Introduction
Homalometron Stafford, 1904 is a species-rich genus of digeneans (with 34 species according to the World Register of Marine Species, 2022) whose species are distributed worldwide. These parasites mature in the digestive tract of actinopterygian fishes with marine, brackish and freshwater habits. Twenty-five out of the 34 described species have been reported in aquatic environments across the Americas. Particularly, six species were described and found as parasites of fish belonging to the family Gerreidae Leach in marine and brackish water environments, namely: Homalometron elongatum Manter, Reference Manter1947 in Gerres cinereus (Walbaum) from the United States (see Manter, Reference Manter1947; see Parker et al., Reference Parker, Curran, Overstreet and Tkach2010 that includes other localities in the Caribbean, Puerto Rico, Jamaica and Bahamas), in Eucinostomus californiensis (Gill) from the Panamanian Pacific (see Sogandares-Bernal, Reference Sogandares-Bernal1959), in Diapterus peruvianus (Cuvier) and in Eucinostomus gracilis (Gill) from Mexico (see Pérez-Ponce de León et al., Reference Pérez-Ponce de León, Garcia-Prieto and Mendoza-Garfias2007); Homalometron longulum Travassos, Teixeira de Freitas & Bührnheim, 1965 in Diapterus rhombeus Cuvier from Brazil (see Travassos et al., Reference Travassos, Teixeira de Freitas and Bührnheim1965), in Diapterus peruvianus and Gerres cinereus from Mexico (see Pérez-Ponce de León et al., Reference Pérez-Ponce de León, Garcia-Prieto and Mendoza-Garfias2007); Homalometron carapevae Amato, Reference Amato1983 in Eugerres brasilianus (Cuvier) from Brazil; Homalometron lesliorum Parker et al., Reference Parker, Curran, Overstreet and Tkach2010 in Eucinostomus currani Zahuranec from Costa Rica;. Homalometron stradbrokense Cribb & Bray, Reference Cribb and Bray1999 in Gerres subfasciatus Cuvier from Australia; and Homalometron palmeri Curran et al., Reference Curran, Tkach and Overstreet2013 was reported in Eucinostomus argenteus Baird & Girard and in various sciaenids and fundulids from the northern Gulf of Mexico (Curran et al., Reference Curran, Tkach and Overstreet2013).
During a field parasitology course at Los Tuxtlas Biological Station (Instituto de Biología, Universidad Nacional Autónoma de México (UNAM)) in tropical rain forest of Los Tuxtlas in Veracruz state in 2018, several marine and brackish water fish species were necropsied and studied for metazoan parasites. Specimens of the stripped mojarra, Eugerres plumieri (Cuvier), were sampled from Sontecomapan lagoon. Specimens of digeneans belonging to the genus Homalometron were found in the intestines of their hosts, studied morphologically. DNA sequence data for two molecular markers were used to corroborate, through a phylogenetic analysis, that they represented an undescribed species. We describe a new species of Homalometron herein as Homalometron avis n. sp., which represents the seventh species of this genus reported in Mexico.
Material and methods
Sample collections
In April 2018, several individuals of E. plumieri were collected in Sontecomapan lagoon, Veracruz, Mexico, on the Gulf of Mexico slope. Fish were collected using cast nets, kept alive and necropsied 4 h after capture. The gastrointestinal tract from each fish was removed and placed in Petri dishes with 6.5% saline solution and examined under a stereomicroscope. For morphological study, specimens were fixed in hot (nearly boiling) tap water, some were kept in vials with 4% formalin for scanning electron microscopy, and some in 96% ethanol for staining and mounting. For molecular study, other specimens were placed in vials with 100% ethanol.
Morphological study
Specimens fixed for morphology were dehydrated in a graded ethanol series, stained with Mayer's paracarmine and Gomori's trichrome, cleared in methyl salicylate and processed as permanent mounts in Canada balsam. Specimens were examined using an Olympus BX51 microscope (Olympus Corporation, Tokyo, Japan). Measurements were taken using an Olympus Quick-Photo Image-Program, and are presented in micrometres (μm) with the range followed by the mean in parentheses. Drawings were made with a drawing tube attached to the microscope. Additionally, for the scanning electron microscopy (SEM) study, two specimens of the new species were dehydrated through a graded ethanol series and then critical point dried with carbon dioxide. Specimens were mounted on a metal stub with carbon adhesive tabs, then gold coated, and examined at 15 kV with a Hitachi Stereoscan SU1510 SEM (Hitachi Ltd., Tokyo, Japan). Specimens were deposited in the Colección Nacional de Helmintos (CNHE), Instituto de Biología, UNAM).
