically from it as much as C. album Rosser, Alberson, Woodyard, Cunningham, Pote & Gri, 2017 does from C. marginatum (Rudolphi, 1819). Several species of Clinostomum have been reported from birds in Australia (e.g. C. wilsoni Matthews & Cribb, 1998, C. australiense Johnston, 1916 or C. hornum Nicoll, 1914; see Matthews & Cribb, Reference Matthews and Cribb1998), for which molecular data are not available yet. Our larval specimens could well belong to one of the adults already known in Australian birds or represent a new species. In addition, two invasive species, the tapeworm S. acheilognathi and the copepod Lernaea sp. (an invasive taxon known to cause multiple pathological effects to their host especially in small fish (Lester & Hayward, Reference Lester, Hayward and Woo2006; Lymbery et al., Reference Lymbery, Hassan, Morgan, Beatty and Doupe2010)), were found in carp gudgeons. Thus, carp gudgeons may serve as reservoirs of these potentially pathogenic parasites. Parasites, along with their hosts and many other free-living animals, are threatened by multiple environmental changes such as urbanization, agriculture, climate change or pollution (e.g. Dušek et al., Reference Dušek, Gelnar and Šebelová1998; Bradley & Altizer, Reference Bradley and Altizer2007; Lagrue et al., Reference Lagrue, Kaldonski, Motreuil, Lefevre, Blatter, Giraud and Bollache2011; Mouritsen et al., Reference Mouritsen, Sørensen, Poulin and Fredensborg2018; Kwak et al., Reference Kwak, Heath and Palma2019). Therefore, it is essential to continue the inventory of Australian freshwater fish parasites before they perish or put other species at threat (Beveridge & Gasser, Reference Beveridge and Gasser2014).
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/S0022149X20000280
Acknowledgements
Thanks are due to Maggie Watson, Charles Sturt University, School of Environmental Sciences, for her initial advice and for providing connections to the key people to assist with our study; Shane Raidal, Charles Sturt University, School of Animal and Veterinary Sciences, for conducting preliminary examination of carp gudgeon parasites; Shokoofeh Shamsi, Charles Sturt University, School of Animal and Veterinary Sciences, for generously providing laboratory space to conduct dissections and for advice; Jan Brabec, Natural History Museum of Geneva and Institute of Parasitology of the Academy of Sciences of the Czech Republic, for his advice regarding molecular analyses; and Jean Mariaux, Natural History Museum of Geneva, for his advice on cestode identification.We would also like to exprenss our gratitude to two anonymous reviewers for their helpful comments that helped us improve the manuscript.
Financial support
This work was supported by the Institute for Applied Ecology, University of Canberra, the Natural History Museum of Geneva, and the Institute of Parasitology, České Budějovice (RVO 60077344).
Conflicts of interest
None.
Ethical standards
The authors assert that all procedures contributing to this work comply with the ethical standards of the University of Canberra ethics committee on animal experimentation (approval number 20180442).








