Introduction
The nematode genus Rhabdochona Railliet, 1916 comprises c. 100 species of intestinal parasites of freshwater fish in all zoogeographical regions except Antarctica (Moravec, Reference Moravec2010). In the Americas, the genus contains 21 species, although they do not comprise a monophyletic assemblage, and their presence in a wide variety of fish species suggests an evolutionary history with extensive ecological host extension and host-switching events (Mejía-Madrid et al., Reference Mejía-Madrid, Choudhury and Pérez-Ponce de León2007a, Reference Mejía-Madrid, Vázquez-Domínguez and Pérez-Ponce de Leónb). Most of the congeneric species are highly host specific, usually at the level of host family (Moravec et al., Reference Moravec2012). In Mexico, 12 species of Rhabdochona have been reported as parasites of freshwater fishes, i.e. R. acuminata Molin, 1860, R. ahuehuellensis Mejía-Madrid and Pérez-Ponce de León, 2003, R. canadensis Moravec and Arai, 1971, R. cascadilla Wigdor, 1918, R. guerreroensis Caspeta-Mandujano, Aguilar-Aguilar and Salgado-Maldonado, 2002, R. ictaluri Aguilar-Aguilar, Rosas-Valdez and Pérez-Ponce de León, 2010, R. kidderi Pearse, 1936, R. lichtenfelsi Sánchez-Álvarez, García-Prieto and Pérez-Ponce de León, 1998, R. mexicana Caspeta-Mandujano, Moravec and Salgado-Maldonado, 2000, R. ovifilamenta Weller, 1938, R. salgadoi Caspeta-Mandujano and Moravec, 2000, and R. xiphophori Caspeta-Mandujano, Moravec and Salgado-Maldonado, 2001 (Garrido-Olvera et al., Reference Garrido-Olvera, García-Prieto and Pérez-Ponce de León2006; Pérez-Ponce de León et al., Reference Pérez-Ponce de León2009, Reference Pérez-Ponce de León2010; Aguilar-Aguilar et al., Reference Aguilar-Aguilar, Rosas-Valdez and Pérez-Ponce de León2010; Moravec et al., Reference Moravec2012). Seven of the 12 species (58%) are endemic and specific to a particular group of freshwater fish. The presence of the remaining five species of Rhabdochona in freshwater fish of Mexico reflects the transitional nature of the area, as the country lies between the Nearctic and Neotropical biogeographical regions (Pérez-Ponce de León, Reference Pérez-Ponce de León2003; Pérez-Ponce de León and Choudhury, Reference Pérez-Ponce de León and Choudhury2005). For instance, R. canadensis, R. ovifilamenta and R. cascadilla are typically associated with Nearctic freshwater fish groups such as catostomids and cyprinids, and a few others (Hoffman, Reference Hoffman1999; Arai and Smith, Reference Arai and Smith2016); meanwhile, R. kidderi and R. acuminata are usually found in Neotropical fishes such as cichlids, heptapterids and characids (Moravec, Reference Moravec1998).
Rhabdochona mexicana is an endemic and highly host-specific species and is morphologically characterized by having 10 anteriorly directed teeth in the prostom, large spicule possessing a small bifurcation at its distal tip, short spicule without a barb at its distal tip, eggs with an irregular flocculent coating lacking filaments, bifurcate deirids, and a conical tail lacking a cuticular spike (Caspeta-Mandujano et al., Reference Caspeta-Mandujano, Moravec and Salgado-Maldonado2000). Rhabdochona mexicana was originally described from the intestine of two species of characids, the Mexican tetra, Astyanax mexicanus (De Filippi), and the banded tetra, A. fasciatus (Cuvier) [ = A. aeneus (Günther)], in four localities of the Panuco River drainage and four localities of the Balsas River drainage, in the Atlantic and Pacific Ocean slopes of Mexico, respectively (Caspeta-Mandujano et al., Reference Caspeta-Mandujano, Moravec and Salgado-Maldonado2000). Currently, the species is widely distributed, having been found in at least 20 localities across Mexico (Garrido-Olvera et al., Reference Garrido-Olvera, García-Prieto and Pérez-Ponce de León2006; Moravec et al., Reference Moravec2012). The presence of this host-specific nematode in separate river basins across Mexico, in close association with Astyanax spp., has resulted in allopatric distribution patterns, raising the possibility of genetic differentiation among populations irrespective of the fact that morphological characters seem to be conserved, with the concomitant possibility of uncovering a complex of cryptic species (Pérez-Ponce de León and Nadler, Reference Pérez-Ponce de León and Nadler2010; Nadler and Pérez-Ponce de León, Reference Nadler and Pérez-Ponce de León2011). In this study, we provide for the first time molecular data for individuals of R. mexicana from different populations across its distributional range; one mitochondrial and two nuclear genes were sequenced to search for the existence of cryptic species through a molecular prospecting approach (Blouin, Reference Blouin2002). The main objectives of this paper were (1) to explore the genetic diversity among specimens of R. mexicana obtained from different hydrological systems across Mexico and Guatemala, (2) to determine if R. mexicana represents a species complex, and (3) to combine information from molecular data and morphology to describe a new species of Rhabdochona.