Molecular study
Each specimen was digested, and its genomic DNA extracted using the REDExtract-N-Amp Tissue PCR kit (Sigma, St Louis, USA) following the manufacturer's instructions. Proteinase was denaturized at 95 °C for 3 min. A fragment of the 28S gene (the domains D1–D3) and the internal transcribed spacer (ITS) (ITS1, 5.8S, ITS2) of the ribosomal DNA were amplified. For the 28S the forward primer 391 5′-AGCGGAGGAAAAGAAACTAA-3′ (Nadler & Hudspeth, Reference Nadler and Hudspeth1998) and the reverse primer 536 5′-CAGCTATCCTGAGGGAAAC-3′ (Stock et al., Reference Stock, Campbell and Nadler2001) were used. For ITS the forward BD1 5′–GTCGTAACAAGGTTTCCGTA–3′, plus the reverse BD2 5′-TATGCTTAAATTCAGCGGGT-3′ (Luton et al., Reference Luton, Walker and Blair1992) were used. The polymerase chain reaction (PCR) was performed in a final volume of 12.5 μl containing 1 μl of each primer, 2 μl of extracted DNA, 2.5 μl of 10× buffer, 1.5 μl of MgCl2 at 25 mm, 0.5 μl of dNTPs at 10 mm and 1 U of Taq DNA polymerase. Thermal cycling conditions for both 28S and ITS consisted of an initial denaturation at 95 °C for 5 min followed by 35 cycles at 94 °C for 1 min, annealing at 50 °C for 1 min and extension at 72 °C for 1 min, with a final extension at 72 °C for 10 min. PCR products for 28S were sequenced with PCR primers plus internal primers 503 5′-CCTTGGTCCGTGTTTCAAGACG-3′ (Stock et al., Reference Stock, Campbell and Nadler2001) and 504 5′-CGTCTTGAAACACGGACTAAGG-3′ (García-Varela & Nadler, Reference García-Varela and Nadler2005). Instead, sequencing reactions for ITS were performed using the two initial primers and with the following internal primers, BD3 5′-GAACATCGACATCTTGAACG-3′ and BD4 5′-ATAAGCCGACCCTCGGC-3′ (Hernández-Mena et al., Reference Hernández-Mena, García-Prieto and García-Varela2014). PCR products were sequenced using an ABI 3730xl Genetic Analyser (Applied Biosystems). The resulting sequences were analysed, and a consensus sequence was obtained for each sequenced specimen, both for 28S and ITS, using the software Geneious Pro 4.8.4 software (Biomatters Ltd., Auckland, New Zealand). Sequences were submitted to GenBank.
Phylogenetic analyses
The identity of the newly generated sequences of 28S and ITS was first checked using the Basic Local Alignment Search Tool (BLAST) (www.ncbi.nih.gov/BLAST/), and sequences were aligned with those available in GenBank of other species of Apocreadiidae Skrjabin, 1942 (the accession numbers of the sequences used in this study are in the phylogenetic trees). Two data sets were constructed, one for each analysed gene, in Mesquite 3.62 (https://www.mesquiteproject.org/), and these data sets were aligned using ClustalW (Thompson et al., Reference Thompson, Higgins and Gibson1994), with the approach ‘SLOW/ACCURATE’ and weight matrix ‘CLUSTALW (for DNA)’ in the web site http://www.genome.jp/tools/clustalw/. Based on previous phylogenetic analyses (Pérez-Ponce de León & Hernández-Mena, Reference Pérez-Ponce de León and Hernández-Mena2019), species of Schistorchis Lühe, 1906, Haintestinum Pulis, Curran, Andres & Overstreet, 2014, Megapera Manter, 1934 and Thysanopharynx Manter, were used in 28S and ITS data sets as outgroups for rooting the trees. Phylogenetic analyses were run under maximum likelihood (ML), using the nucleotide evolution models selected with jModelTest v2 (Darriba et al., Reference Darriba, Taboada, Doallo and Posada2012). The ML trees, model parameters and bootstrap (Bt) support (1000 repetitions) were estimated with RAxML v. 7.0.4 (Stamatakis, Reference Stamatakis2006). The phylogenetic trees obtained from both analyses were visualized in FigTree v. 1.4.2. Genetic distances were calculated as uncorrected P-distance using MEGA v6 (Tamura et al., Reference Tamura, Stecher, Peterson, Filipski and Kumar2013).
