Introduction
The genus Galactosomum Looss, 1899 (Plagiorchiida: Heterophyidae) is a widespread group of parasite species that infect the gastrointestinal tract of fish-eating birds and marine mammals. Their life cycle includes gastropod and fish intermediate hosts, and all species so far reported inhabit marine or brackish environments. Species of Galactosomum have been recorded from all continents except Antarctica, and five of the 22 species are found in Australia (Fischthal & Kuntz, Reference Fischthal and Kuntz1972; Pearson, Reference Pearson1973; Dailey et al., Reference Dailey, Demaree and Critchfield2002).
Adults of Galactosomum spp. have only once been recorded from New Zealand: specimens of an unnamed species were recovered from a little blue penguin on Tiritiri Matangi Island, off North Island (McKenna, Reference McKenna2009). However, larval stages (rediae and cercariae) have been known for some years to be infecting the intertidal mud snail, Zeacumantus subcarinatus (Sowerby, 1855), and Martorelli et al. (Reference Martorelli, Fredensborg, Leung and Poulin2008) concluded from morphology that the cercariae belonged to a species closely related to Galactosomum bearupi Pearson, 1973 (Martorelli et al., Reference Martorelli, Fredensborg, Leung and Poulin2008).
During a survey of the helminth parasites of birds in South Island, New Zealand, we found specimens of adult Galactosomum sp. in the intestines of Caspian terns (Hydroprogne caspia (Pallas, 1770)), red-billed gulls (Chroicocephalus scopulinus (Forster, 1844)), black-backed gulls (Larus dominicanus Lichtenstein, 1823) and little blue penguins (Eudyptula novaehollandiae (Stephens, 1826)). The birds all came from the same geographical and ecological region as the snails, so it was suspected that, if these specimens represented a single species, it was likely to be that found as larval stages in Z. subcarinatus from Otago Harbour.
The aim of this study is to give a scientific name, describe and add to our knowledge of the life cycle of the Galactosomum sp. previously known to infect New Zealand Z. subcarinatus. We use molecular tools to compare the cercarial stage of Galactosomum sp. infecting Z. subcarinatus with the adult infecting fish-eating birds, and present a preliminary phylogeny for the genus based on both 28S and ITS2 available sequences. We use morphological methods to compare the adult Galactosomum specimens from New Zealand to other described Galactosomum species, and conclude that the specimens belong to a hitherto undescribed species, which we here describe and name.
Materials and methods
Bird collection and trematode sampling
A total of 87 birds of four species, two Caspian terns (H. caspia), 30 red-billed gulls (C. scopulinus), 20 black-backed gulls (L. dominicanus) and 35 little blue penguins (E. novaehollandiae), were examined for gastrointestinal helminths between October and November 2018. Birds were found dead, or donated after death or euthanasia by the Dunedin Wildlife Hospital, and were frozen less than 12 h after death. Birds were defrosted, the intestines removed and examined under a dissecting microscope, and the worms preserved in 70% ethanol for whole-mount and 96% ethanol for genetic analyses.
All little blue penguins were collected from around the Otago coast; therefore, we have assigned these specimens to E. novaehollandiae (Otago and Australian little blue penguin) as opposed to E. minor (Forster, 1781) (New Zealand little blue penguin) (see Grosser et al., Reference Grosser, Burridge, Peucker and Waters2015, Reference Grosser, Scofield and Waters2017).
Morphological data
Trematodes fixed for whole mounts were stained using acetic acid iron carmine stain, dehydrated through a graded ethanol series, cleared in clove oil and mounted in permanent preparations with Canada balsam. A few specimens were used to trial Nile blue and haematoxylin stains, which proved useful for highlighting muscle fibres and vitellaria. Measurements were made using ImageJ software (Wayne Rasband, NIH, USA) from photographs taken on an Olympus BX51 compound microscope mounted with DP25 camera attachment. All measurements are in micrometres unless otherwise indicated, with the mean followed by the range. Drawings were made with the aid of a drawing tube mounted on an Olympus compound microscope. For scanning electron microscopy (SEM), five specimens were fixed overnight in 2.5% aldehyde in 0.1 M cacodylate buffer. They were then post-fixed in 1% osmium tetroxide for 1 h prior to being dehydrated through a gradient series of ethanol, critical-point dried in a CPD030 BalTec critical-point dryer (BalTec AG, Balzers, Liechtenstein) using carbon dioxide, mounted on aluminium stubs using double-sided adhesive carbon tape and sputter coated with gold/palladium (60:40) to a thickness of 10 nm in an Emitech K575X Peltier-cooled high-resolution sputter coater (EM Technologies, Ashford, Kent, UK). The specimens were viewed with a JEOL 6700 F field emission scanning electron microscope (JEOL Ltd, Tokyo, Japan) at the Otago Centre for Electron Microscopy (OCEM, University of Otago, New Zealand). Type specimens were deposited in Te Papa Museum, Wellington (accession numbers W.003498–W.003502) and the Otago Museum, Dunedin (accession numbers IV107611–IV107616). Comparative material examined comprised paratype and vouchers of G. angelae (South Australia Museum; accession numbers 20107–20112, 41029) and G. bearupi paratype (South Australia Museum; accession number AHC41030).
Molecular data and analysis
Genomic DNA was extracted from five ethanol-fixed specimens (one from each of black-backed gull, red-billed gull, Caspian tern and two from little blue penguins) using the DNeasy® Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. To compare the sequence of larval Galactosomum sp. infecting Z. subcarinatus, we amplified a partial fragment of cytochrome c oxidase subunit 1 gene (cox1) of an adult Galactosomum sp. from little blue penguin using primers JB3 (5′-TTT TTT GGG CAT CCT GAG GTT TAT-3′) (Bowles et al., Reference Bowles, Hope, Tiu, Liu and McManus1993) and Trem.Cox.1 (5′AAT CAT GAT GCA AAA GGT A-3′) (Králová-Hromadová et al., Reference Králová-Hromadová, Špakulová and Horáčková2008). Polymerase chain reactions (PCRs) were run in 25 µl reaction mixtures using an Eppendorf Mastercycler Pro thermal cycler (Eppendorf, New York) and conditions consisted of an initial denaturation phase (2 min at 95°C); 40 cycles of denaturation (30 s at 95°C), primer annealing (40 s at 48°C), extension (1 min at 72°C) and a 10 min final extension (72°C).
