Introduction
The Franciscana, Pontoporia blainvillei Gervais and d'Orbigny 1844, is a small dolphin endemic to the western South Atlantic, whose occurrence ranges from Espírito Santo, Brazil (18°25′S), to Golfo San Matias, Argentina (42°35′S) (Siciliano et al., Reference Siciliano, Di Beneditto and Ramos2002). The greatest threat to P. blainvillei is its incidental capture in coastal gillnets throughout most of its distribution and it is one of the most threatened small cetacean species in the western South Atlantic (IBAMA, 2001; Secchi et al., Reference Secchi, Danilewicz and Ott2003). However, the comprehensive impact of the by-catch on populations is still unknown due to uncertainties about stock structure and lack of abundance estimates for most of the areas. These topics have been considered research priorities for this species in several meetings, workshops and action plans during the past decades (Secchi et al., Reference Secchi, Wang, Murray, Rocha-Campos and White1998, Reference Secchi, Danilewicz, Ott, Ramos, Lazaro, Marigo and Wang2002; Mendez et al., Reference Mendez, Rosenbaum and Bordino2007, Reference Mendez, Rosenbaum, Subramaniam, Yackulic and Bordino2010; Barbato et al., Reference Barbato, Secchi, Di Beneditto, Ramos, Bertozzi, Marigo, Bordino and Kinas2011). Secchi et al. (Reference Secchi, Danilewicz and Ott2003) proposed four management stocks (known as Franciscana Management Areas or FMAs), based on data available on life history, parasite load and genetics. Recently, the subdivision of Franciscanas off the coast of Santa Catarina into two stocks (Ott et al., Reference Ott, Oliveira, Barreto, Secchi, Almeida, Moreno, Danilewicz and Bonatto2008) and from Argentina and Uruguay into at least two genetically recognizable populations each (Mendez et al., Reference Mendez, Rosenbaum and Bordino2007; Costa-Urrutia et al., Reference Costa-Urrutia, Abud, Secchi and Lessa2011) was verified. This demonstrates a significant genetic subdivision at regional levels and fine-scale structure within P. blainvillei populations.
Multidisciplinary approaches have been used to investigate host population structure, using a variety of parameters, including biological tags such as parasites. Parasites can often be used to identify subpopulations even where genetic studies fail to do so (e.g. MacKenzie, Reference MacKenzie2002). The term ‘ecological stock’ is used to describe subpopulations which are distinguished by behavioural differences, but there is still a considerable amount of gene flow among them (MacKenzie, Reference MacKenzie2002). It differs from the ‘genetic or biological stock’ that would represent reproductively isolated units that are genetically different from each other (Altukhov, Reference Altukhov1981; Wang, Reference Wang, Perrin, Würsig and Thewissen2002). Up to the present moment, parasite load has been recommended (Reeves & Leatherwood, Reference Reeves and Leatherwood1994; IBAMA, 2001) and used to identify ecological stocks of P. blainvillei (Aznar et al., Reference Aznar, Raga, Corcuera and Monzón1995; Andrade et al., Reference Andrade, Pinedo and Pereira1997; Marigo et al., Reference Marigo, Rosas, Andrade, Oliveira, Dias and Catão-Dias2002; Secchi et al., Reference Secchi, Danilewicz, Ott, Ramos, Lazaro, Marigo and Wang2002), corroborating the hypothesis of segregated populations.
Recently, studies based on parasite genes have been used to elucidate the history or demography of host populations, revealing useful markers to indicate the source of host populations. Given that parasites are closely linked to their hosts, it might be expected that hosts would have similar phylogeographical patterns. Indeed, there is a relationship between host and parasite genetic differences related to their population historical biogeography (for a review see Criscione et al., Reference Criscione, Poulin and Blouin2005) and this is influenced by transmission and which host's life cycle is analysed. Also, parasite populations can be more structured than their hosts, and through one parasite's molecular data it could be possible to identify its host population better than using the host's genotypes (Criscione et al., Reference Criscione, Cooper and Blouin2006).