Results
Family: Apocreadiidae Skrjabin, 1942
Genus: Homalometron Stafford, 1904
Homalometron avis Hernández-Mena, Cabañas-Granillo, Medina-Hernández & Pérez-Ponce de León n. sp.
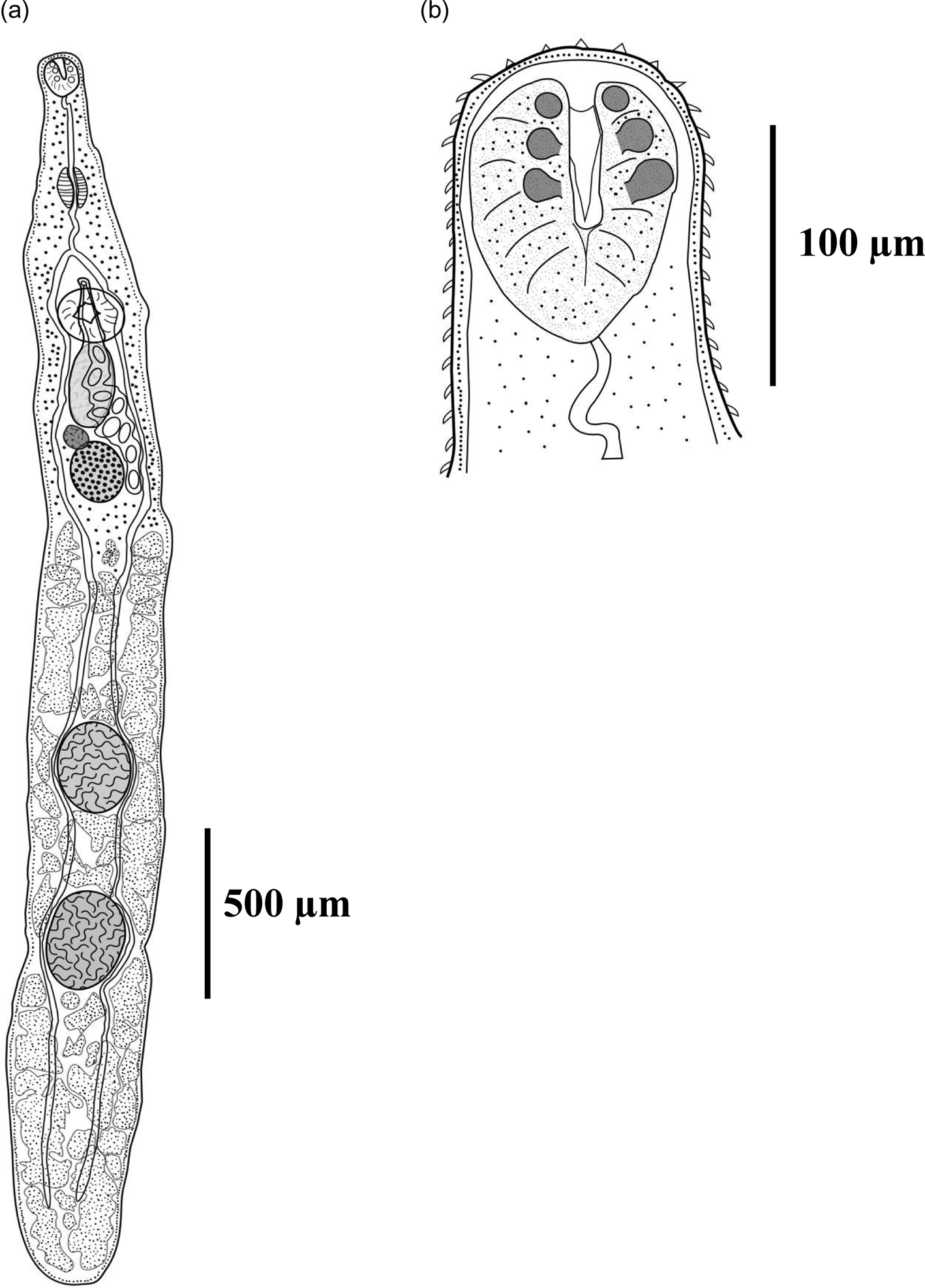
Fig. 1. Homalometron avis n. sp. (a) ventral view of holotype; and (b) ventral view of ventral sucker.

