Introduction
Trichuris trichiura (family Trichuridae) nematode parasite or whipworm causes trichuriasis disease in humans and occurs worldwide, mostly in warm humid tropical countries. It is among the soil-transmitted helminth (STH) infections categorized as the neglected tropical diseases (NTDs) and targeted for elimination (WHO, 2010, 2017). Trichuris trichiura infects an estimated 450 million people worldwide, mostly school-age children (G/hiwot et al., Reference G/hiwot, Degarege and Erko2014). Chronic trichuriasis causes anaemia and poor nutrition status, especially in children, while heavy worm burdens can result in ulcerative colitis and rectal prolapse (Njiru et al., Reference Njiru, Muhoho, Simbauni and Kabiru2016).
Diagnosis of trichuriasis, like other STH infections, relies on the microscopic detection of parasite eggs in faecal samples prepared using the Kato–Katz technique (Katz et al., Reference Katz, Chaves and Pellegrino1972) or the formol-ether concentration method (Allen & Ridley, Reference Allen and Ridley1970). While these procedures are relatively simple to perform and do not require expensive equipment, they have limited sensitivity and specificity, require expertise to make a correct diagnosis and can be tedious when processing large samples (George & McCulloch, Reference George and McCulloch2011), leading to increased cases of misdiagnosis (Njiru et al., Reference Njiru, Mikosza, Matovu, Enyaru, Ouma, Kibona and Ndung'u2016).
DNA-based diagnostic tests such as polymerase chain reaction (PCR) have proven to out-perform microscopy in their reliability to detect gastro-intestinal parasitic infections (Phuphisut et al., Reference Phuphisut, Yoonuan, Sanguankiat, Chaisiri, Maipanich, Pubampen and Adisakwattana2014; Acosta Soto et al., Reference Acosta Soto, Santisima-Trinidad, Bornay-Llinares, Martin Gonzalez, Pascual Valero and Ros Munoz2017). However, despite its high-throughput screening potential, PCR requires costly equipment (Kaisar et al., Reference Kaisar, Brienen, Djuardi, Sartono, Yazdanbakhsh, Verweij and Van Lieshout2017), and, therefore, has not been able to replace microscopy in the diagnosis of STH infections in large-scale epidemiological surveys or in disease control programs.
The loop-mediated isothermal amplification (LAMP) technique, developed by Notomi et al. (Reference Notomi, Okayama, Masubuchi, Yonekawa, Watanabe, Amino and Hase2000), offers a viable alternative to microscopy or PCR as it amplifies DNA with high specificity, efficiency and rapidity under isothermal conditions. This nucleic acid amplification technique requires only one enzyme (Bst DNA polymerase), and is able to amplify large amounts of DNA within 30–60 min by auto-strand displacement DNA synthesis (Notomi et al., Reference Njiru, Muhoho, Simbauni and Kabiru2000). The advantages of this approach include the use of simple equipment, since the reactions take place at isothermal temperatures between 60°C and 65°C, and amplification products can easily be visualized by the naked eye after staining with SYBR green, a DNA-binding dye (Leal et al., Reference Leal, Green, Allen, Humble and Rott2005). LAMP has emerged as a powerful tool for point-of-care diagnostics, and among the parasitic infections, LAMP diagnostic tests have been developed for the human malaria parasite Plasmodium falciparum (Han et al., Reference Han, Watanabe, Sattabongkot, Khuntirat, Sirichaisinthop, Iriko and Tsuboi2007), Schistosoma haematobium (Abbasi et al., Reference Abbasi, King, Muchiri and Hamburger2010), African trypanosomiasis (Kuboki et al., Reference Kuboki, Inoue, Sakurai, Di Cello, Grab, Suzuki, Sugimoto and Igarashi2003; Njiru et al., Reference Njiru, Mikosza, Matovu, Enyaru, Ouma, Kibona and Ndung'u2008), hookworm Necator americanus (Mugambi et al., Reference Mugambi, Agola, Mwangi, Kinyua, Shiraho and Mkoji2015), Ascaris lumbricoides (Shiraho et al., Reference Shiraho, Agola, Mwangi, Maina, Kinuthia, Mutuku and Mkoji2016) and Schistosoma mansoni (Mwangi et al., Reference Mwangi, Agola, Mugambi, Shiraho and Mkoji2018). Rapid and reliable diagnosis of trichuriasis is central to its control and elimination, as well as disease surveillance and monitoring. In the present study, we developed a LAMP test for the diagnosis of whipworm (T. trichiura) infections in human faecal samples.
Materials and methods
Study approval and ethical considerations
This study was approved by the Scientific and Ethical Review Unit (SERU) of the Kenya Medical Research Institute (KEMRI/SERU/CBRD/160/3397). Faecal samples used in this study were collected from school-going children in the endemic locality of Mwea, central Kenya. Faecal sample collection for diagnosis of trichuriasis and other common STH infection is a routine procedure and does not pose any risks or cause harm to participating children. The parents/guardians of the participating children gave written consent for their children to participate in the study, and children aged 12 years and above assented to participate. The samples of the study participants were coded and data stored in a password-protected personal computer. All the children found to be infected with T. trichiura or any other soil-transmitted intestinal helminth infection received treatment with a single dose of 400 mg albendazole and/or 40 mg/kg Paziquantel for Schistosomiasis, under the supervision of a clinician.
Parasite material
Faecal samples were collected from a total of 137 school-going children, aged 6–12 years from Mwea, Kirinyaga County, who were attending Mianya, Ngurubani and Mukou Primary Schools. Overall, the prevalence of T. trichiura infection among the children examined was 28%. Other faecal-based intestinal parasites present among the study children were hookworm (N. americanus), S. mansoni and A. lumbricoides.