Molecular studies on helminths have increased over the past decade, especially among parasites of medical importance and also those of fish and other marine organisms (e.g. Semyenova et al., Reference Semyenova, Morozova, Chrisanfova, Asatrian, Movsessian and Ryskov2003; Théron et al., Reference Théron, Sire, Rognon, Prugnolle and Durand2004; Morgan et al., Reference Morgan, Dejong, Adeoye, Ansa, Barbosa, Brémond, Cesari, Charbonnel, Corrêa and Coulibaly2005; Shrivastava et al., Reference Shrivastava, Gower, Balolong, Wang, Qian and Webster2005; Mattiucci & Nascetti, Reference Mattiucci and Nascetti2007; Zhu et al., Reference Zhu, Podolska, Liu, Yu, Chen, Lin, Luo, Song and Lin2007; Attwood et al., 2008; Mattiucci et al., Reference Mattiucci, Farina, Campbell, Mackenzie, Ramos, Pinto, Abaunza and Nascetti2008; Klimpel et al., Reference Klimpel, Busch, Kuhn, Rohde and Palm2010). From the Brachycladiidae family (Trematoda: Digenea), the marine mammal parasites, only the 18S rDNA gene and mtDNA NADH dehydrogenase subunit 3 (ND3) gene sequences are available and have been used in phylogenetic studies (Fernández et al., Reference Fernández, Aznar, Latorre and Raga1998a, Reference Fernández, Littlewood, Latorre, Raga and Rollinsonb). In a recent phylogenetic study using nuclear 18S and mitochondrial ND3 genes, the genus Synthesium, represented by Synthesium pontoporiae (Raga, Aznar, Balbuena and Dailey, 1994) and S. tursionis (Marchi, 1873), was positioned among brachycladids (Marigo et al., Reference Marigo, Thompson, Santos and Iñiguez2011), but no genetic intraspecific studies have been performed up to the present moment.
Synthesium pontoporiae is a small intestinal trematode parasite with adult worms exclusively in P. blainvillei which has been suggested as a biological marker for the species and shows differences in prevalence and mean intensity along its distribution (Aznar et al., Reference Aznar, Raga, Corcuera and Monzón1995; Marigo et al., Reference Marigo, Rosas, Andrade, Oliveira, Dias and Catão-Dias2002). Marigo et al. (Reference Marigo, Rosas, Andrade, Oliveira, Dias and Catão-Dias2002) reported significant differences in the prevalence and mean intensity of S. pontoporiae among three areas with a small sample size (12–17 intestines) as preliminary data; however, with a larger number of intestines analysed, these numbers greatly changed (Marigo, pers. obs.), possibly due to seasonality. The next step was to investigate the parasite morphometrics, and no significant difference was found among areas that could indicate different ecological parasite stocks (Marigo et al., Reference Marigo, Vicente, Valente, Measures and Santos2008) Thus, based on the main aim of this study, which was to identify Franciscana stocks through parasites, we decided to use the molecular approach. For the investigation of large-scale evolutionary patterns and the internal structure of this species, molecular markers have been used with increased frequency and robustness. Since studies on the intraspecific level usually deal with recent evolutionary processes, the most useful markers, in these cases, are rapidly evolving DNA regions, such as mitochondrial DNA (mtDNA) (Avise, Reference Avise2008).