Fig. 2. Scanning electron micrographs of Homalometron avis n. sp. (a) ventral view of oral sucker with three lateral papillae; (b) ventral sucker surrounded by scale-like spines; (c) posterior end of the body where the excretory pore is located surrounded by small tegumental spines; (d) ventral view of the forebody and the anterior part of the hindbody showing the decrease in tegumental spines as they approach the posterior end of the body; (e) ventral view of the hindbody where the external relief of the posterior testicle is observed; and (f) scale-like spines formed by a solid base and pectinate minute spines.
ZooBank Life Science Identifier: urn:lsid:zoobank.org:act:03B5E209-A416-4F43-B8F6-E067A32D09D3
Taxonomic summary
Type host: Eugerres plumieri (Cuvier, 1830) (Gerreidae)
Type locality: Laguna de Sontecomapan, Veracruz, Mexico (18°33′18.3″N, 94°59′28.2″W).
Site in host: Intestine.
Prevalence: Five of 15 (33.3%).
Type material: Holotype (CNHE 11667); XX paratypes: (CNHE 11668).
Representative DNA sequences: 28S rDNA (ON814857–ON814858) ITS rDNA (ON815018–ON815019).
Etymology: The epithet of the species refers to the Latin word ‘avus,’ which means ‘grandparents’ (or ‘abuelos’ in Spanish), since the name of the new species is dedicated to the relatives of the first author.
Description
Based on five adult specimens, and on additional observations of two specimens through SEM. Body elongated, 1907–3665 (2921) long, narrow at oral sucker level; maximum width 212–443 (286) at testes level. Body tegument covered by scale-like spines denser at anterior half of body, decreasing their numbers towards posterior end. Each spine formed by a solid base covered by minute pectinate spines. Forebody length 580–925 (703), 18–35% (25%) of body length; unobservable eyespots. Oral sucker subterminal, 89–117 (102) long, 66–97 (81) wide, with three pairs of well-developed opposing papillae surrounding mouth opening. Ventral sucker 110–192 (155) long, 100–186 (147) wide. Oral sucker/ventral sucker length ratio 1:1.22–2.13 (1:1.56). Oral sucker/ventral sucker width ratio 1:1.41–2.64 (1:1.85). Mouth subterminal. Prepharynx long, elongated, 158–357 (249), with anterior end curved and posterior end slightly enveloping anterior end of pharynx; pharynx muscular, subglobular, 65–133 (102) long, 49–84 (63) wide, slightly surrounded by gland cells. Oesophagus shorter than prepharynx, 83–187 (132) long. Intestinal bifurcating 526–775 (627) from anterior body end, approximately two-thirds of distance between oral and ventral suckers and approximately one-half between pharynx and ventral sucker, ending blindly 140–241 (189) from posterior body end. Postcaecal space representing 5.4–8.1% (6.3%) of body length.
Two testes of similar size (slightly smaller anterior testis), subspherical, entire, in tandem, median, post-ovarian, in posterior half of body; anterior testis 122–301 (226) long, 106–310 (207) wide; posterior testis 135–352 (251) long, 127–352 (230) wide. Inter-testicular space 34–245 (131) long. The post-testicular space 307–881 (612), representing 14.1–24.5% (19.5%) of body length. Male terminal genitalia consisting of seminal vesicle, pars prostatica, and ejaculatory duct; seminal vesicle saccular, median, dorsal to ventral sucker, extending into hindbody, 169–279 (231) long, 71–144 (106) wide, opening into spindle-shaped prostatic region; pars prostatica opening into slender elongated ejaculatory duct; ejaculatory duct merging with muscular elongated female duct forming a muscular dorsoventral hermaphroditic duct; hermaphroditic duct opening through ventral genital pore, median, immediately anterior to ventral sucker.
Ovary globular, 83–179 (142) long, 67–159 (125) wide, median, pre-testicular, approximately one-third the distance between ventral sucker and anterior testis. Distance between ovary and anterior testis 251–844 (599). Seminal receptacle globular, anterior to ovary, opening into oviduct near posterior margin of ovary. Mehlis’ gland posterior to ovary and seminal receptacle. Laurer's canal not observed. Vitellarium consisting of one confluent field of large follicles in hindbody; anteriorly, vitelline follicles post-ovarian, and at 50–60 (57) from posterior margin of ovary; posteriorly, follicles extend to posterior end of body; follicles intercaecal and extracaecal and dorso-ventral in post-ovarian region and in intertesticular space. Vitelline reservoir ventral to Mehlis’ gland. Ootype immediately post-ovarian. Uterus short, pre-testicular, intercaecal, between posterior end of ovary and genital pore. Eggs 52–84 (71) long, 35–51 (44) wide. Excretory vesicle I-shaped, terminating in post-testicular region; excretory pore terminal, surrounded by spines.