Materials and methods
Sample collection and molecular methods
Nematodes were recovered from 15 populations of Astyanax spp. in 13 locations of Mexico and two of Guatemala (table 1, fig. 1). In addition, specimens of three other species of Rhabdochona and one species of Spinitectus were sampled from their freshwater fish hosts (table 1). Hosts were collected using seine nets and maintained alive until parasitological analyses. Individual fish were euthanized by putting them in ice water (4°C), dissected, internal organs obtained, placed in Petri dishes with 0.65% saline, and examined using a stereomicroscope. Nematodes were recovered from the intestine of their hosts; some were fixed in 100% ethanol for molecular analysis, and a few others in hot 4% formalin and stored in 70% ethanol for morphological studies. Genomic DNA was extracted from the mid-section of individual nematodes. Each worm was incubated for 8 h in a solution containing 10 mm Tris–HCl (pH 7.6), 20 mm NaCl, 100 mm Na2EDTA (pH 8.0), 1% Sarkosyl, and 0.1 mg/ml proteinase K. DNA was extracted with DNAzol reagent (Invitrogen) following the manufacturer's instructions. The anterior and posterior ends of each individual nematode (hologenophores sensu Pleijel et al., Reference Pleijel2008) were used to contrast the molecular data with the morphology of the paragenophores that were used for the species description. The mitochondrial gene cytochrome c oxidase subunit 1 (cox1) and two nuclear genes, 28S and 18S rRNA genes, were amplified for partial sequences using polymerase chain reaction (PCR) and the primers listed in table 2. The PCR reactions (25 μl) consisted of 2 μl of genomic DNA, 1 μl of each primer (10 pmol), 0.125 μl Taq DNA Polymerase (Vivantis), 2.5 μl dNTP (2 mm), 1.5 μl MgCl2, and 14.375 μl ddH2O. The PCR conditions were as follows: 94°C for 2 minutes; 30 cycles of 94°C for 1 minute (denaturalization), annealing temperature varied according to the primer combination (table 2), 72°C for 2 minutes (extension) and 72°C for 7 minutes (final extension). The amplicons were purified using ExoSAP-IT (Affymetrix). Sequencing reactions were performed using a BigDye Terminator Cycle Sequencing Kit (Applied Biosystems). Samples were sequenced on an ABI 3730 capillary DNA sequencer.

Fig. 1. Collecting sites of Rhabdochona from Astyanax mexicanus and A. aeneus. The locality code corresponds with table 1.
Table 1. Sampling localities, host species and nematode species. Locality code corresponds with those in fig. 1.

*Nematode sample size
Table 2. Sequences of primers used in this study.
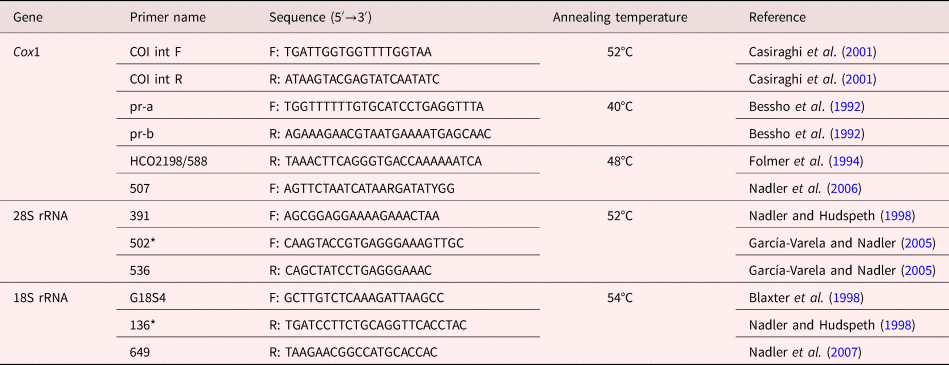
F: forward; R: reverse; Asterisk indicates primers used only in sequencing reaction
Phylogenetic analyses and genetic divergence
The forward and reverse sequences were assembled using Geneious v7 (Kearse et al., Reference Kearse2012), aligned with the algorithm implemented in Clustal Omega, with gaps treated as missing data (McWilliam et al., Reference McWilliam2013), and edited in Mesquite v3.10 (Maddison and Maddison, Reference Maddison and Maddison2016). For the cox1 dataset, sequences obtained for the same individual with several primer combinations were assembled to obtain the sequence for each individual. New sequences for the three molecular markers were generated for species of Rhabdochona, i.e. R. acuminata, R. salgadoi and R. canadensis. Also, a sequence of Spinitectus mexicanus Caspeta-Mandujano, Moravec and Salgado Maldonado, 2000 was generated and used as outgroup for rooting the trees, based on previous phylogenetic analyses that showed that Spinitectus and Rhabdochona were sister groups (Černotíková et al., Reference Černotíková, Horák and Moravec2011; Choudhury and Nadler, Reference Choudhury and Nadler2018). Sequences of other species of Rhabdochona available in GenBank were downloaded and used in phylogenetic analyses, particularly for the 18S rRNA gene (supplementary table S1). The cox1 dataset was trimmed after alignment. The best substitution model for the aligned datasets was obtained using jModelTest v2.1.7 (Posada, Reference Posada2008) and selected under Akaike Information Criterion (AIC). The model GTR+ G+I was used for Maximum Likelihood (ML) and Bayesian Inference (BI) analysis. The phylogenetic reconstructions were performed for each molecular marker separately, and for a dataset containing all genes (cox1 + 28S + 18S rRNA). ML analyses were performed using RaxML v1.5 (Stamatakis, Reference Stamatakis2014) with 10,000 bootstrap replicates. Bayesian inference analyses were performed in MrBayes v3.2.5 (Ronquist and Huelsenbeck, Reference Ronquist and Huelsenbeck2003), including two parallel runs for 10 million generations; the trees in each chain were sampled every 1000 generations, with a burn-in of 25%. The generated trees were visualized in FigTree v1.4.2 and edited in Illustrator. To assess the level of variation among isolates of each lineage, the uncorrected (p) pairwise genetic distance was calculated in MEGA v7 (Kumar et al., Reference Kumar, Stecher and Tamura2016) for the cox1 and the 28S rRNA datasets.