Here, the first internal transcribed sequence (ITS) and cytochrome oxidase I (COI) sequences of the genus Synthesium were recovered from S. pontoporiae from different geographical areas. Also, the presence of polymorphisms in these markers and ND3 was investigated in order to relate them to intraspecific variation that could indicate stock differentiation of their host populations. This is the first study on population genetics of a marine mammal trematode.
Materials and methods
Sample collection, DNA extraction, amplification and sequencing
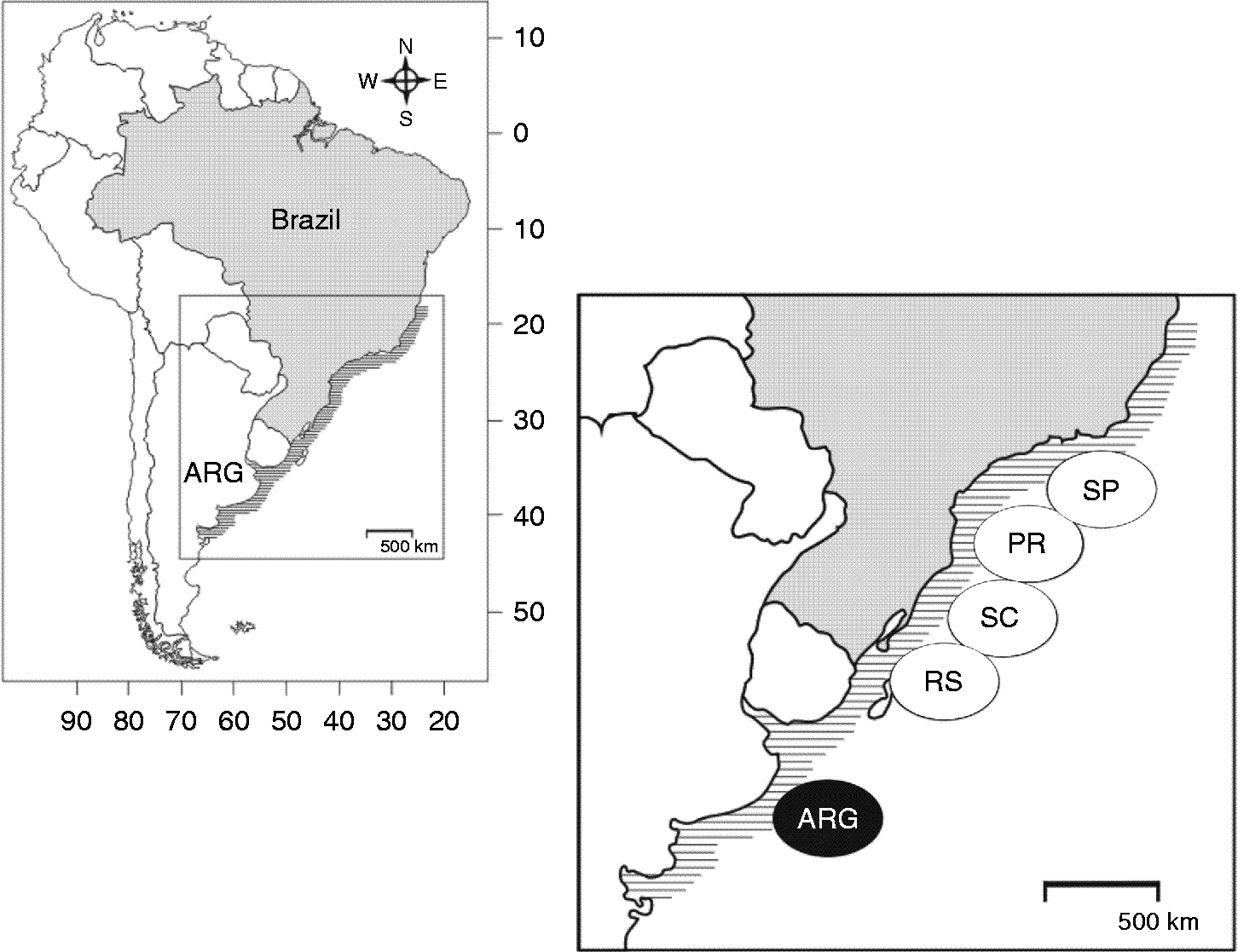
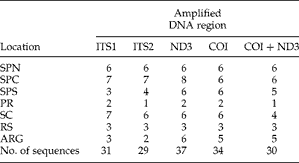
Synthesium pontoporiae specimens were collected from the intestine of P. blainvillei individuals stranded or incidentally captured off the south-eastern and southern Brazilian and Argentinian coasts (fig. 1). Their collection and transport for research were authorized by local environmental agencies. Intestines were frozen after necropsy and thawed for parasite inspection. Trematodes were fixed and conserved in 70% ethanol for up to 10 years. Each parasite used was collected from a different individual host and named after host number and location (N= 131). Seven areas were sampled: São Paulo North (SPN); São Paulo Central (SPC); São Paulo South (SPS); Paraná (PR); Santa Catarina (SC); Rio Grande do Sul (RS) and Argentina (ARG) (table 1).

Fig. 1 Sampling sites of Synthesium pontoporiae from the endangered Franciscana dolphin off the coast of Brazil and Argentina: SP, São Paulo North, Central and South; PR, Paraná; SC, Santa Catarina; RS, Rio Grande do Sul; ARG, Argentina.
Table 1 The occurrence of Synthesium pontoporiae sequences in each amplified DNA region, relative to geographical location; see fig. 1 for full names of locations.
DNA was extracted according to the QIAamp DNA Mini Kit (Qiagen, Los Angeles, California, USA) protocol. ITS regions were amplified using two sets of primers, for ITS1: 5′-GACGACCAAACTTGATCATT-3′ and 5′-TGCGCTCTTCATCGACACACGA-3′ (this study); and for ITS2: NC13F 5′-ATCGTGAAGAACGCAGC-3′ and NC2R 5′-TTAGTTTCTTTTCCTCCGCT -3′ (Zhu et al., Reference Zhu, Gasser, Jacobs, Hung and Chilton2000).
The ND3 gene was amplified using two sets of primers:
(a) ND3A 5′-GCGTTAGCAGGATCCTGTGATATAG-3′ and ND3B 5′-CCAAAGCTTAAATCATCGTTAGCAG-3′; and
(b) ND3C 5′-CTACTAGTGAGATTGATCTYCGTCGGT-3′ and ND3D 5′-CTACTAGTCCCACTCAACRTAACCYT-3′ (Fernández et al., Reference Fernández, Aznar, Latorre and Raga1998a).
The COI gene was amplified by polymerase chain reaction (PCR) using primers COIPRA 5′-TGGTTTTTTGTGCATCCTGAGGTTTA-3′ and 5′-AGAAGAACGTAATGAAAATGAGCAAC-3′ (Bessho et al., Reference Bessho, Ohama and Osawa1992).