To compare the identity of Galactosomum specimens among hosts sampled, two partial gene fragments were amplified; 28S rRNA gene (28S), using primers T16 (5′-GAG ACC GAT AGC GAA ACA AGT AC-3′) and T30 (5′-TGT TAG ACT CCT TGG TCC GTG-3′) (Harper & Saunders, Reference Harper and Saunders2001) and ITS2 rDNA region (ITS2), using primers 3s (5′-GGT ACC GGT GGA TCA CGT GGC TAG TG-3′) (Bowles et al., Reference Bowles, Hope, Tiu, Liu and McManus1993) and ITS2.2 (5′-CCT GGT TAG TTT CTT TTC CTC CGC-3′) (Cribb et al., Reference Cribb, Adlard and Bray1998). The 28S marker was selected for its previous use in heterophyid phylogenies (e.g. Thaenkham et al., Reference Thaenkham, Nawa, Blair and Pakdee2011). The ITS2 region was selected because although not suitable at family-level analyses (Nolan & Cribb, Reference Nolan and Cribb2005; Thaenkham et al., Reference Thaenkham, Blair, Nawa and Waikagul2012), it has been useful for distinguishing species and population-level diagnostics between digenean species (e.g. Nolan & Cribb, Reference Nolan and Cribb2005). The amplification protocol for 28S consisted of an initial denaturation phase (2 min at 94°C); 38 cycles of denaturation (30 s at 94°C), annealing (30 s at 50°C), extension (2 min at 72°C) and a 7 min final extension (72°C). The amplification protocol for ITS2 region consisted of an initial denaturation phase (3 min at 95°C); one cycle of annealing (2 min at 45°C) and extension (1 min 30 s at 72°C); followed by four cycles of denaturation (45 s at 95°C), annealing (45 s at 50°C) and extension (1 min 30 s at 72°C); followed by 30 cycles of denaturation (20 s at 95°C), annealing (20 s at 52°C) and extension (1 min 30 s at 72°C) and a 5 min final extension (72°C). All PCR products were cleaned using EXOSAP-IT™ Express PCR Product Cleanup Reagent (USB Corporation, Cleveland, OH, USA), following manufacturer's instructions. Sanger sequencing by capillary electrophoresis was performed by the Genetic Analysis Service, Department of Anatomy, University of Otago (Dunedin, New Zealand).
All sequences were imported into Geneious v8.1.9 (Kearse et al., Reference Kearse, Moir and Wilson2012), trimmed using the trim function with default parameters and manually edited for incorrect or ambiguous base calls. The generated sequences were aligned together with published heterophyid sequences, as found on GenBank. The datasets were aligned using MAFFT algorithm implemented in Geneious v8.1.9 (auto algorithm using default settings). For outgroups, an echinostomatid outgroup was selected based on previous phylogenetic studies of Heterophyidae (e.g. Thaenkham et al., Reference Thaenkham, Nawa, Blair and Pakdee2011) for the 28S dataset, and an opisthorchiid outgroup was used