Remarks
Homalometron avis n. sp. conforms to the diagnosis of Homalometron and Apocreadiidae as given by Cribb (Reference Cribb, Jones, Bray and Gibson2005). The new species can be easily distinguished from 30 of the 34 congeneric species by having an elongated body and possessing three pairs of oral papillae. Four species in the genus share these morphological characters, namely Homalometron carapevae, Homalometron elongatum, Homalometron lesliorum and Homalometron papilliferum (Szidat, 1956) Ostrowski de Nuñez et al., Reference Ostrowski de Nuñez, Brugni and Viozzi2000. Interestingly, the first three species are also found in fish of the family Gerreidae, whereas H. papilliferum parasitizes Percichthys trucha (Valenciennes) (Ostrowski de Nuñez et al., Reference Ostrowski de Nuñez, Brugni and Viozzi2000; Shimazu et al., Reference Shimazu, Urawa and Coria2000). Homalometron avis n. sp. can be readily distinguished from these congeneric species by having a smaller oral sucker (89–116 × 66–97 vs. 120–140 × 108–136 in H. carapevae, 156–223 × 162–218 in H. elongatum, 125–207 × 162–213 in H. lesliorum, and 189–254 × 185–263 in H. papilliferum). The new species further differs from H. elongatum and H. lesliorum by the anterior distribution of the vitelline follicles, and by the oral to ventral sucker length and width ratios; the vitelline follicles in the new species are post-ovarian, and they have a ventral sucker larger than the oral sucker of 1:1.21–2.13 × 1:1.40–2.64, whereas in H. elongatum and H. lesliorum the vitelline follicles reach into pre-ovarian zone, and the ventral sucker is smaller than the oral sucker (1:0.76–0.88 × 1:0.79–0.88 and 1:0.90–1.05 × 1:0.81–0.98, respectively). The new species is also readily distinguished from H. papilliferum because in this species the oral and ventral suckers are almost similar in size, although with a tendency for the oral sucker to be larger (1:0.71–1.21 × 1:0.67–1.20), the prepharynx is shorter (34–143 in H. papilliferum vs. 158–357 in the new species), the eggs of H. papilliferum are larger (77–110 × 48–65 vs. 52–84 × 35–51 in the new species), and finally, the vitelline follicles of H. papilliferum reach the posterior margin of the ovary and are absent in the inter-testicular space, whereas in the H. avis n. sp. follicles never extend beyond the ovary and occupy the inter-testicular space.
The new species closely resembles H. carapevae which presents more similar oral/ventral sucker ratios, as well as pre-pharynx and eggs sizes. The new species also share with H. carapevae the anterior distribution of the vitelline follicles; however, they can be distinguished because in H. carapevae follicles at the anterior end reach the posterior margin of the ovary, whereas in H. avis n. sp. follicles never extend to reach the ovary. There are also differences in host association and geographical distribution since H. carapevae was described by Amato (Reference Amato1983) from the intestine of the Brazilian mojarra, Eugerres brasilianus in Brazil, whereas the new species was found in Eugerres plumieri further north, in the Gulf of Mexico.
Phylogenetic affinities
The aligned data set for the 28S gene was 1414 base pairs (bp) long and consisted of 25 species of Apocreadiidae. The substitution model selected for this data set used to infer the ML phylogenetic hypothesis was GTR + I, nucleotide frequencies were A = 0.221, C = 0.222, G = 0.313 and T = 0.243, and a likelihood value of ln = −5535.184152. The phylogenetic tree resolved three clades which are consistent with subfamilies accepted within Apocreadiidae, that is, Schistorchiinae Yamaguti, 1942, Megaperinae Manter, 1934 and Apocreadiinae Skrjabin, 1942, with high bt support value (fig. 3a). Within the Apocreadiinae clade, the genus Homalometron was resolved as not monophyletic since three species were grouped separately, that is, Homalometron mexicanum + Homalometron sp. (which was the sister group of the other representative genera of Apocreadiinae), and Homalometron synagris (Yamaguti, 1953) Cribb & Bray, Reference Cribb and Bray1999 (grouped in an independent clade along with Neoapocreadium Siddiqi & Cable, 1960 and Callohelmis Cribb & Bray, Reference Cribb and Bray1999). Moreover, species of Crassicutis Manter, 1936 were recovered as the sister clade of Homalometron, with the exception of Crassicutis archosargi Sparks & Thatcher, Reference Sparks and Thatcher1960, a species that nested within the Homalometron clade.