Amplification reactions contained 400 ng of DNA, 0.2 mm of deoxynucleoside triphosphates (dNTPs), 200 ng of each primer, 3 mm of MgCl2 and 2 U of Taq DNA polymerase, in a final volume of 50 μl. PCR conditions were: initial denaturation at 94°C for 5 min, followed by 40 cycles at 94°C for 30 s, 47°C for 30 s; 72°C for 30 s. PCR amplicons were purified using Perfect Cleanup (Eppendorf, Hamburg, Germany) and directly sequenced on both strands in an ABI PRISM 3730 Genetic Analyzer (Applied Biosystems, Foster City, California, USA) from both strands. Raw sequences were transferred to DNASTAR software (Lasergene software, Madison, Wisconsin, USA) and the consensus sequences were determined. Sequences were aligned on CLUSTAL W (Thompson et al., Reference Thompson, Higgins and Gibson1994). The ND3 amplicon included a tRNA sequence prior to the ND3 gene sequence; the analysis was performed using only the ND3 gene sequence.
Data analysis
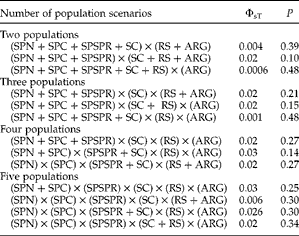
Haplotypes were identified and the genetic diversity within populations was estimated using the DnaSP software (Rozas et al., Reference Rozas, Sánchez-Delbarrio, Messeguer and Rozas2003). COI and ND3 sequences were concatenated for population analyses. The Arlequin 4.5 software (Excoffier et al., Reference Excoffier, Laval and Schneider2005) was used for population structure analyses. Pairwise fixation indexes based on the haplotype diversity (FST) were estimated and their significance was ascertained by 10,000 random permutations. The variation distribution of the genetic diversity within and between populations was inferred and tested for different scenarios (two to five populations, see table 3) using an analysis of molecular variance (AMOVA; Excoffier et al., Reference Excoffier, Smouse and Quattro1992). Spatial AMOVA (SAMOVA, Dupanloup et al., Reference Dupanloup, Schneider and Excoffier2002) was also used to search groupings of geographically adjacent locations that had the highest amount of variation among them.
The S. pontoporiae population size oscillations over time were also investigated through a Bayesian skyline plot method implemented in the BEAST v.1.3 software (Drummond & Rambaut, Reference Drummond and Rambaut2007) using COI sequences. Demographic reconstruction used the above-mentioned evolutionary rate, the HKY +G mutation model as selected using ModelTest (Posada, Reference Posada2008) and 10 group intervals. Ten million Markov chain Monte Carlo (MCMC) steps and a burning of 1,000,000 were used to achieve effective sample size (ESS) values > 200 for all parameters.
Statistical parsimony networks for each mitochondrial gene and the concatenated sequence were built using TCS 1.21 (Clement et al., Reference Clement, Posada and Crandall2000). Ambiguities in the network of concatenated COI and ND3 haplotypes were solved according to criteria of Crandall & Templeton (Reference Crandall and Templeton1993). The new nucleotide sequences reported in this paper for S. pontoporiae are available in GenBank under accession numbers: JX644084 (ITS1); JX6440845 (ITS2); JX644086–JX644122 (ND3) and JX644086–JX644156 (COI).
Results
All 31 ITS1 (534 bp) and 29 ITS2 sequences (447 bp) of S. pontoporiae were identical (monomorphic) in all locations studied, so they were not used for population analyses. A total of 37 sequences of the mtDNA ND3 of S. pontoporiae, spanning 331 bp, were analysed, revealing ten haplotypes defined by 13 variable sites. The G+C content was 0.351. Haplotype (Hd) and nucleotide diversity (per site, π) were 0.577 and 0.003, respectively. Thirty-four 416 bp long COI sequences were also analysed, revealing 24 haplotypes and 30 variable sites. The G+C content was 0.381. Haplotype diversity (Hd) was 0.932 and nucleotide diversity (π) was 0.007.
For population analyses, the ND3 and COI sequences of 30 single parasites were concatenated (747 bp). The unique sequence from PR was joined with sequences from SPS (N= 5), the closest locality (77 km). The highly divergent BP06SPC sequence was excluded during population differentiation analysis.
The FST pairwise values were non-significant for all site comparisons (table 2). Population differentiation scenarios based on stocks previously proposed for Franciscana dolphins in the literature were tested using AMOVA. No significant separation of populations was supported by AMOVA in any scenario proposed, suggesting the absence of geographical structuring and no restriction of gene flow among the parasites, despite their different locations. Hypothetical population divisions were also evaluated, but none of them was statistically significant (table 3). SAMOVA also failed to detect any significant grouping of geographically adjacent sampling localities. However, due to the small sample size, there is insufficient evidence to infer population structure.