Morphological analysis
Photomicrographs and measurements of nematode specimens were obtained with an Olympus BX51 inverted microscope. The anterior and posterior regions of hologenophores preserved in ethanol, and complete individuals fixed in formalin, were mounted and cleared in glycerol-alcohol, and examined using differential interference contrast (DIC). Some individuals were prepared for scanning electron microscopy (SEM) observations; specimens were dehydrated in a graded series of ethanol, dried, and mounted on a strip of carbon conductive tape. Samples were sputter coated with gold and observed in a Hitachi Stereoscan Model SU1510 (Hitachi Ltd, Tokyo, Japan). Drawings were made with the aid of a drawing tube attached to a light microscope (Olympus BX51). Measurements are presented in micrometers, unless specified otherwise. Observations and data in this study were compared with the original description by Caspeta-Mandujano et al. (Reference Caspeta-Mandujano, Moravec and Salgado-Maldonado2000). Voucher specimens were deposited in Colección Nacional de Helmintos (CNHE), Universidad Nacional Autónoma de México, Mexico City, with the accession numbers CNHE 10866–10869 and CNHE 10966.
Results
Phylogenetic analyses
Thirty-eight individuals of R. mexicana were sequenced from 15 localities. In addition, we sequenced eight individuals from four other species of Rhabdochona and one individual of S. mexicanus used as outgroups. The datasets generated in our study were analysed separately for each molecular marker. A subsample of individuals was sequenced for the 18S rRNA gene, the less informative, and analysed through BI and ML (supplementary fig. S1). The 10 species of Rhabdochona available for the analysis appeared as a monophyletic assemblage with high posterior probability support (although bootstrap support value was moderate), and specimens of R. mexicana sequenced in our study were recovered forming two independent lineages, one of them including the type and host locality of R. mexicana. Furthermore, 28S rDNA sequences were analysed through BI and ML (supplementary fig. S2). As expected, the 28S rRNA tree seems to possess a higher phylogenetic signal than the 18S rRNA tree; sequenced individuals also formed two well-supported and reciprocally monophyletic lineages. Both ribosomal genes yielded a phylogenetic tree showing R. mexicana as paraphyletic.
The topologies derived from cox1 were the most well resolved and informative, contained the largest number of individuals, and were analysed through ML and BI inference methods; both recovered similar topologies with high bootstrap and posterior probability support values, respectively (fig. 2). This tree showed that R. mexicana consisted of two well-supported and reciprocally monophyletic lineages; Lineage I comprised three subgroups, each with high nodal support. One of them (subgroup 1) contained samples from seven localities, including some from the type host and locality (Astyanax mexicanus; Río Jalpan, Querétaro [RJ]) (see fig. 1); subgroup 2 included individuals from two geographically disjunct populations found in A. aeneus in Jalisco, Mexico (RRT) and in A. cf aeneus from Guatemala (RAG and RCG), in river basins draining to the Pacific Ocean slope. Individuals from subgroup 3 were collected only from the Mexican tetra, A. mexicanus in two localities of San Luis Potosí and Tamaulipas, in northern Mexico. As samples from the type locality were included in Lineage I, this was considered as R. mexicana sensu stricto. Lineage II was highly divergent and reciprocally monophyletic, was collected from A. aeneus in two localities of the state of Veracruz (PM and RPO) and one of Chiapas (ME), and was recovered as the sister group of Lineage I. In this phylogenetic tree, R. acuminata was recovered as the sister species of the two lineages of R. mexicana (fig. 2).