as one of the closest to the ITS2 heterophyid sequences. GenBank accession numbers of sequences used in each phylogenetic analysis are shown in tables 1 and 2, respectively. The cox1 sequence was used only to confirm identity with the Galactosomum sp. infecting Z. subcarinatus. Genetic divergences were calculated in MEGA v7 (Kumar et al., Reference Kumar, Stecher and Tamura2016) using uncorrected pairwise genetic distances and, as there was no genetic difference between the sequences from each different host, a single representative sequence of adult Galactosomum for 28S and ITS2 was included in the final datasets.
Table 1. Taxa included in the dataset for partial 28S phylogenetic analysis, including ID, GenBank accession numbers and references.

Table 2. Taxa included in the dataset for ITS2 phylogenetic analysis, including ID, GenBank accession numbers and references.

To infer the phylogenetic positioning of the new Galactosomum specimens within the Heterophyidae, Bayesian inference was conducted in MrBayes version 3.2.6 (Huelsenbeck & Ronquist, Reference Huelsenbeck and Ronquist2001) using the online interface, Cyberinfrastructure for Phylogenetic Research (CIPRES) Science Gateway (Miller et al., Reference Miller, Pfeiffer and Schwartz2010). The analyses used random starting trees for two runs each (one cold and three heated chains), employing a Markov Chain Monte Carlo approach for sampling the joint posterior probability distribution across 10,000,000 generations, sampling every 1000 generations. A heating chain value of 0.01 was selected and the first and last 25% of samples were discarded as burn-in. The resulting phylogenies were summarized in a 50% majority-rule consensus tree with clade credibility support values (Bayesian posterior probability (BPP)) and branch length information. BPP higher than 0.95 was considered strong support for nodal positions.
Results
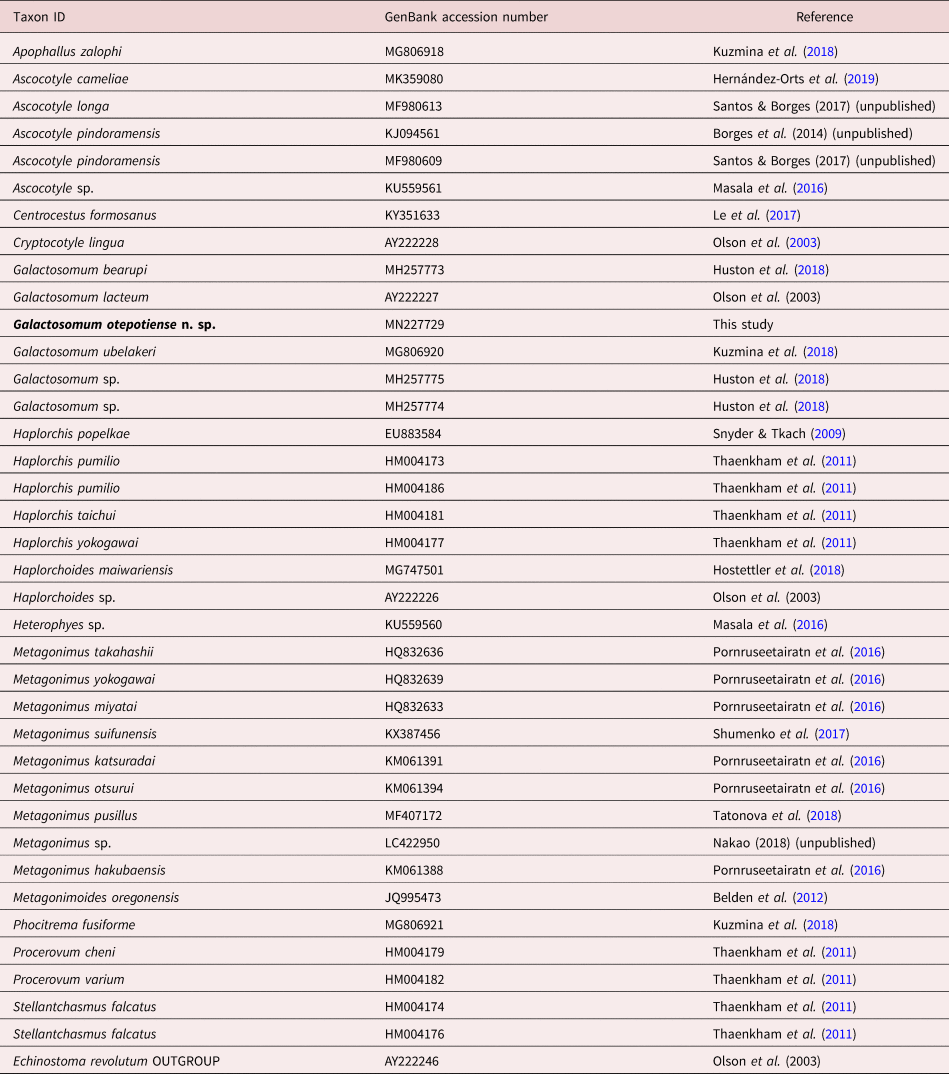
Galactosomum otepotiense n. sp. (figs 1 and 2; table 3)
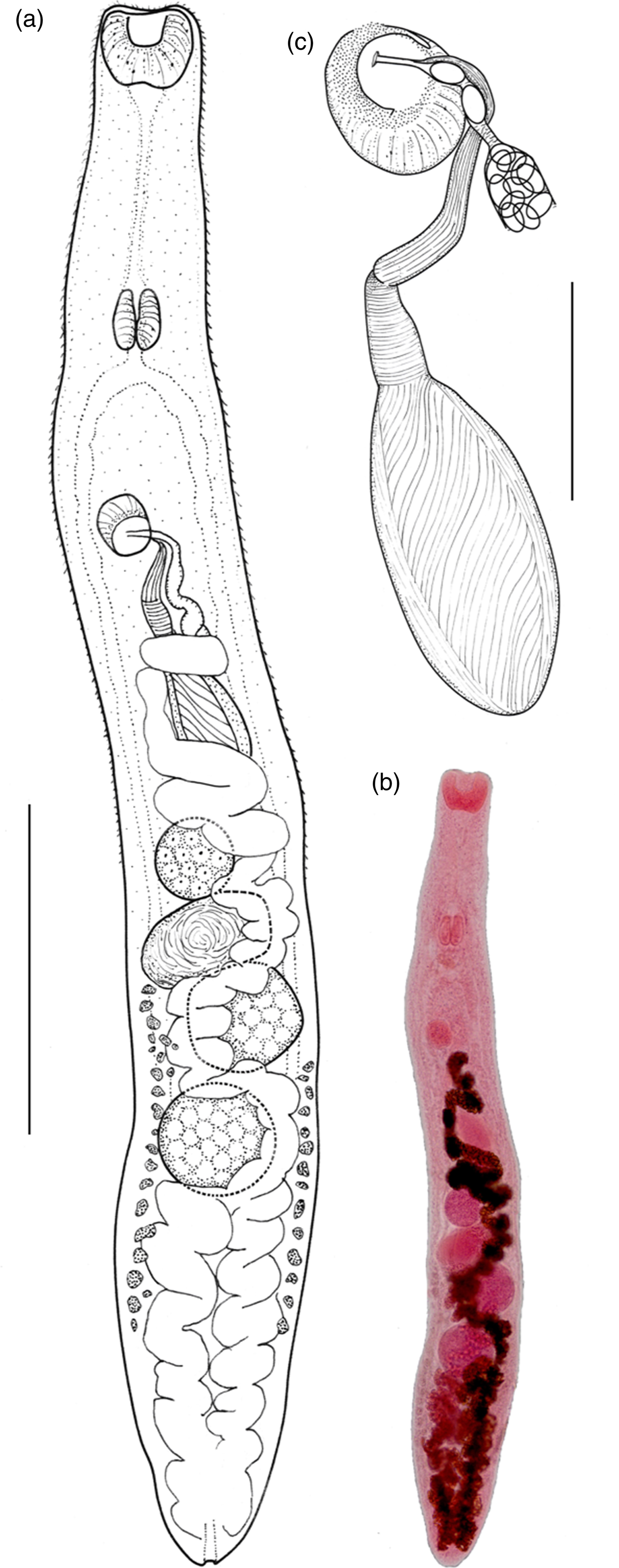
Fig. 1. Galactosomum otepotiense n. sp. (a) Holotype (Te Papa W.003498) ex Caspian tern; (b) photomicrograph of holotype; (c) ventrogenital complex. Scale bars: (a) 500 µm; (c) 100 µm.
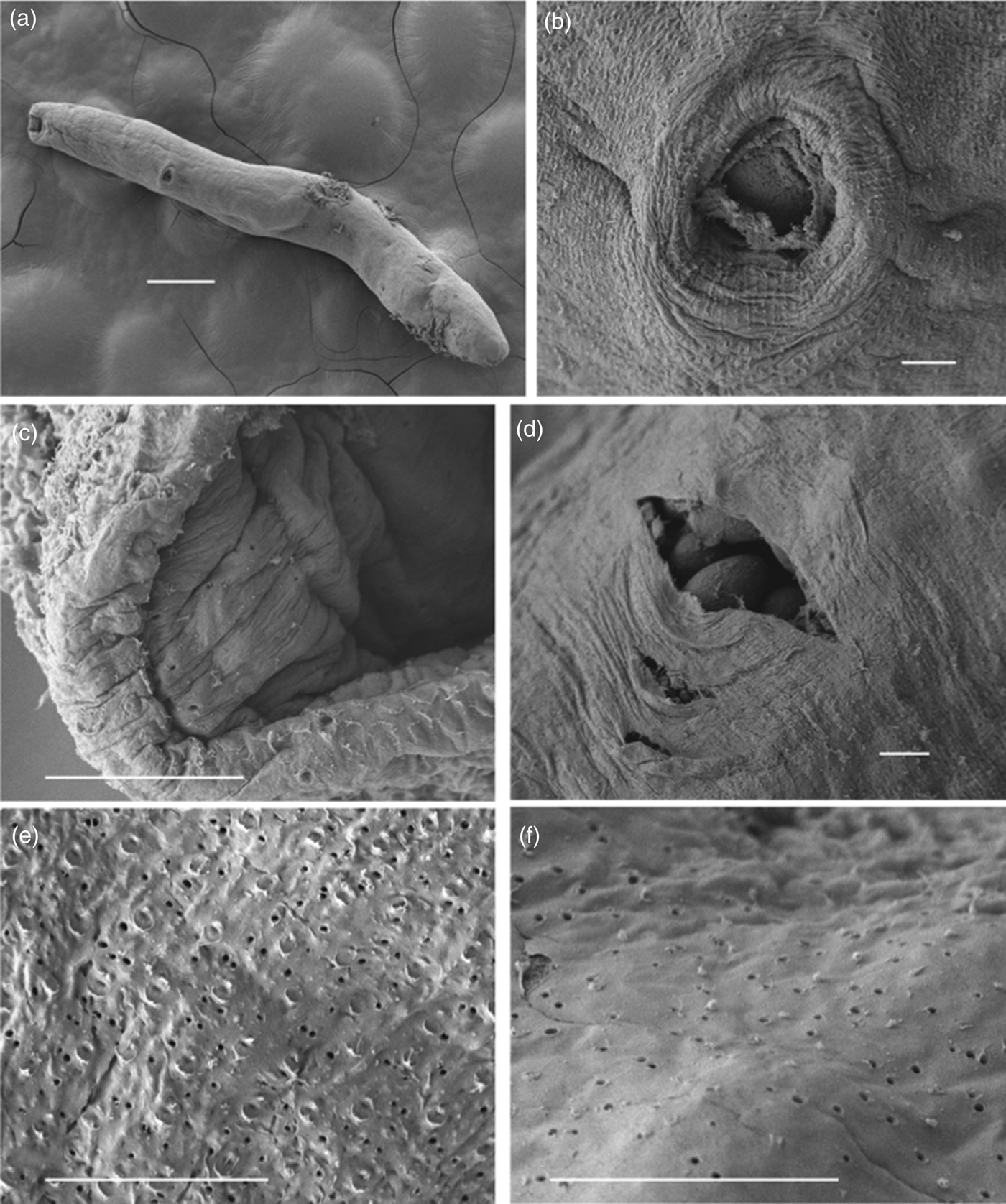
Fig. 2. Galactosomum otepotiense n. sp. ex Caspian tern, SEM photomicrographs. (a) Entire worm having lost tegumental spines; (b) ventrogenital complex; (c) mouth and oral sucker showing pores around lip and inside oral opening; (d) a split in hindbody wall showing mature eggs inside; (e) tegument at testicular level showing pores and scars of spine placements; (f) similar tegument from posterior end showing pores but no spine scars. Scale bars: (a) 200 µm; (b–f) 10 µm.