Table 2 Pairwise FST values of Synthesium pontoporiae for COI and ND3 concatenated sequences relative to geographical location (SPN, SPC, SPS+PR, SC, RS, ARG; see fig.1 for the full names of locations).

Table 3 Analysis of genetic population subdivisions (ΦsT) through AMOVA, using COI and ND3 concatenated sequence data, for each population differentiation scenario (P values indicate the proportion of random values larger than observed values).
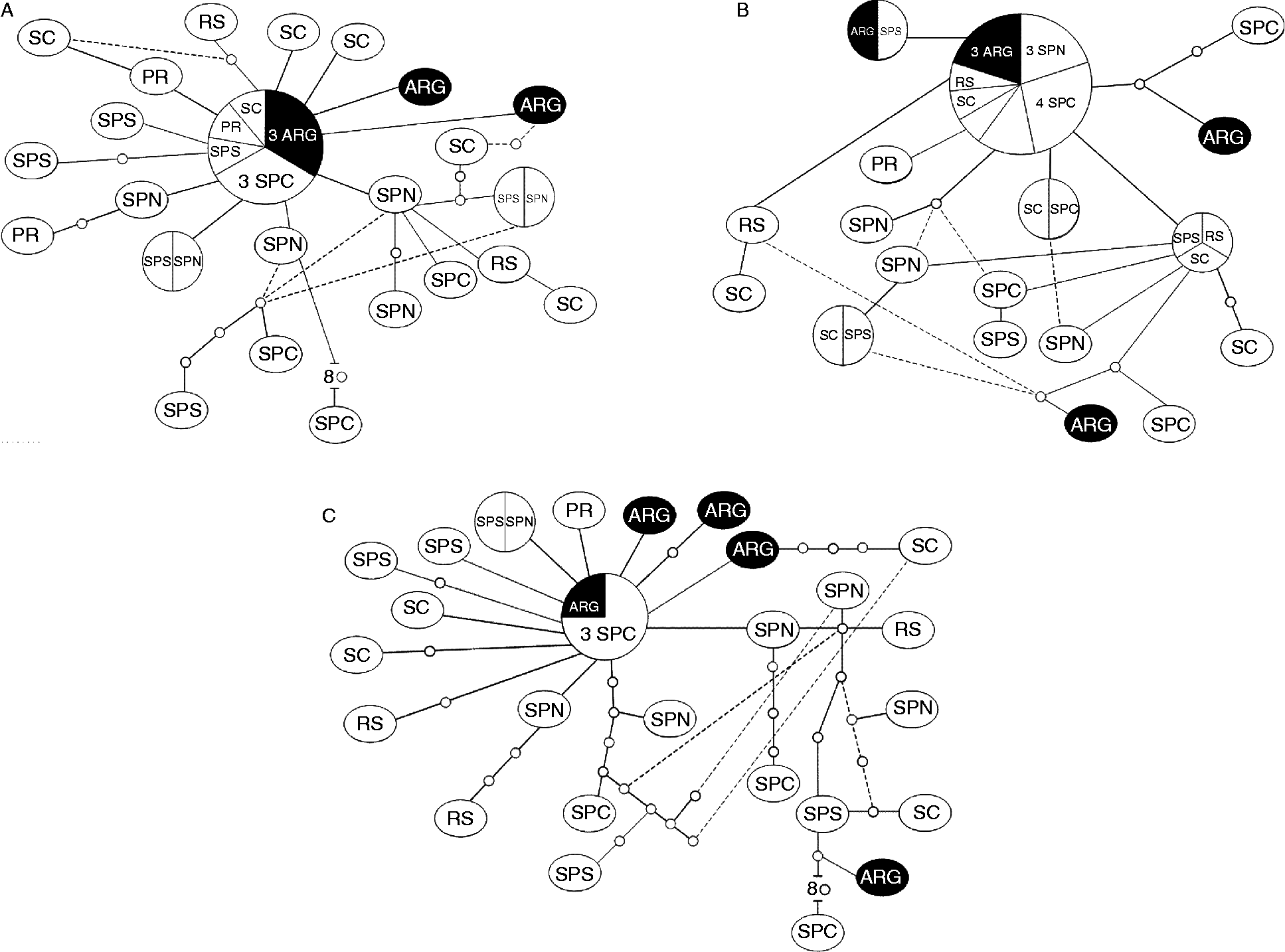
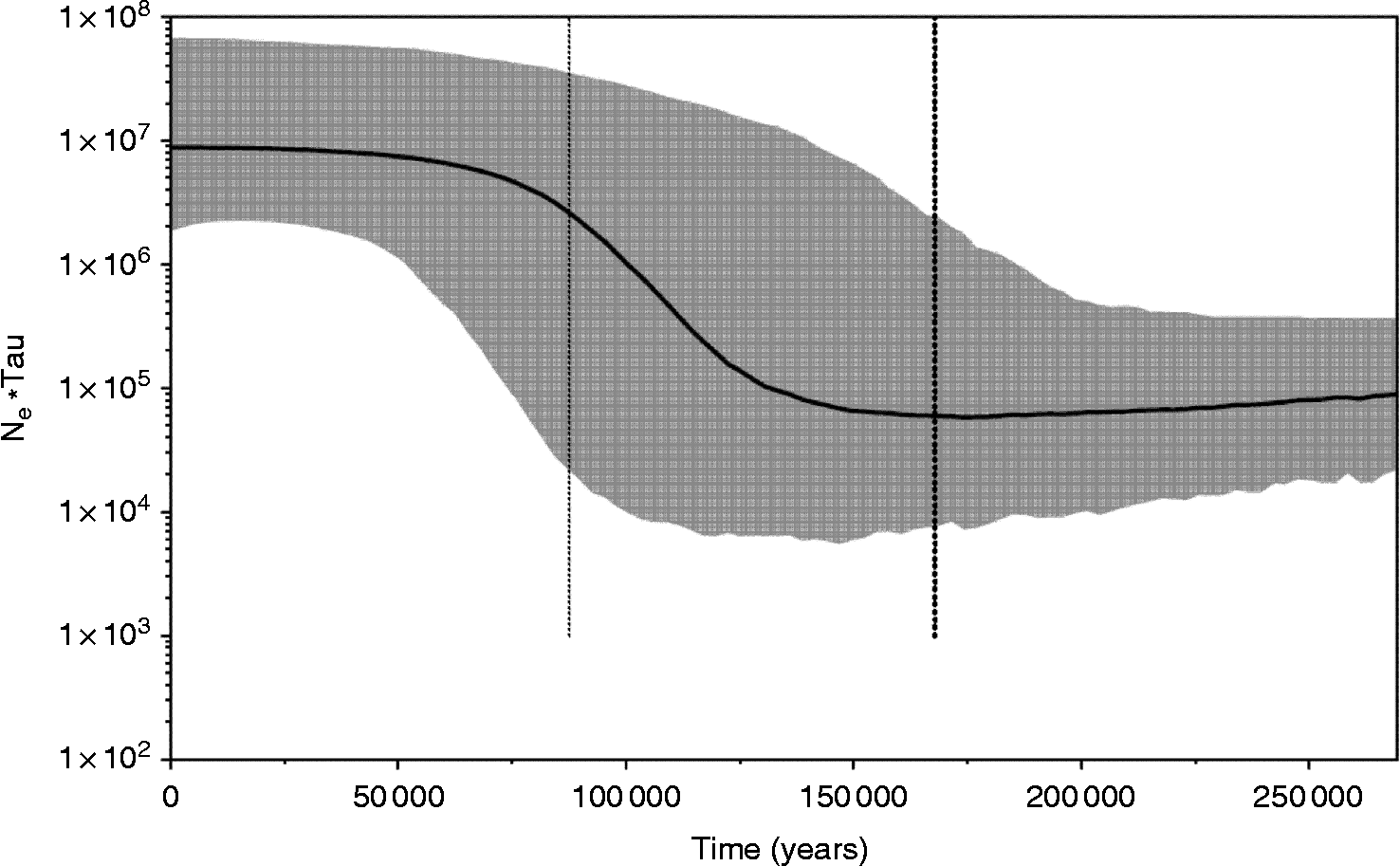
In the TCS haplotype network, each connection is a single mutational step, with white circles representing inferred haplotypes (extinct/unsampled), and coloured circles representing the actual observed haplotypes. The sizes of circles are proportional to the number of individuals that have that haplotype. The networks of both markers and the concatenated sequence also suggest a lack of population structure, since samples from all localities are scattered throughout the networks. However, another pattern was observed in the haplotype networks: a star-like conformation usually attributed to recent population expansion in both markers studied, with the exception of COI (fig. 2). The Bayesian skyline plot (fig. 3) also indicates population expansion, and estimates that all S. pontoporiae sequences coalesced around 167,840 (87,590–269,360) years ago.

Fig. 2 Haplotype network drawn from (A) 331 bp of ND3; (B) 416 bp of CO1 and (C) 747 bp of ND3 and CO1 gene sequences from seven geographical locations: SP, São Paulo North, Central and South; PR, Paraná; SC, Santa Catarina; RS, Rio Grande do Sul; ARG, Argentina. Each cross bar along network connectors represents one mutational step, and small white circles represent extinct/unsampled haplotypes.

Fig. 3 Bayesian skyline plot with COI Synthesium pontoporiae sequences for the entire species (Ne, effective population size; tau, generation length in years).