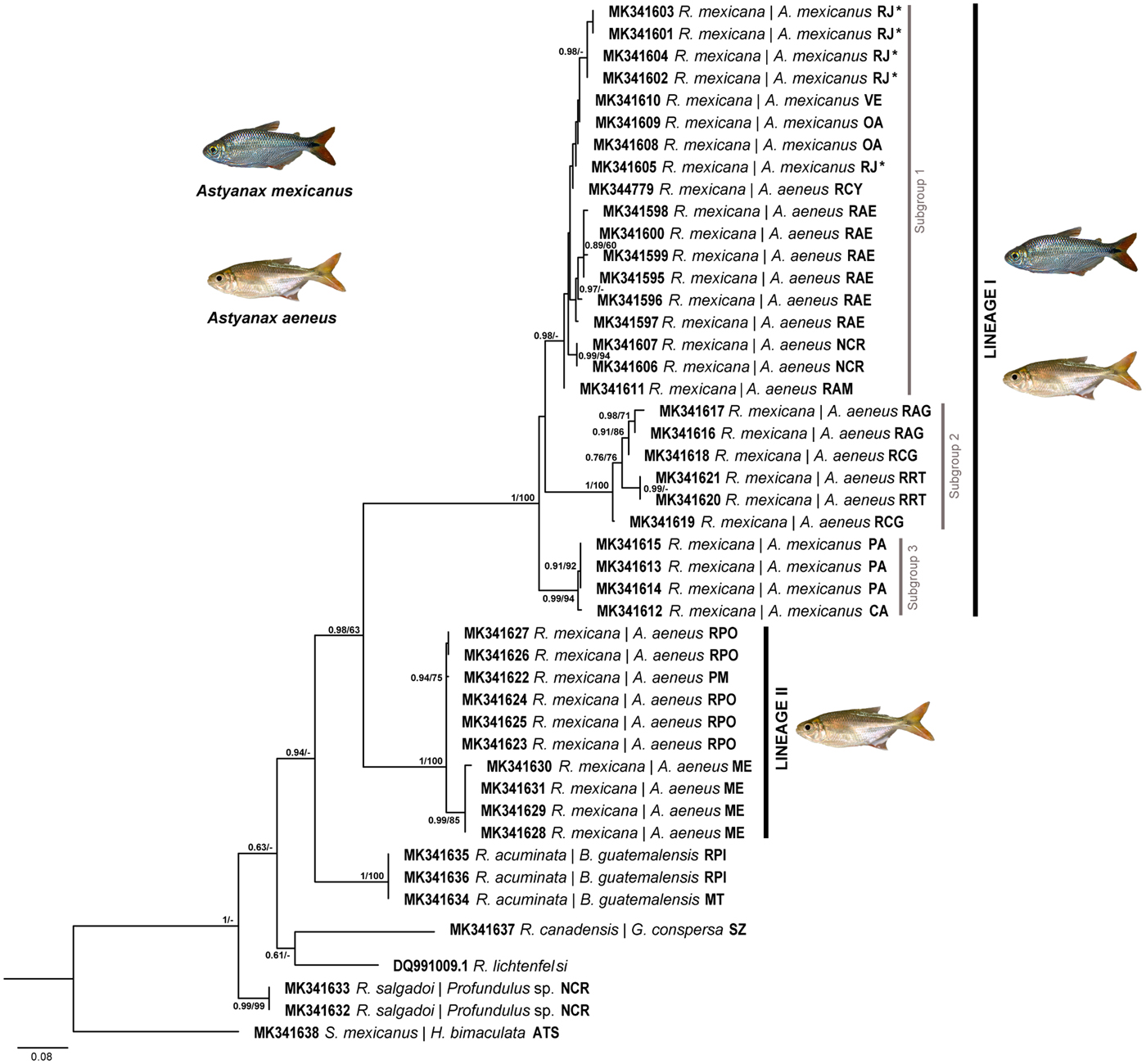
Fig. 2. Maximum likelihood phylogenetic reconstruction recovered from sequences of cox1. Posterior probability support value from the BI analysis and bootstrap support value (above 60%) are shown in the internodes. The scale bar refers to branch length. An asterisk indicates the type locality.
The concatenated tree from the three molecular markers (18S + 28S + cox1) recovered the same topology as the cox1 tree showing two reciprocally monophyletic lineages for R. mexicana, suggesting one of them as a separate species; relationships were supported by high bootstrap and posterior probability values (supplementary fig. S3). Also, in the concatenated tree, the sister group of the species complex of Rhabdochona in Astyanax was R. acuminata, a species recovered from the Macabi tetra, Brycon guatemalensis Regan.
Genetic divergence
The genetic divergence (uncorrected p-distance) among the three subgroups that comprise Lineage I varied from 4.51 to 6.24%. However, the genetic divergence between the three subgroups of Lineage I, and Lineage II varied between 10.94 and 11.51%. Furthermore, the divergence between Rhabdochona acuminata and the two lineages of R. mexicana varied between 8.32 and 12.73%. With respect to the remaining species of Rhabdochona depicted in fig. 2, the lineages of R. mexicana diverged by 8.32–14.43%. The cox1 intraspecific variation among individuals of each of the lineages and subgroups uncovered in the present study varied between 0.24 and 1.53% (table 3). Additionally, the genetic divergence for the 28S rRNA gene among the three subgroups that form Lineage I, and Lineage II varied from 0.23 to 6.99%, with the highest divergence also exhibited by Lineage II (table 3). Intraspecific divergence among individuals of the lineages of R. mexicana for the 28S rRNA gene was nil.
Table 3. Genetic divergence among species of Rhabdochona and S. mexicana (outgroup).
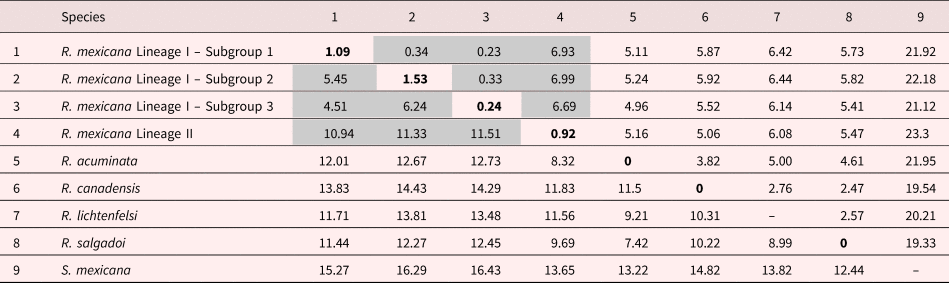
The numbers on the diagonal are the intraspecific genetic variation of each taxon. The numbers below the diagonal indicate the genetic distance calculated with the cox1 matrix, and those above the diagonal correspond to the genetic distance from the 28SrDNA matrix. The distances are shown as percentages.