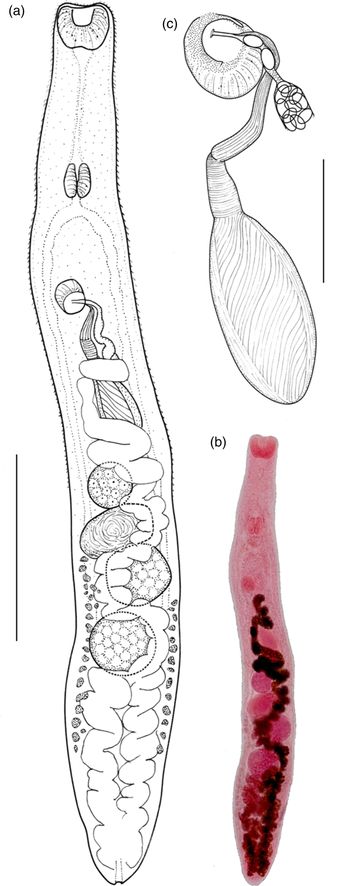
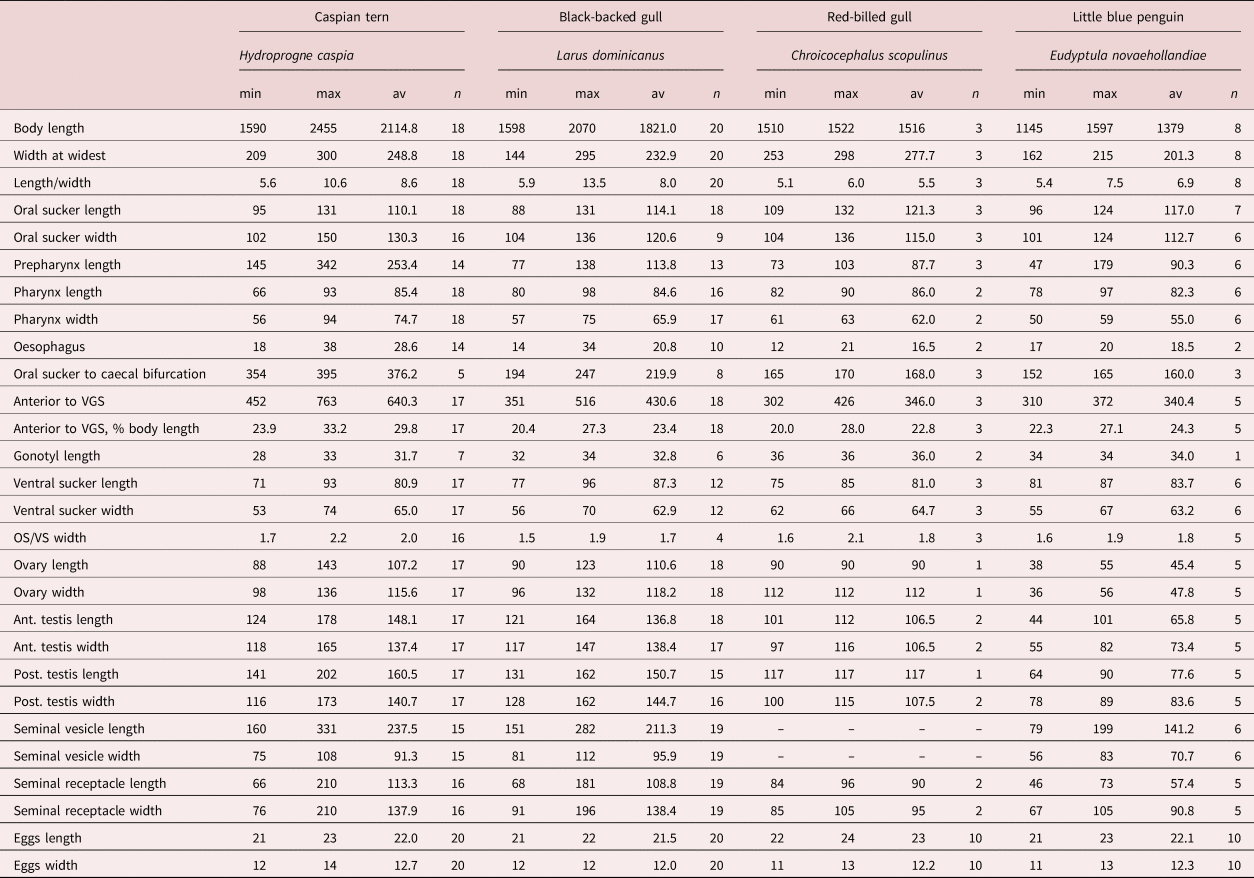
Table 3. Comparative metrics for Galactosomum otepotiense n. sp. from four different hosts.
min, minimum; max, maximum; av, average; VGS, ventrogenital sac; OS, oral sucker; VS, ventral sucker.