Fig. 3. Maximum likelihood phylogenetic trees for Homalometron species, showing the position of the new species that is highlighted in bold red font. (a) phylogenetic tree of the 28S gene; and (b) phylogenetic tree of the internal transcribed spacer (ITS) gene. Purple rectangle = schistorchiinae; green rectangle = megaperinae; blue rectangle = apocreadiinae. Numbers near internal nodes indicate bootstrap support values.
Particularly, H. avis n. sp. was resolved as a member of Homalometron, as the sister species of C. archosargi, with high bt support value (98). In addition, within the Homalometron clade other subclades were formed albeit some with low nodal support values: a subclade grouping Homalometron mesoamericanum + (. Homalometron octopapillatum + . Homalometron cupuloris); Homalometron pseudopallidum separated independently as the sister group to the other species of the genus; another subclade grouping Homalometron elongatum + Homalometron lesliorum; and finally, a subclade formed by. Homalometron armatum + ((Homalometron manteri + . Homalometron palmeri) + (Homalometron frocioneae + (Homalometron robisoni + Homalometron pallidum))). Homalometron elongatum and H. lesliorum are morphologically similar species to the new species as mentioned above, due to the elongated body and the presence of three pairs of oral papillae, and they all infect gerreid fishes, but interestingly they were not grouped together in the molecular phylogenetic analysis.
Furthermore, the aligned data set for the ITS was 1111 bp long and had 20 species of apocreadiids. The selected model for the ML analysis was GTR + G + I, and nucleotide frequencies were A = 0.200, C = 0.256, G = 0.271 and T = 0.273. The ML tree had a likelihood value of ln = −5592.505050. In this data set as in that of 28S, it also included representative species of Schistorchiinae and Megaperinae. The phylogenetic relationships of the species with the ITS were similar to those of 28S, where H. mexicanum was not grouped with the rest of the species of its genus, since Crassicutis cichlasomae remained as the sister species of the Homalometron clade. Instead, C. archosargi was grouped within the Homalometron clade. Within the Homalometron clade, H. pseudopallidum remained the sister species of the remaining sequenced species, and 3 subclades were formed (fig. 3b): one containing H. mesoamericanum + (H. octopapillatum + H. cupuloris); another containing H. elongatum + H. lesliorum; and finally another containing (((H. avis + C. archosargi) + (H. frocioneae + (H. robisoni + H. pallidum))) + (H. manteri + H. palmeri)) + H. armatum. Therefore, the new species also turned out to be sister to C. archosargi, and as in the 28S tree, the ITS tree topology indicated that H. avis n. sp. was not a sister taxon of H. elongatum and H. lesliorum.
In terms of genetic variation, the 28S gene showed H. avis n. sp. and C. archosargi, its sister species, differing by 1.91%, whereas the genetic divergence between the new species and those of Homalometron within the same clade varied from 1.64 to 4.45% (table 1). The divergence between the new species and the remaining species of Homalometron grouped in independent clades was very high and varied between 6.32 and 7.15%. Intergeneric genetic divergence was also high among genera of the subfamily Apocreadiinae, with values ranging from 5.67 to 6.59% (table 1). For ITS sequences, the genetic variation between H. avis n. sp. and C. archosargi was 1.37%, and with the congeneric species within the same clade ranged from 2.47 to 5.27% (table 1); the new species differed between 6 and 6.42% from H. elongatum and H. lesliorum, respectively. Finally, variation between the new species and the remaining species was from 6.94 to 13.06%; the highest variation was found with respect to H. mexicanum (table 1).
Table 1. Genetic distances between the new species and congeneric species used for phylogenetic analyses. In the centre column is the comparison with the 28S gene and in the right column is the comparison with the internal transcribed spacer (ITS) gene. DA = data not available.