Morphological analyses
The three subgroups of R. mexicana lineage I are reciprocally monophyletic and reached some level of genetic divergence; however, the three subgroups recovered within Lineage I are morphologically similar in that all possess 10 teeth in the prostom, bifurcate deirids, and eggs without filaments (see supplementary fig. S4). They represent a cryptic species complex. Instead, a detailed observation of the specimens of R. mexicana Lineage II allowed us to corroborate that they represented a different species, characterized by having a prostom with 14 teeth instead of 10, a simple deirid that is not bifurcate, and a large number of preanal papillae. The morphological description of Lineage II is presented next.
Family Rhabdochonidae Skrjabin, 1946
Genus Rhabdochona Railliet, 1916
Rhabdochona osorioi n. sp. (figs 3–5)
Description
(Based on the measurements of the holotype (CNHE 10866), the allotype (CNHE 10966), and 13 paratypes (CNHE 10867–10869); measurements taken from adult worms.) Medium-sized nematodes with smooth cuticle. Oval oral aperture surrounded by four cephalic papillae and two amphids; prostom with 14 teeth, four dorsal, four ventral and three lateral on either side (figs 3C and 4A). Simple and large deirids, sometimes asymmetrically disposed near cephalic end (figs 3D and 4C), usually not exceeding the end of prostom. Conical tail in both sexes without cuticular extensions (figs 3J, 4E and 5F).
Male (based on eight individuals). Body length 7.8–10.5 (9.5) mm, maximum width 163–200 (181). Prostom 23–34 (29) long, 15–23 (19) wide; length of vestibule including prostom 112–128 (117). Muscular oesophagus 221–0.342 (0.305) long; glandular oesophagus 1.8–3.2 (2.5) mm long. Nerve ring surrounding muscular oesophagous, 125–169 (150) from anterior end. Deirids and excretory pore 39–49 (42) and 202–249 (220) from anterior end, respectively (fig. 5A–D). Area rugosa absent. Fifteen pairs of preanal papillae. Subventral postcloacal papillae present, 7 + 7 and occasionally 7 + 6, plus one pair of lateral papillae between subventral papillae one and two (figs 3L and 4F). Terminal end of left spicule scoop-shaped, 414–451 (430) long (fig. 3G, H); shaft 251–273 (258) long, representing 60.2–60.9% of entire spicule length; right spicule 117–138 (125) long (fig. 3F). Tail 151–347 (272) long.
Female (based on seven individuals). Body length 11.6–27.3 (16.9) mm. Maximum width 154–327 (218). Prostom 24–41 (34) long, 21–28 (24) wide; length of vestibule including prostom 112–128 (117). Muscular oesophagus 173–436 (300) long. Nerve ring 147–227 (185) from anterior end; deirids and excretory pore 39–45 (42) and 173–240 (204) from the anterior end, respectively. Deirids large and simple. Vulva 4.6–14.1 (7.9) mm from anterior end (figs 3K and 5E). Eggs numerous, with no filaments (fig. 4D). Tail 218–364 (311) long (fig. 4E). Fourth-stage larva (based on two individuals) (fig. 3E). Body length 3.2–3.7 mm, maximum width 81–83. Prostom 15 long, 8–10 wide: length of vestibule including prostom 84–85. Muscular oesophagous 180–206 long, maximum width 23. Glandular oesophagous 1.1–1.3 mm long, 49–54 wide. Deirids, nerve ring and excretory pore 17, 115–121 and 154, respectively from anterior end. Tail conical 112–121 long (figs 3I and 5H). Vulva opening not observed.

Fig. 3. Line drawings of Rhabdochona osorioi n. sp. (A) Lateral view from anterior end, (B) lateral view of prostom from an adult, (C) apical view of the anterior end, (D) deirid, (E) anterior end of a fourth-stage larva, lateral view, (F) right spicule, (G) distal termination of left spicule, (H) complete left spicule, (I) tail of fourth-stage larva, (J) tail of female, lateral view, (K) vulvar opening of gravid female, lateral view, and (L) posterior end of male, lateral view.

Fig. 4. SEM microphotographs of Rhabdochona osorioi n. sp. (A) Apical view from anterior extremity, (B) lateral view from anterior end, (C) deirid, (D) egg, (E) posterior end of female, and (F) posterior end of body, lateral view. Abbreviations: p, papillae; a, amphid.