Synonyms. Galactosomum sp. adult ex little blue penguin from Tiritiri Matangi Island, of McKenna (Reference McKenna2009); Galactosomum sp. rediae and cercariae ex Z. subcarinatus from Otago Harbour, of Martorelli et al. (Reference Martorelli, Fredensborg, Leung and Poulin2008); Leung et al. (Reference Leung, Donald, Keeney, Koehler, Peoples and Poulin2009); Lloyd & Poulin (Reference Lloyd and Poulin2011); Studer & Poulin (Reference Studer and Poulin2012); MacLeod & Poulin (Reference MacLeod and Poulin2015); Guilloteau et al. (Reference Guilloteau, Poulin and MacLeod2016); Lawrence & Poulin (Reference Lawrence and Poulin2016).
Description
Based on 18 specimens from Caspian terns: Heterophyidae Odhner, 1914; Galactosominae Looss, 1899. Body elongate, more-or-less cylindrical, lateral margins parallel or slightly wider in hindbody; 2.11 (1.59–2.46) mm long by 249 (209–300) wide at widest point. Tegument spinose, although spines often lost; spines c. 7 µm long around anterior end, becoming smaller posteriorly, extending approximately to level of first testis; density c. 115 per 50 µm2 on forebody. Oral sucker subterminal, 110 (95–131) long by 130 (102–150) wide; tegumental spines surrounding edges of sucker. Prepharynx long, 253 (145–342) in length. Pharynx 85 (66–93) long by 75 (56–94) wide. Oesophagus short, 29 (18–38) in length. Caecal bifurcation 376 (354–395) from oral sucker; caeca extend almost to posterior extremity, usually longer on right side, usually obscured by eggs. Pigment granules scattered in area between ventrogenital complex and oral sucker. Ventrogenital sac median or slightly dextral, 24–33% distant from anterior end, unarmed, without lateral pocket; tegumental spines missing in area around sac mouth. Ventral sucker dextral, often overlapping right branch of caeca; oval, 81 (71–93) by 65 (53–74), long axis oblique, tilted posteriorly on the left, lip enlarged ventrally on right, armed with complete circle of c. 340 min spines, ten rows wide on raised dextral lip, narrowing to 3–5 rows on sinistral lip; widest part of band c. 10 µm. Ratio oral sucker to ventral sucker (widths) 1:2.0 (1:1.7–2.2). Gonotyl unarmed, roughly circular in outline, axis oblique, 31 (28–33) long, arises sinistrally and posteriorly, overlies ventral sucker ventrally; traversed by genital atrium, genital pore a subterminal transverse slit or circular. Seminal vesicle fusiform, single-chambered, unconstricted, 237 (160–331) by 91 (75–108), with thick wall of diagonal fibres, except at anterior end where muscle fibres are strongly circular; terminal papilla not observed. Prostatic ejaculatory duct elongate, with conspicuous outer longitudinal muscle-fibres, at junction with seminal vesicle sharply twisted ventrally. Ejaculatory duct opens into genital atrium dorsal to uterus; muscular papilla and prostatic gland cell bodies not observed. Testes 2, entire, usually tandem, occasionally slightly offset, in which case anterior, located slightly to left of posterior; both testes posterior to midbody; round to transverse or longitudinal oval, anterior testis 148 (124–178) long by 137 (118–165) wide, posterior testis 161 (141–202) long by 141 (116–173) wide. Ovary rounded, dextral, located between posterior of seminal vesicle and anterior of seminal receptacle, 107 (88–143) by 116 (98–136). Seminal receptacle very large and circular when full of sperm, transversely oval when not full; contiguous with ovary and anterior testis when full, with gaps between when not full, 113 (66–201) by 138 (76–210). Course of uterus typical (see Pearson, Reference Pearson1973, p. 351), except that loops of ascending arm lie ventral to seminal vesicle in cylindrical specimens. Vitellaria follicular; reaching anteriorly to level of seminal receptacle or ovary, posteriorly about level with ends of caeca; right and left fields often unequal in length; rosettes discernible in less mature specimens, distributed irregularly over ventral side of ovary to posterior testis. Eggs 22 (21–23) by 13 (12–14). Excretory pore terminal; excretory bladder does not reach posterior margin of posterior testis.
Taxonomic summary
Type host. Caspian tern, H. caspia (Pallas, 1770) (Charadriiformes: Sternidae).
Other hosts. Red-billed gull, C. scopulinus (Forster, 1844) (Charadriiformes: Laridae); southern black-backed gull, L. dominicanus Lichtenstein, 1823 (Charadriiformes: Laridae); little blue penguin, E. novaehollandiae (Stephens, 1826) (Sphenisciformes: Spheniscidae).
Site of infection in definitive host. Intestine.
Prevalence in definitive hosts. 2/2 Caspian terns (100%); 4/30 red-billed gulls (13%); 1/20 black-backed gulls (5%); 3/35 little blue penguins (9%).
Intensity in definitive hosts. Caspian terns, nine and 50; red-billed gulls, 1–8; black-backed gull, 20; little blue penguins, 15–40.
Intermediate hosts. Marine intertidal mud whelk, Z. subcarinatus (Sowerby, 1855) (Prosobranchia: Batillariidae).
Type locality. Portobello Bay, Otago Harbour, South Island, New Zealand (45°52′S, 107°42′E).
Other localities. Hampden Beach, Otago (45°19′S, 170°49′E); Dunedin, Otago (45°52′S, 170°30′); Aramoana, Otago (45°46′S, 170°40′E).
Deposited specimens. Museum of New Zealand Te Papa Tongarewa, holotype W.003499, paratypes W.003499–W.003502; Otago Museum, paratypes IV107611–IV107616.
Zoobank registration. urn:lsid:zoobank.org:act:ED1CB047-8272-4F77-B0D3-E968C794D491.
Etymology. The species name is inspired by the Maori name for Dunedin, Ōtepoti, and is in recognition of the Dunedin Wildlife Hospital, which plays such an invaluable role in the welfare and conservation of New Zealand birds, and the staff of which were kind enough to donate their dead birds to the cause of parasitology.
Remarks
In having an elongate, spinose body, an oval, dextral ventral sucker with spines on external face, a bipartite seminal vesicle, expulsor, permanent ventro-genital sac with unspined gonotyl bearing the genital pore and a tubular excretory vesicle, the specimens described herein clearly belong to the genus Galactosomum (sPearson, Reference Pearson1964, Reference Pearson, Bray, Gibson and Jones2008).
Pearson (Reference Pearson1973) assigned the species of Galactosomum to four morphological groups. Galactosomum otepotiense n. sp. morphologically falls into his ‘bearupi-group’, in having a one-chambered seminal vesicle with an additional layer of diagonal fibres over the major proximal portion and prominent circular fibres in the distal portion, and a short excretory bladder. There are ten species in this group, of which eight (G. darbyi Price, 1934; G. dollfusi Pearson, 1973; G. fregatae Prudhoe, 1949; G. johnsoni Price, 1934; G. puffini Yamaguti, 1941; G. timondavidi Pearson & Prévot, 1971 G. ussuriense Oshmarin, 1963; and G. yehi (Dissanaike, 1961)) are distinguishable from G. otepotiense n. sp. by a number of features, including the form and spination of the gonotyl and ventrogenital sac, and the form of the seminal vesicle. In particular, all of these species have zero, two or three patches of spines on the ventral sucker, as opposed to G. otepotiense n. sp., which has a single circle of spines. The new species is unique among this group in having tegumental spines from the anterior tip to the level of the testes, as opposed to anterior scales grading to posterior spines.