Discussion
Molecular studies on digenean trematodes with an indirect life cycle are more often motivated by the need to understand the epidemiology of parasites of medical and economical importance, such as Schistosoma (Shrivastava et al., Reference Shrivastava, Gower, Balolong, Wang, Qian and Webster2005; Attwood et al., 2008). Additionally, other studies have focused on the genetic structure of digeneans from marine habitats (Vilas et al., Reference Vilas, Paniagua and Sanmartin2003; Prugnolle et al., Reference Prugnolle, Liu, de Meeus and Balloux2005a; Criscione et al., Reference Criscione, Cooper and Blouin2006, Reference Criscione, Vilas, Paniagua and Blouin2011; Hansen & Poulin, Reference Hansen and Poulin2006; Criscione & Blouin, Reference Criscione and Blouin2007). The present study analysed S. pontoporiae, aiming to reveal polymorphism labelling the previously recognized stocks of its definitive host, the endangered P. blainvillei dolphin.
Preliminary data on infection levels of S. pontoporiae along its host distribution (Marigo et al., Reference Marigo, Rosas, Andrade, Oliveira, Dias and Catão-Dias2002; Secchi et al., Reference Secchi, Danilewicz, Ott, Ramos, Lazaro, Marigo and Wang2002) corroborated the hypothesis of different P. blainvillei stocks (Pinedo, Reference Pinedo1991; Secchi et al., Reference Secchi, Wang, Murray, Rocha-Campos and White1998; Mendez et al., Reference Mendez, Rosenbaum and Bordino2007) and the putative stock subdivision proposed by Secchi et al. (Reference Secchi, Danilewicz and Ott2003). However, this conclusion changed later when more intestinal samples were analysed (Marigo, pers. obs.) and the morphometry of the parasite was assessed. We searched for possible genetic differences among these morphologically homogeneous parasites that exhibited latitudinal variation in infection level. The ITS, COI and ND3 sequences of S. pontoporiae from three Franciscana management stocks did not show any polymorphism that could be associated with the genetic structure of the species.
Host vagility should be a major determinant of parasite gene flow because many parasites have low dispersal capacity in their free-living stages, comparing the limited mobility of molluscs to that of rats as intermediate hosts for Schistosoma mansoni, for instance (Prugnolle et al., Reference Prugnolle, Theron, Pointier, Jabbour-Zahab, Jarne, Durand and de Meeûs2005b). As a consequence, the most mobile host can control the gene flow in a parasite with a complex life cycle (Jarne & Théron, Reference Jarne and Théron2001; Criscione et al., Reference Criscione, Poulin and Blouin2005; Prugnolle et al., Reference Prugnolle, Theron, Pointier, Jabbour-Zahab, Jarne, Durand and de Meeûs2005b). Although dolphins are considered highly mobile, P. blainvillei individuals satellite-tracked in Argentina exhibited very restricted and localized movements, with small home ranges in bay areas (Bordino et al., Reference Bordino, Wells and Stamper2008). Furthermore, there are at least two genetically recognizable local populations of Franciscana dolphins in Argentina, and also in Uruguay (Mendez et al., Reference Mendez, Rosenbaum, Subramaniam, Yackulic and Bordino2010; Costa-Urrutia et al., Reference Costa-Urrutia, Abud, Secchi and Lessa2011). If the life cycle of S. pontoporiae was restricted to P. blainvillei, or if intermediary hosts had lower vagility compared to genetically structured Franciscana, the parasite population would also be subdivided. However, it is important to mention that ecological stocks do not always reflect their differences of behaviour, seasonality, maturity and also morphology in their genes, consequently they will not be recognized as genetic stocks (Meyer et al., Reference Meyer, Kocher, Basasibwaki and Wilson1990).
In this context, taking into account only the genetics of the parasites of Franciscana dolphins, our results showed no structure due to: (1) the existence of apparently a single parasite population; or (2) the small sample size that would have caused lack of power in pairwise comparisons and hierarchical AMOVA. When discussing stock identification results and inadequate statistical power, Wang (Reference Wang, Perrin, Würsig and Thewissen2002) mentioned that in this case, ‘the appropriate conclusion would be that differences in the characters examined were not detected rather than differences do not exist between the units being studied’. Therefore, we would have to improve the sample size of our parasites to meet the first explanation. However, if the first explanation was right (a single parasite population) we suggest that this lack of structure could be due to some prey of P. blainvillei involved in the S. pontoporiae life cycle that is more mobile than the definitive host and therefore could carry the parasites across FMAs.