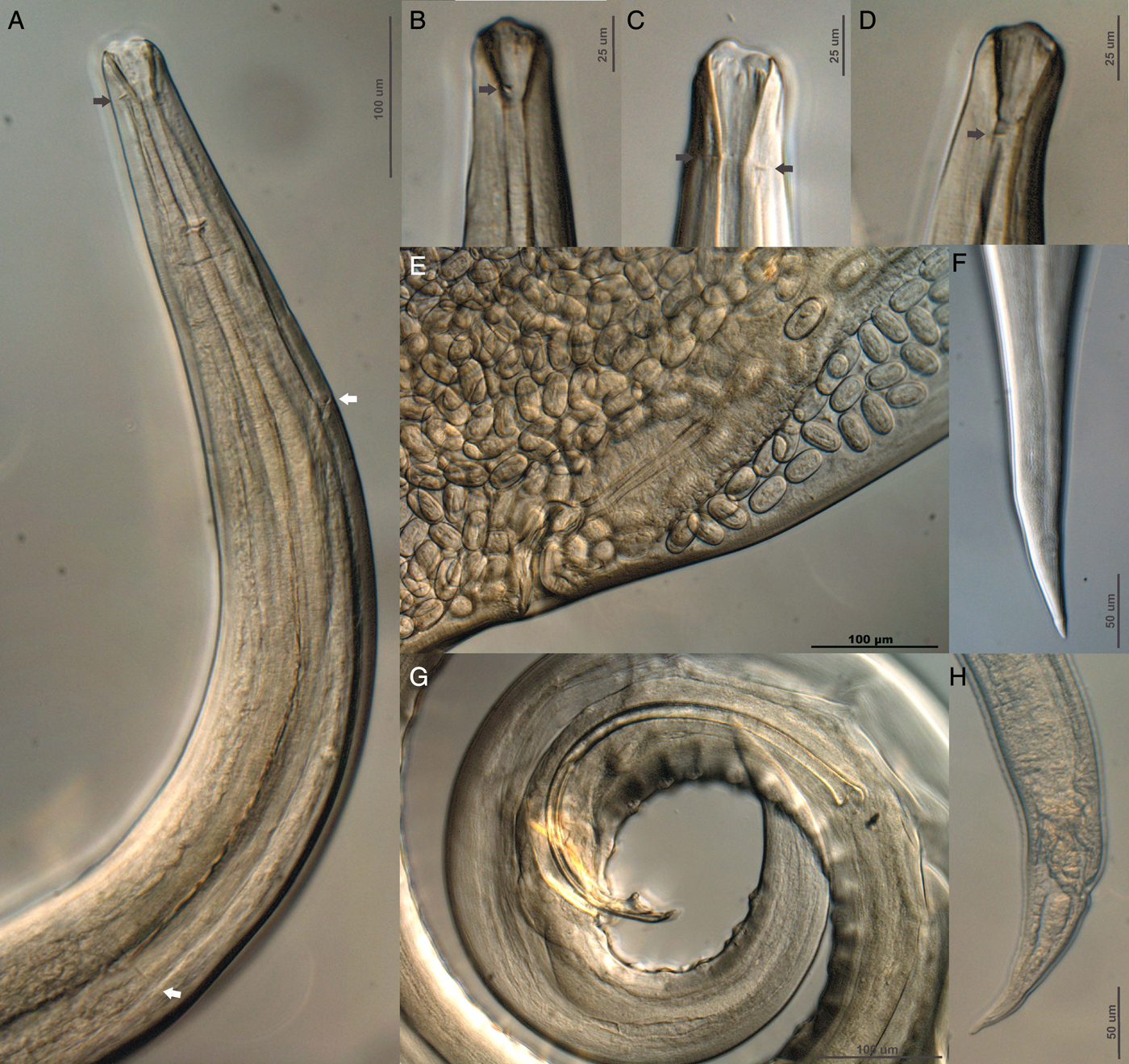
Fig. 5. Differential interference contrast microphotographs of Rhabdochona osorioi n. sp. (A) Lateral view of anterior end (black arrow indicates the position of the deirids, white arrow indicates the division of the muscular and glandular oesophagous), (B, C and D) lateral view of anterior end of different male specimens (black arrow indicates the position of deirids), (E) vulvar opening, lateral view, (F) posterior end of female, (G) male posterior end, lateral view, and (H) tail of fourth-stage larva, lateral view.
Taxonomic summary
Type host. Astyanax aeneus (Günther, 1860).
Type locality. Río Paso de Ovejas, Pueblo el Crucero, Veracruz, Mexico (19°19′1.35″N, 96°32′11.59″W).
Other localities. Atoyac, Paso de Macho, Veracruz (18°58′18.80″N, 96°43′53.32″W); and Metzabok, Chiapas (17°7′3.41″N, 91°37′54.61″W).
Type material. CNHE 10866 (holotype); CNHE 10966 (allotype); CNHE 10867–10869 (paratypes).
ZooBank registration. To comply with the regulations set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN, 2012), details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) for Rhabdochona osorioi n. sp. is urn:lsid:zoobank.org:act: 1D9F9E2A-371E-4028-A9EF-00272EF15059.
Etymology. The epithet osorioi is for Professor David Osorio in recognition of his long contribution (40 years!) in describing the nematode fauna of Mexican vertebrate wildlife, and training students in nematode taxonomy.
Representative DNA sequences in GenBank. MK341595–MK341638, MK344779 (cox1), MK341639–MK341652 (18S rRNA), MK341653–MK341687 (28S rRNA).
Taxonomic remarks
Rhabdochona osorioi n. sp. is diagnosed by having simple and large deirids, 14 anterior prostomal teeth and by the number and arrangement of papillae in males. This combination of characters clearly sets it apart from R. mexicana. In the Americas, nine Rhabdochona species possesses 14 teeth in the prostom: seven in the Nearctic biogeographical region (i.e. R. cascadilla Wigdor, 1918; R. cotti Gustafson, 1949; R. decaturensis Gustafson, 1949; R. milleri Choquette, 1951; R. canadensis Moravec et Arai, 1971; R. catostomi Kayton, Kritsky et Tobias, 1979; and R. ictaluri Aguilar-Aguilar, Rosas-Valdez and Pérez-Ponce de León, 2010) and two in the Neotropical biogeographical region (i.e. R. acuminata Molin, 1860 and R. kidderi Pearse, 1936). Rhabdochona osorioi n. sp. is the 10th species in the Americas with 14 teeth. Even though Mejía-Madrid et al. (Reference Mejía-Madrid, Choudhury and Pérez-Ponce de León2007a) argued that the species of Rhabdochona could not be grouped consistently by number of teeth in the prostom, we compared the new species with all the species of the Americas with the same number of teeth. Rhabdochona osorioi n. sp. differs from four of the Nearctic species (i.e. R. canadensis, R. milleri, R. catostomi and R. cotti) in having eggs with no filaments or any other ornamentation. The new species differs also from R. decaturensis, a parasite of the freshwater drum, Aplodinotus gruniens Rafinesque, 1819, in the USA and Canada, and from R. cascadilla, a parasite of several fish species across the USA and Canada, in that the eggs of these two species are covered with a thin gelatinous layer, although this character might be overlooked during processing; additionally, the new species differ from these two species by having a larger number of preanal papillae (see Gustafson, Reference Gustafson1949). From R. ictaluri, a species described from ictalurid catfishes in northern Mexico (Aguilar-Aguilar et al., Reference Aguilar-Aguilar, Rosas-Valdez and Pérez-Ponce de León2010), the new species differs by having a simple deirid, and a prostom without basal teeth. In R. ictaluri deirids are bifurcated and the prostom possesses six basal teeth.