The new species is morphologically closest to G. bearupi and G. angelae Pearson, 1973, with which it shares a symmetrical ventral sucker armed with a complete circle of minute spines, the absence of a lateral pocket, similar distribution of vitellaria and a one-chambered seminal vesicle in two parts, the smaller with circular muscle fibres and the larger with spiral outer fibres. It differs from G. bearupi in having a larger, oval, ventral sucker (length 81(71–93)–versus–51(43–56)) and a larger pharynx (length 85(66–93)–v–51(45–60)). In addition, G. bearupi has ventral sucker spines in a circular band of uniform number (8–9 rows), whereas that in G. otepotiense n. sp. is narrower on the right (reduced to 2–3 rows). The new species differs from G. angelae in having a smaller gonotyl (length 32(28–33)–v–53(39–68)), and a narrower ventral sucker (width 65(53–74)–v–113(110–120)). Where G. angelae has no scales at the anterior tip but a row of three scales on the anterior margin of the oral sucker, G. otepotiense n. sp. has spines that extend to the anterior tip of the worm. Additionally, G. otepotiense n. sp. can be distinguished from these two similar species by overall size (maximum length 2455–v–1900 and 1740), by the long forebody, exemplified by the very long prepharynx (2.2–3.7 times length of pharynx, as opposed to the same length as pharynx), and by the size of the ovary and testes, which are all larger in the new species than in G. bearupi and G. angelae. The genetic evidence confirms that G. otepotiense n. sp. is not conspecific with G. bearupi, but no genetic data are available for G. angelae.
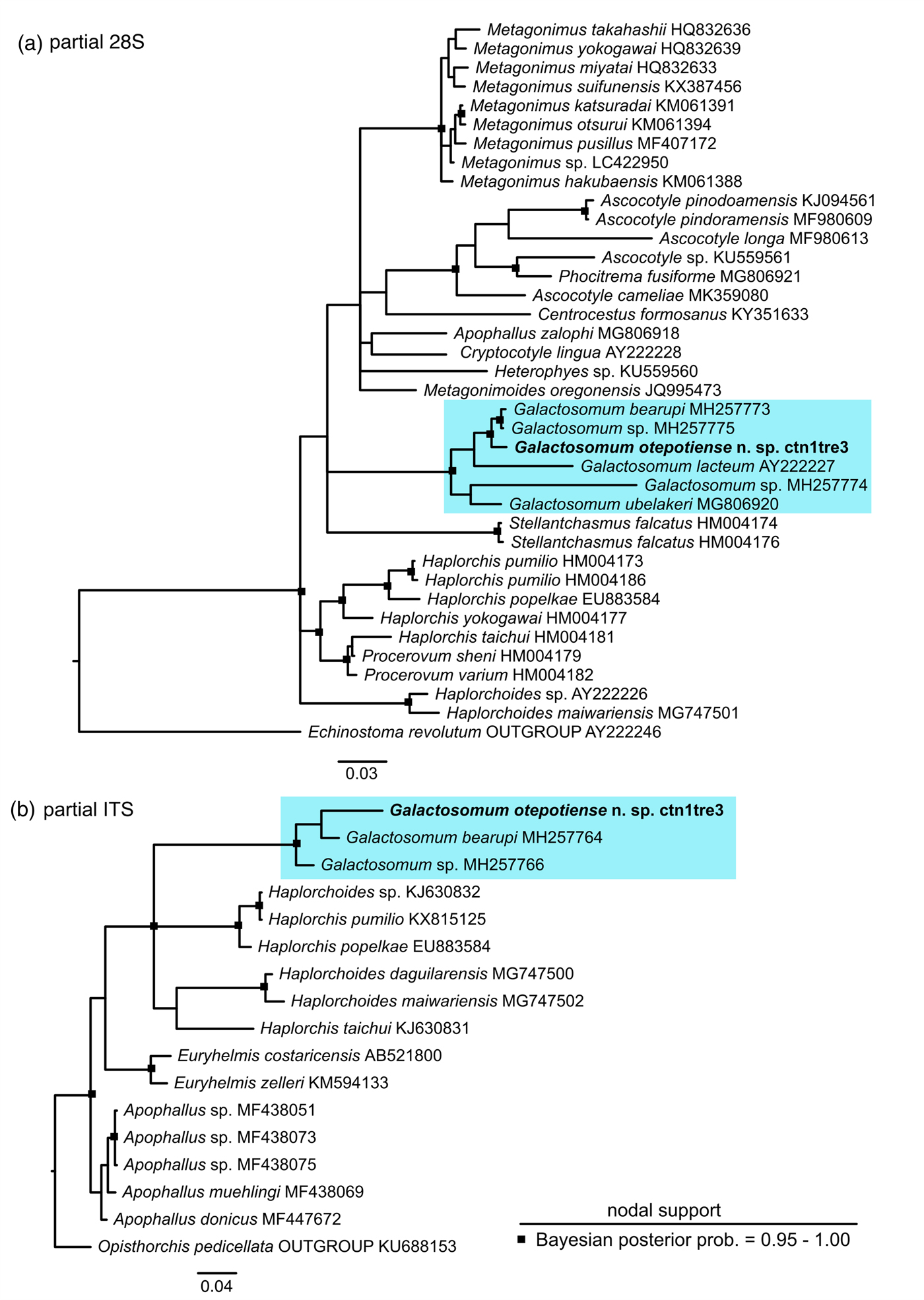
The specimens recovered from all four piscivorous birds were found to be genetically identical for the partial 28S and ITS2 markers, confirming that they were the same species. In addition, a short (230 bp), but variable, region of the cox1 was sequenced (GenBank accession number MN233790), which confirmed identity with the cercarial and redial stages of Galactosomum sp. from Z. subcarinatus (sequence provided by Leung et al., Reference Leung, Donald, Keeney, Koehler, Peoples and Poulin2009; Genbank accession number FJ765489). A phylogeny inferred using all available heterophyid ITS2 sequences confirmed the close relationship between the specimens herein and G. bearupi from Australia (fig. 3), with strong support (BPP > 0.95). A larger number of heterophyid sequences were available for 28S, and the phylogeny placed the new species as sister to both G. bearupi and a species of Galactosomum that inhabits the same snail in the Great Barrier Reef (fig. 3) (BPP > 0.95), with G. lacteum (from Ukraine) as sister to these species. Genetic divergence was relatively low between G. otepotiense n. sp. and both closely related Australian species in the ‘bearupi-group’ (uncorrected p-distance 1.6–1.8%) compared to divergence between G. otepotiense n. sp and the other three representatives of Galactosomum (uncorrected p-distance 5.5–11.6%) for the 28S marker.