The life cycle of Synthesium is unknown and, for digeneans in marine mammals, larval stages including metacercariae are inadequately described. The digenean life cycle involves two asexual generations in a molluscan and a sexual one in a vertebrate host and, in the majority of cases, one or more intermediary hosts are needed to complete the life cycle (Gibson, Reference Gibson2002). The Franciscana diet includes cephalopods, crustaceans and fish, and many of those are species that spend most of their adult lives offshore and migrate to estuarine and protected areas to spawn (Sardiña & Lopez Cazorla, Reference Sardiña and Lopez Cazorla2005; Rodrigues & Gasalla, Reference Rodrigues and Gasalla2008). Any of these animals may be part of the cycle, although no brachycladiid cercaria or metacercariae have been found inside them so far. In coastal areas, S. pontoporiae adults present in Franciscanas, as regular digenean trematodes, shed their eggs and the cycle is completed in the presence of these molluscs and fish.
Considering that the possible intermediary hosts are more mobile than the Franciscana dolphin, our data would support the hypothesis that the most mobile host can influence the gene flow in a parasite with complex life cycle (Jarne & Théron, Reference Jarne and Théron2001; Criscione et al., Reference Criscione, Poulin and Blouin2005; Prugnolle et al., Reference Prugnolle, Theron, Pointier, Jabbour-Zahab, Jarne, Durand and de Meeûs2005b). In a similar study, Baldwin et al. (Reference Baldwin, Rew, Johansson, Banks and Jacobson2011) found that anisakid nematodes were not applicable as markers to identify population subdivision of Pacific sardines, possibly due to the migration of both fish and cetacean hosts mixing the haplotypes. Therefore, in view of the assumed vagility of intermediary hosts and/or the small sample size, precise conclusions on the structure of single populations of S. pontoporiae cannot be made.
In relation to the population expansion scenario, since S. pontoporiae occurs exclusively in Franciscanas, one potential explanation would be that the expansion reflected the colonization of the species by the parasite. Within the Brachycladiidae, Synthesium is a genus that exhibits a worldwide distribution and occurs in a number of odontocete families (Pontoporiidae, Delphinidae, Monodontidae and Phocoenidae), suggesting a long period of host–parasite interactions, with many potential host captures among different odontocete groups (Fernández et al., Reference Fernández, Aznar, Latorre and Raga1998a). However, when and which Synthesium species adapted to its first odontocete host cannot be defined. It is possible to suggest that the ancestor of the Synthesium species could adapt first to other odontocete families and later to Pontoporia. The evolutionary and biological peculiarities of Franciscanas may be responsible for S. pontoporiae exclusivity as a parasite found only in P. blainvillei. The anatomical characteristics of P. blainvillei would have resulted in adaptations of the ancestral S. pontoporiae, and the reproductive isolation that followed caused the speciation.
Acknowledgements
This work is part of a PhD dissertation of J.M. developed at Instituto Oswaldo Cruz, FIOCRUZ, RJ, Brazil. We thank Katia Groch and Heloisa M. Diniz (Laboratório de Produção e Tratamento de Imagem-IOC) for technical assistance and the PDTIS/FIOCRUZ genomic platform for DNA sequencing. We are particularly grateful to those who provided intestines for parasite sampling: Projeto BioPesca, SP (V. Ruoppolo, J.A.Ribeiro, J.V.S. Lima, M.B. Alonso, F. Marcatto and B. Henning); Instituto Terra & Mar, São Paulo; Instituto de Pesquisas de Cananéia (IPeC); Centro de Estudos do Mar (CEM-UFPR); Universidade da Região de Joinville (UNIVILLE); Universidade do Vale do Itajaí (UNIVALI); Grupo de Estudos de Mamíferos Aquáticos do Rio Grande do Sul (GEMARS) (D. Danilewicz, I.B. Moreno, P.H. Ott, M. Tavares, R. Machado, S.B. Nakashima, C.C. Trigo); and Museo Argentino de Ciencias Naturales (MACN-CONICET) (M.F. Negri, M.N. Paso Viola, M.V. Panebianco, F. Pèrez).
Financial support
The project ‘Research and Conservation of Aquatic Mammals and Sea Turtles in the Rio Grande do Sul’ (‘Pesquisa e Conservação de Mamíferos Marinhos e Tartarugas Marinhas no Litoral Norte do Rio Grande do Sul’) was supported by Fundação O Boticário de Proteção à Natureza and Fundo Nacional do Meio Ambiente – FNMA/MMA (Convenio 094/2001), which provided financial support especially for sampling Franciscanas along the coast of Rio Grande do Sul. We also thank the Cetacean Society International, the Society for Marine Mammalogy, Yaqu Pacha, Project AWARE-PADI, MacArthur Foundation, Fundação O Boticário de Proteção à Natureza and PROBIO/FNMA for the financial support of the collaborating institutions.
Conflict of interest
The authors declare no conflict of interest.
Ethical standards
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional guides on the care and use of laboratory animals.