Discussion
The new species described in this study is the seventh described as parasite of the Gerreidae, and the seventh for Mexico. The newly discovered species is readily distinguishable from most congeners excepting a group of four species that possess three pairs of prominent oral papillae on the oral sucker. Still, the new species can be diagnosed by the combination of characters such as the distribution of the vitelline follicles, the size of the oral sucker, the oral sucker/ventral sucker length ratio, the size of the eggs and the length of the pre-pharynx. Amato (Reference Amato1983), Ostrowski de Nuñez et al. (Reference Ostrowski de Nuñez, Brugni and Viozzi2000), and Parker et al. (Reference Parker, Curran, Overstreet and Tkach2010) distinguished the species they described possessing three pairs of oral papillae from their congeners using these characters. These authors also pointed out that the tegument of H. carapevae, H. elongatum and H. lesliorum were completely covered by small spines, whereas Ostrowski de Nuñez et al. (Reference Ostrowski de Nuñez, Brugni and Viozzi2000) described that H. papilliferum possessed scale-like spines distributed only on the anterior region of the parasite. Our observations of the tegument of the new species through SEM revealed that specimens possessed scale-like spines distributed along most body surfaces, more abundant in the forebody and particularly in the region of the ventral sucker, but their number decreasing towards the posterior end of body. Furthermore, SEM photomicrographs showed that each one of the scale-like structures comprises a pectinate structure of smaller spines at their distal end (see fig. 2f). Unfortunately, we cannot corroborate the existence of these types of spines on H. carapevae, H. elongatum, H. lesliorum and H. papilliferum. We emphasize here the need to use SEM as a common practice in future descriptions of trematodes, and we look forward for researchers to show the spines of the tegument of these species in the future in case they are sampled in the localities where they have been reported.
Sequence data available for two of the four species allows us to explore their phylogenetic relationships with respect to the new species herein described. The 28S and ITS phylogenetic trees revealed that H. elongatum and H. lesliorum are sister taxa, they both are parasites mainly of Gerreidae and one of them, H. elongatum occurs in localities nearby the type locality of the new species in the Gulf of Mexico (Veracruz), including Tortugas, Florida, and other areas of the Caribbean Sea (see Parker et al., Reference Parker, Curran, Overstreet and Tkach2010). Interestingly, H. elongatum and H. lesliorum are not close relatives to the new species. They do not conform a monophyletic group; therefore, the presence of three pairs of oral papillae cannot be considered as a synapomorphy for the group of species, at least the ones for which sequences are available. Sequence data for H. papilliferum and H. carapevae are needed to corroborate this hypothesis. The new species is morphologically very similar to H. carapevae and both are found in gerreid fishes, although the latter was described from Brazil; these species possess qualitative and quantitative characters that allow us to differentiate them, but the lack of molecular data for H. carapevae limits our understanding of their potential phylogenetic relationships. One possible hypothesis is that due to their morphological similarities and their host affinities (Eugerres plumieri for the new species and Eugerres brasilianus for H. carapevae) they might be sister species as in the case of H. elongatum and H. lesliorum even though they are found in the Gulf of Mexico/Caribbean Sea, and Pacific Ocean, respectively (Manter, Reference Manter1947; Amato, Reference Amato1983; Parker et al., Reference Parker, Curran, Overstreet and Tkach2010). As in the case of SEM, molecular data are also required to confirm this hypothesis.
Overall, the phylogenetic trees also show that the genus Homalometron needs a detailed taxonomic revision. The species sequenced up to now show that they do not conform a monophyletic assemblage. The morphological concept of the genus Homalometron requires further evaluation, incorporating not only genetic but also host association and geographical data. Species identified as Homalometron sp. 1, Homalometron mexicanum and H. synagris are not resolved as monophyletic in relation to the remaining sequenced species of Homalometron. They were originally allocated as belonging to the genus Apocreadium Manter, Reference Manter1947 but later transferred to Homalometron by Cribb & Bray (Reference Cribb and Bray1999) and validated in Cribb (Reference Cribb, Jones, Bray and Gibson2005). Homalometron sp. 1 (GenBank No. MK648264) represents a potential new species found in Balistes vetula L. from Puerto Morelos reef in the Caribbean Sea of Mexico (Pérez-Ponce de León & Hernández-Mena, Reference Pérez-Ponce de León and Hernández-Mena2019); H. mexicanum was described from several species of Balistidae, Labrisomidae and Monacanthidae in the Gulf of Mexico, Caribbean Sea and Pacific coast of Mexico (see records in Cribb & Bray, Reference Cribb and Bray1999); finally, H. synagris was described as a parasite of Scolpis monogramma (Cuvier) (Nemapteridae) from Malaysia, Indonesia and Australia (Cribb & Bray, Reference Cribb and Bray1999). Clearly, more sequence data of apocreadiines are required to further understand the interrelationships of members of the Apocreadiidae; that includes adding data on other species of Homalometron as well as data on other genera of the family.