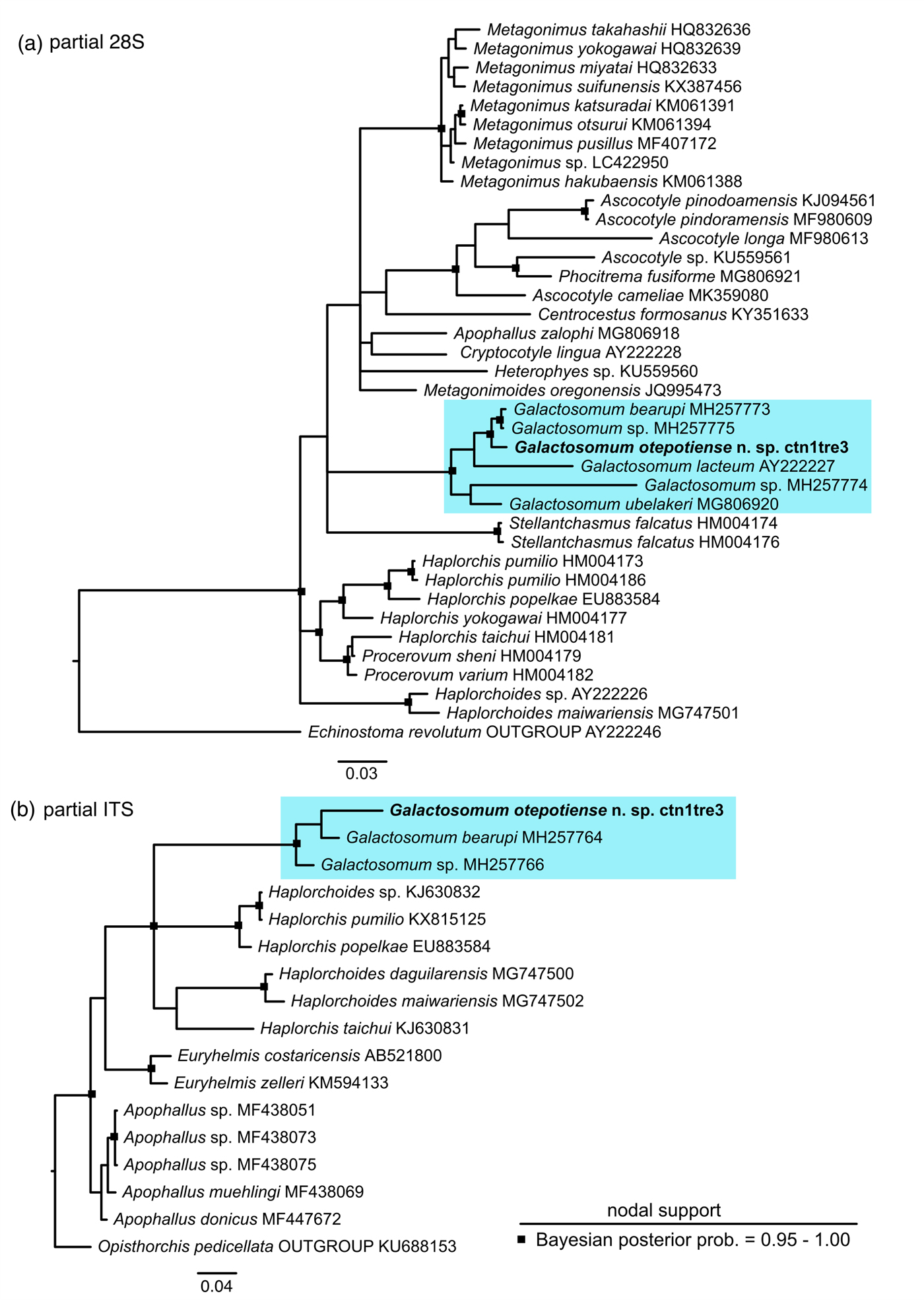
Fig. 3. Bayesian 50% majority-rule inference tree for (a) partial 28S dataset and (b) ITS2 dataset. Scalebars indicate the number of substitutions per site. Outgroup for the 28S phylogeny is Echinostoma revolutum (Echinostomatidae) and outgroup for the ITS2 phylogeny is Opisthorchis pedicellata (Opisthorchiidae).
Discussion
Intermediate hosts
This study provides a morphological description of a new species of Galactosomum, G. otepotiense n. sp., and matches two lifecycle stages (adult and redia/cercaria) using molecular tools, following the approach encouraged by Blasco-Costa & Poulin (Reference Blasco-Costa and Poulin2017). The redial and cercarial stages of this parasite were described by Martorelli et al. (Reference Martorelli, Fredensborg, Leung and Poulin2008) and compared morphologically to other cercariae described in the literature, and those authors concluded that the cercaria from the intertidal mud snail, Z. subcarinatus, in New Zealand, was a close relative of G. bearupi. We have herein confirmed their speculation with both our molecular phylogeny and morphometric comparison of the adults. In other species of Galactosomum, the large, worm-like cercariae are actively ingested by a fish, where they encyst as metacercariae (Kearn, Reference Kearn1998). Although Z. subcarinatus is the only known first intermediate host for G. otepotiense n. sp. and it is unknown at this stage what fish species fill the role of second intermediate host, we have shown that at least four species of fish-eating bird act as definitive hosts (Caspian tern, red-billed gull, black-backed gull and little blue penguin). All four of these birds prey on small inshore fish species (New Zealand Birds Online: www.nzbirdsonline.org.nz). In the case of little blue penguins at Oamaru, Graham's gudgeon (Grahamichthys radiata) and slender sprat (Sprattus antipodum) are predominant in the diet (Fraser & Lalas, Reference Fraser and Lalas2004; Flemming et al., Reference Flemming, Lalas and van Heezik2013), and these fish species may be a good starting point in a search for the second intermediate host.