Rhabdochona osorioi n. sp. most closely resembles the two Neotropical Rhabdochona species (R. kidderi and R. acuminata) in having 14 teeth in the prostom and eggs with smooth surface; however, it can easily be differentiated from R. kidderi because in this species deirids are bifurcate and males possess 5–7 pairs of preanal papillae. Instead, the new species possesses simple deirids, and 15 pairs of preanal papillae in males. Even though R. kidderi and the new species are sympatrically distributed in southern Mexico, host associations are different; the new species is found only in the characid Astyanax aeneus, whereas adults of R. kidderi are found in different species of cichlids and in heptapterids (Garrido-Olvera et al., Reference Garrido-Olvera, García-Prieto and Pérez-Ponce de León2006). Rhabdochona acuminata was recovered in our analyses as the sister species of the R. mexicana species complex and R. osorioi n. sp. (fig. 1; supplementary fig. S3); R. acuminata is morphologically very similar to the new species and is also found in characiforms. A morphometric comparison between the new species and R. acuminata is presented in supplementary table S2. Both species possess 14 teeth in the prostom and single deirids. However, the new species can be readily distinguished from R. acuminata by the number of preanal papillae in males (15 pairs vs 9–11 pairs), by the size and position of deirids (large deirids near cephalic end, usually not exceeding the end of prostom in the new species vs small deirids in the first third of vestibular length in R. acuminata), in the arrangement of the 14 teeth in the prostom (4 dorsal, 4 ventral and 3 lateral on either side in the new species vs 3 dorsal, 3 ventral and 4 on each lateral side in R. acuminata), and by the fact that teeth in R. acuminata are entire, whereas in the new species teeth are bifurcated. In terms of host association, even though R. acuminata was originally described as a parasite of the bryconid Brycon falcatus Müller and Troschel in the Paraná River in Brazil (Molin, Reference Molin1860), it has been additionally recorded in a wide array of distantly related hosts, including five species of characiforms, three species of pimelodids and one species of cichlid in Brazil, but also from one species of diplomystid (Siluriformes), one species of anablepid and one species of percychthid in Argentina, and one species of cichlid, one of characid and one of pimelodid in Ecuador (Cremonte et al., Reference Cremonte2002; Ramallo, Reference Ramallo2005; Pinto et al., Reference Pinto2010). Based on host association pattern, it seems likely that most of these records need to be further verified, and whether or not R. acuminata in South America represents a species complex needs to be determined by further molecular phylogenetic analyses. In Mexico, R. acuminata has been reported only as a parasite of the bryconid Brycon guatemalensis Regan in the Usumacinta River basin (Caspeta-Mandujano et al., Reference Caspeta-Mandujano2005), in southern Mexico. Rhabdochona osorioi n. sp. is also found in the same geographical area, occurring in water bodies of Chiapas and Veracruz. However, they are clearly differentiated, and both are valid species. Rhabdochona osorioi n. sp. is the fifth species of Rhabdochona known to parasitize characiform freshwater fishes in the Neotropical biogeographical region. The other three species are found in South America, R. acuminata and R. fabianae Ramallo, 2005 from Bryconamericus iheringi Boulanger, 1887 (in Argentina), and R. uruyeni Díaz-Ungría, 1968 from the characiform Piabucina sp. (and also from the freshwater scianid Pachiurus squamipennis Agassiz, 1831) (Cremonte et al., Reference Cremonte2002; Ramallo, Reference Ramallo2005; Pinto et al., Reference Pinto2010).
Discussion
Our analyses uncovered two genetic lineages within R. mexicana. Two lines of evidence allowed us to recognize the existence of these two genetic lineages. Firstly, genetic divergence for the mitochondrial cytochrome c oxidase subunit I (cox1) gene was high and allowed us to recognize them as independent evolutionary entities, with divergence levels between 10.94 and 11.51%. Secondly, sequenced individuals belonging to both lineages were reciprocally monophyletic (fig. 2). The molecular results prompted us to conduct a detailed morphological examination of the specimens and, as a result, Rhabdochona mexicana Lineage II was described as a new species, for which the name Rhabdochona osorioi n. sp. was coined; however, the lack of morphological differences, genetic divergence values of cox1, and the tree topology led us to further investigate a potential case of cryptic species for the subgroups formed within Rhabdochona mexicana Lineage I.