Our phylogenetic analyses based on the 28S rRNA gene unequivocally demonstrated that Crassicutis archosargi, a species described from a sparid fish in the Gulf of Mexico (Archosargus probatocephalus Walbaum) (Sparks & Thatcher, Reference Sparks and Thatcher1960; Overstreet, Reference Overstreet1976), is resolved as the sister taxon of the new species. This may indicate the species is not a member of Crassicutis, but a species of Homalometron. Previous phylogenetic studies have shown that the genus Crassicutis represents a monophyletic assemblage (see Razo-Mendivil et al., Reference Razo-Mendivil, Vázquez-Domínguez, Rosas-Valdez, Pérez-Ponce de León and Nadler2010; Pantoja et al., Reference Pantoja, Scholz, Luque and Perez-Ponce de León2021). Some morphological traits indicate that C. archosargi falls within the concept of Homalometron. We refrain now from considering the synonymy of C. archosargi with Homalometron, as H. archosargi until new samples are available for morphological comparison that includes SEM observations of the tegument. In C. archosargi the tegument is highly modified, bearing refractile bodies and was described by Sparks & Thatcher (Reference Sparks and Thatcher1960) (and later redescribed by Overstreet, Reference Overstreet1976) as lacking tegumental spines. Additionally, the fact that vitelline follicles in C. archosargi extend to the forebody make their inclusion in the current concept of Homalometron incorrect. The synonymy requires further morphological verification that spines might be present on the tegument, or probably lost after modification of a rugose and thick tegument, and a probable modification of the diagnosis of the genus Homalometron to include species with vitelline follicles extending into the forebody.
To conclude, it is important to emphasize that the comparison of morphological, genetic and phylogenetic information has allowed us to discover a new species in a coastal lagoon of Veracruz, Mexico. The discovery expands our knowledge about the biodiversity of brackish water fishes, although some species of Homalometron were previously reported or described from freshwater habitats. This is the seventh species of the genus currently reported from Mexico, along with H. pallidum Stafford, 1904, H. mesoamericanum Pérez-Ponce de León, Razo-Mendivil, & García-Magaña, 2012; H. octopapillatum Pérez-Ponce de León, Razo-Mendivil, & García-Magaña, 2012; H. caballeroi (Bravo-Hollis, 1954) Cribb & Bray, Reference Cribb and Bray1999; H. verrunculi (Lamothe-Argumedo, 1965) although see Cribb & Bray (Reference Cribb and Bray1999) who suggested this was a nom. nov. for H. caballeroi; and H. mexicanum (Manter, 1937) Cribb & Bray, Reference Cribb and Bray1999 (see Cribb & Bray, Reference Cribb and Bray1999; Cribb, Reference Cribb, Jones, Bray and Gibson2005; Pérez-Ponce de León et al., Reference Pérez-Ponce de León, Mendoza-Garfias, García-Prieto, Del Moral, Martínez, Franco, Ramírez and Tello2012; Pérez-Ponce de León & Aguilar-Aguilar, Reference Pérez-Ponce de León, Aguilar-Aguilar, Álvarez and Ojeda2019). Therefore, seven species of Homaloletron sensu lato have been reported from Mexico. Our study also emphasizes the view that current taxonomic studies must use various sources of information that allow us to recognize and find support for the discovery of new taxa.
Acknowledgements
We thank Luis García-Prieto, Alejandro Oceguera-Figeroa, David Osorio-Sarabia and Martín García-Varela, for their help during field work, Berenit Mendoza, Laboratorio Nacional de Biodiversidad (LANABIO), for obtaining scanning electron microscopy photomicrographs, and Laura Márquez and Nelly López (LANABIO) for their help with the use of the automatic sequencer.
Financial support
This project was partially funded by the Consejo Nacional de Ciencia y Tecnología (CONACyT) A1-S-21694, and by the Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT-UNAM) IN212621 to G.P.P.L.
Conflict of interest
None.
Ethical standards
Specimens in Mexico were collected under the Cartilla Nacional de Colector Científico (FAUT 0202 and 0057) issued by the Secretaría del Medio Ambiente y Recursos Naturales (SEMARNAT), to M.G.V. and G.P.P.L., respectively.