Comments on closely related species
In this study, both morphometric and DNA sequence data suggest that G. otepotiense n. sp. is most closely related to species from Australia. This pattern is seen in several species of parasitic helminth (Presswell et al., Reference Presswell, Poulin and Randhawa2012; Cribb et al., Reference Cribb, Miller, Bray and Cutmore2014b; Georgieva et al., Reference Georgieva, Blasco-Costa and Kostadinova2017; Huston et al., Reference Huston, Cutmore and Cribb2018), and is perhaps unsurprising when considering that Australia is the closest large land mass and much of New Zealand's marine avifauna has evolved from Australian immigrant species or vice versa (e.g. Diamond, Reference Diamond1984; Given et al., Reference Given, Mills and Baker2005). It is interesting that the two closest species to G. otepotiense n. sp. (G. angelae and G. bearupi) have also been described from Caspian terns in Australia; indeed, the Caspian tern is type host for all three species. The Australian G. angelae has also been found in the little blue penguin and both Australian Galactosomum species have been found in the silver gull (Chroicocephalus novaehollandiae (Stephens, 1828)), which is sister species to the New Zealand red-billed gull (Given et al., Reference Given, Mills and Baker2005). Two further species of Galactosomum are found on the Queensland coast, G. ussuriense Oshmarin, 1963 and G. renincola Pearson, 1973, the former sharing a host with G. bearupi, G. angelae and G. otepotiense n. sp. (Caspian tern), and all of which have hosts with a diet of small inshore fish. With several species using the same hosts, there has obviously been strong selection pressure for speciation in this genus. It is not within the remit of this study, but elucidating the life cycles of each of the Australasian species and the historical biogeography of their hosts, may clarify the driving force for this level of speciation.
Although we have been able clearly to distinguish G. otepotiense n. sp. from G. bearupi, we do not have genetic data for G. angelae, which appears morphologically close to the new species. A combination of morphological similarity and host preference, added to the fact that G. angelae was described from South Australia, leads us to speculate that the two species will be found to be very closely related genetically. Genetic data for G. angelae, and a description of the cercariae, ideally genetically identified, will demonstrate the relationships between these three closely related Australasian species.
Variability in different hosts
We have chosen H. caspia as the type host for this species because similar members of the genus (G. angelae, G. bearupi and G. ussuriense) are found in the same host in the Australasian region, and because the specimens recovered from terns were in the best condition. In addition, one of the Caspian terns was found at Portobello Bay, the locality of the larval stages from Z. subcarinatus. We note, however, that there appears to be considerable morphological variability between the specimens found in the four different bird hosts (fig. 4). A range of comparative measurements is given in table 3 to illustrate the variability between specimens from different hosts. In particular, those from gulls appeared more flattened than cylindrical when recovered, and the length of the forebody (exemplified by the prepharynx length and distance between the anterior tip and the ventrogenital complex) was much shorter than in the specimens from the Caspian tern. The little blue penguin parasites were considerably smaller, although all were ovigerous (considered ‘mature’), and in body shape and proportions resembled the gull specimens more than those from the tern (see fig. 4). These differences appeared to be consistent within the different hosts, rather than an artefact induced by the state of preservation of each individual bird. It is true that some birds were in better condition of preservation than others, and the length of time between death and freezing is often a factor in the shape and condition of parasites recovered from their intestines. But, as worms from different individuals of the same host seem consistent, we suggest that this is an example of host-induced variability, where the worms are genetically identical in the genes sampled. Although there are plenty of examples in the literature of cryptic species of trematode (i.e. morphologically indistinguishable but genetically divergent) (e.g. León-Régagnon et al., Reference León-Régagnon, Brooks and Pérez-Ponce de León1999; Georgieva et al., Reference Georgieva, Selbach, Faltýnková, Soldánová, Sures, Skírnisson and Kostadinova2013; Cribb et al., Reference Cribb, Adlard, Bray, Sasal and Cutmore2014a), and reports of host-induced variability within species (e.g. Stunkard, Reference Stunkard1957; Blankespoor, Reference Blankespoor1974; Pérez Ponce de León, Reference Pérez Ponce de León1995), records of host-induced variability supported by proof of genetic identity are rarer (Cutmore et al., Reference Cutmore, Bennett and Cribb2010; Hildebrand et al., Reference Hildebrand, Adamczyk, Laskowski and Zaleśny2015). During this study we examined slides of G. angelae from five different host species, and found that they, too, showed considerable variability. These observations lend weight to the argument that apparent host specificity is not a reliable criterion with which to delineate a species, and that both morphological and molecular analyses should be used in combination when investigating parasites infecting multiple hosts.

Fig. 4. Galactosomum otepotiense n. sp. from four different hosts showing morphological variation: (a) Ex Caspian tern; (b) ex black-backed gull; (c) ex red-billed gull; (d) ex little blue penguin. Scale bar = 500 µm.
Comments on specimens
As Pearson (Reference Pearson1973) observed, we found that the tegumental spines were frequently lost after death, even on the most well-preserved specimens (measurements noted herein were taken from a few stained specimens that had retained their spines). The scanning electron micrograph shows that not a single tegumental spine seems to persist on the specimen from the Caspian tern, but we interpret the small tears in the outer tegument to be the scars of the spines, based on their distribution and frequency when compared to other specimens where the spines were in situ. Also visible in the SEMs are tegumental pores, or pits, that are arranged in apparently random fashion on the visible ventral surface. Such pores have been observed on the surface of other trematodes, and have been assumed to be secretory (Bakke, Reference Bakke1976; Smales & Blankespoor, Reference Smales and Blankespoor1984; Otubanjo, Reference Otubanjo1985; El Abdou et al., Reference El Abdou, Betalgy, Heckmann and Ashour2001).
Importance of Galactosomum species
Galactosomum spp. are potentially of future interest because, although they probably do little harm to their definitive hosts, the metacercarial stages of many species inhabit the optic lobe of the second intermediate host's brain, where they affect the behaviour of the fish, causing it to flip and whirl at the water surface, thus making the host highly visible to piscivorous birds, and more likely to be predated (Kimura & Endo, Reference Kimura and Endo1979; Bartoli & Boudouresque, Reference Bartoli and Boudouresque2007; Ogawa, Reference Ogawa2015). Concern has been expressed that the parasite, causing ‘trematode whirling disease’, could have a marked negative effect on the mariculture of food fish, as it has been found in farmed populations of Seriola quinqueradiata (Japanese amberjack), Takifugu rubripes (Japanese puffer fish), Oplegnathus fasciatus (barred knifefish) and Chrysophrys major (red seabream) (Kimura & Endo, Reference Kimura and Endo1979; Yasunaga, Reference Yasunaga1981; Ogawa, Reference Ogawa2015). Currently, aquaculture provides over half of all seafood consumed globally, and mariculture makes up one-third of this production (Liu et al., Reference Liu, Molina, Wilson and Halpern2018). With an ever-growing human population and ever-decreasing natural marine fish stock this can only increase, and any threat to production, including parasites, will need to be taken seriously.
Acknowledgements
The authors would like to thank Leslie Chisholm at South Australia Museum for the loan of specimens, Olivia McPherson for her help with bird dissections and Robert Poulin for his support. The authors would also like to thank the Dunedin Wildlife Hospital for their kind donation of bird specimens. We would also like to thank two anonymous reviewers for their helpful feedback.
Financial support
This work has been supported indirectly by the Marsden Fund (Royal Society of New Zealand: Grant No: UOO1718) and a Zoology Department PBRF Research Enhancement grant to Professor Robert Poulin.
Conflicts of interest
None.
Ethical standards
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional guides on the care and use of laboratory animals.