Lineage I was composed of three genetic subgroups and was considered as a cryptic species complex based on the fact that they represent reciprocally monophyletic groups, with moderate genetic divergence for the mitochondrial gene cox1, and with no clear morphological differentiation. To shed light on the genetic diversity of R. mexicana Lineage I, our study followed a molecular prospecting approach, as suggested by Blouin (Reference Blouin2002); we used sequence data and a genetic yardstick to search for populations that could represent a cryptic species, assuming the null hypothesis that individuals represented a single species, i.e. R. mexicana (see Pérez-Ponce de León and Nadler, Reference Pérez-Ponce de León and Nadler2010). By comparing species-pairs of different genera of nematodes, Blouin (Reference Blouin2002) suggested that if two groups show genetic divergence values around 10% (with a range between 6.9 and 13%) for the cox1 gene, we might consider that they are not conspecific. Overall, cryptic species of nematodes have been detected using nuclear and mitochondrial molecular markers (e.g. Derycke et al., Reference Derycke2005; Ristau et al., Reference Ristau, Steinfartz and Traunspurger2013; Jorge et al., Reference Jorge2013; Chilton et al., Reference Chilton2016), although mitochondrial genes have been shown to be more informative because of the higher substitution rate (Blouin, Reference Blouin2002).
The three subgroups within R. mexicana Lineage I exhibited a genetic divergence varying from 4.41 to 6.24%; it seems plausible then to postulate they may represent a recent diversification event (see Fišer et al., Reference Fišer, Robinson and Malard2018). Nadler and Pérez-Ponce de León (Reference Nadler and Pérez-Ponce de León2011) discussed the use of pair-wise distance threshold for initial cryptic species prospecting as a reasonable exploratory approach; however, whether or not 6.9% is the minimum distance threshold to evaluate if two putative taxa are different enough to merit recognition as separate species in nematodes is debatable, given the heterogeneity of the evolutionary patterns in different groups. For instance, Miranda et al. (Reference Miranda2008) reported a genetic divergence of 7% among populations of Ancylostoma caninum Ercolani, 1859 in Brazil, suggesting the existence of a cryptic species; St-Onge et al. (Reference St-Onge, LaRue and Charpentier2008) reported cox1 divergence between 12.9 and 15.7% in the mermithid Mesomermis flumenalis Welch, 1962 in Canada, and considered that the species represented four clearly distinguished species. In contrast, the genetic divergence of the congeneric species Rhabdochona lichtenfelsi, a parasite of endemic freshwater fishes in central Mexico, was evaluated by Mejía-Madrid et al. (Reference Mejía-Madrid, Vázquez-Domínguez and Pérez-Ponce de León2007b). In that study, genetic divergence values for cox1 were lower than 4.9% among populations from different localities and no genetic structure associated either with host species or geographical distribution was found; in this case, the genetic divergence pattern was not in agreement with the intricate evolutionary history of their host, which experienced a large diversification process in central Mexico resulting from the complex geological and hydrographical history. Our results also showed that the three subgroups of the Rhabdochona mexicana cryptic species complex exhibit a well-defined geographical distribution pattern, and currently represent allopatric populations. Subgroup 1 is distributed in river basins of central Mexico; subgroup 2 exhibits a wider geographical distribution range, although restricted along the Pacific Ocean slope, between Jalisco, in western Mexico and Guatemala; and subgroup 3 is found only in locations of northern Mexico (San Luis Potosí and Tamaulipas), and exclusively infects the intestine of the Mexican tetra, A. mexicanus, the species of characiform that reach the most northern distributional range in the Americas (see Ornelas-García et al., Reference Ornelas-García, Domínguez-Domínguez and Doadrio2008).
The taxonomic identification of Rhabdochona species in Mexican freshwater fishes has been sometimes based on the host association pattern, without a rigorous analysis of the morphological traits; this practice might have resulted in inaccuracies in species identification, even though we acknowledge that most species are host-specific and are part of the biogeographical core helminth fauna of freshwater fishes, especially at family level (see Pérez-Ponce de León and Choudhury, Reference Pérez-Ponce de León and Choudhury2005). Our study also showed that in addition to host association, a detailed morphological study is necessary to establish the species’ identity, and that an accurate estimate of diversification in the genus Rhabdochona requires scrutiny through DNA sequences. The generation of DNA sequences for representative species of Rhabdochona from across the world will increase our understanding of the current diversity within the genus and will help to elucidate their evolutionary and biogeographical history. We also suggest that an integrative taxonomy approach must be followed to fully understand the diversity of this speciose group of nematodes, adding as many sources of information as possible, as recently discussed by De Sousa et al. (Reference De Sousa2018) for the study of nematode taxonomy.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/S0022149X19000014
Author ORCIDs
G. Pérez-Ponce de León, 0000-0001-6472-5113
Acknowledgements
We thank David Hernández-Mena, Carlos Pedraza Lara and Brenda Solórzano-García for their help during field work, and Martin García-Varela and Leopoldo Andrade Gómez for donating specimens of Rhabdochona from Guatemala. Thanks are also due to Luis García for the loan of specimens from Colección Nacional de Helmintos (CNHE, Mexico City), Berenit Mendoza-Garfias for her assistance with the scanning electron microscope, and Laura Márquez for technical assistance with the automatic sequencer.
Financial support
This work was partially supported by the Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT-UNAM) IN202617 to GPPL.
Conflict of interest
None.
Ethical standards
Specimens were collected under the Cartilla Nacional de Colector Científico (FAUT 0057) issued by the Secretaría del Medio Ambiente y Recursos Naturales (SEMARNAT), to GPPL, and collecting permit issued by SGPA/DGVS/05464/, Secretaría de Agricultura, Ganadería, Desarrollo Rural y Pesca (SAGARPA) to CPOG.