Faecal sample collection and parasitological examination
Each participating child provided a single faecal sample in a capped plastic polypot, which were transported to the field laboratory in an ice box within 4 h of collection, and a double-slide Kato–Katz sample was prepared on glass microscope slides according to the standard procedure and examined under a compound microscope at X400 magnification within 1 hr of sample preparation (Katz et al., Reference Katz, Chaves and Pellegrino1972). Trichuris trichiura-positive samples were used to isolate ova for DNA extraction used for optimizing the LAMP technique. Similarly, faecal samples that were positive for N. americanus, S. mansoni and A. lumbricoides provided parasite ova for DNA extraction for evaluation of the LAMP test for specificity. Negative samples were also collected and used for the test of agreement between the Kato–Katz technique and LAMP.
Isolation of parasite eggs from faecal samples
To optimize the technique, parasite ova were isolated from faecal samples to avoid contamination with other materials in faecal samples. This was done using the Modified Wisconsin Sugar Flotation Method (Goodman et al., Reference Goodman, Haji, Bickle, Stoltzfus, Tielsch, Ramsan, Savioli and Albonico2007). Briefly, Sheather's solution was prepared by adding 454 gm of table sugar to 355 ml of very hot water and stirred until it dissolved. This was then allowed to cool before use. Approximately 6 gm of faecal sample was then placed in a 3.5-inch diameter petri dish and 25 ml of the Sheather's solution was added. The mixture was stirred into a homogenate using a wooden tongue depressor and filtered through layered wet cheesecloth into a 50 ml centrifuge tube. The filtrate was centrifuged at 4000 rpm for 15 min, and the solution was left to stand for 1 h to allow the parasite eggs to float. The presence of parasite ova was confirmed under the microscope by placing a cover slip over the solution and transferring it onto a microscope slide. Approximately 5 ml of the supernatant was transferred into a 15 ml centrifuge tube, and two volumes of water added to dilute the sugar solution to reduce the specific gravity. This allowed the eggs to settle at the bottom after a 5 min spin in the centrifuge at 4000 rpm. The resulting pellet was re-suspended in 2 m distilled water, vortexed and then transferred into 1.5 ml centrifuge tubes.
DNA extraction from isolated parasite eggs and from crude faecal samples
DNA extraction from isolated parasite eggs was performed using the modified alkaline lysis DNA extraction (HotSHOT) method (Truett et al., Reference Truett, Heeger, Mynatt, Truett, Walker and Warman2000). Briefly, 10 μl of concentrated eggs was added to 30 μl of lysis buffer (sodium hydroxide, ethylenediaminetetraacetic acid and distilled water), followed by sample incubation at 95°C for 1 h. After incubation, the sample was rapidly cooled to 4°C followed by the addition of 30 μl of lysis buffer (Tris-hydrochloride and distilled water).
Genomic DNA was extracted from crude freshly collected human faecal samples using the QIAamp DNA Stool Mini Kit (QIAGEN, Massachusetts, USA; catalogue no. 51504) as per the manufacturer's instructions.
Primer design and selection
LAMP primers were designed, based on the internal transcribed spacer 2 (ITS-2) ribosomal DNA of T. trichiura, using Primer Explorer version 5.0 software (Fujitsu Ltd., Tokyo, Japan). This region was chosen due to its conserved nature and specificity to T. trichiura. Five generated primer sets were subjected to computation analysis by the basic local alignment search tool (BLAST; https://blast.ncbi.nlm.nih.gov/Blast.cgi) to verify their suitability and close relatedness to the T. trichiura from GenBank with sequence accession number GQ301555.1 (T. trichiura from Homo sapiens internal transcribed spacer 1, partial sequence; 5.8S ribosomal RNA gene, complete sequence; and ITS-2, partial sequence of a length of 1404 bp.). The one primer set (having FIP, BIP, F3 and B3) that generated reproducible results in optimization tests was chosen for subsequent experiments (table 1).
Table 1. Primer sequences used for the amplification of Trichuris trichiura ova.

Optimization of the LAMP reaction conditions
The LAMP assay was optimized to determine the optimum incubation temperature, reaction time and primer ratios. Temperatures from 60°C to 65°C, reaction time from 30 min to 120 min and primer ratios of inner (FIP,BIP):outer (F3,B3) were tested. In this assay, optimum LAMP reaction conditions were found to be temperatures of 63°C for 60 min, and primer ratios were 4:2.5 (FIP,BIP:F3,B3), respectively.
LAMP reaction
LAMP reaction was performed as previously described (Notomi et al., Reference Njiru, Muhoho, Simbauni and Kabiru2000). Initial optimization reactions were done using a thermal cycler (GeneAmp PCR System 9700, Applied Biosystems, Singapore), but all other subsequent experiments (sensitivity, specificity and evaluation) were performed on a water bath and the reaction stopped in boiling water to inactivate the Bst enzyme. The reaction mix had a final volume of 27 μl containing 1.48 pmoles (FIP and BIP) and 0.93 pmoles (F3 and B3), 1 μl of 8 U/μl of Bst DNA polymerase (New England BioLabs, Ipswish, Massachusetts, USA), 10× Bst buffer and 1 μl(10 ng/μl) target DNA. Amplification was carried out at 63°C for 60 min, and Bst DNA polymerase activity stopped by boiling in water for 10 min.
LAMP product detection
LAMP products were detected using two methods: (1) electrophoresis on a 2.0% agarose gel stained with SYBR Safe DNA gel stain (InVitrogen by Thermo Fisher Scientific, Massachusetts, USA) and photographed on a gel documentation system (UVP VisiDoc-It Imaging System, CA, USA); and (2) staining the amplified products with SYBR green nucleic acid stain and visualization with the naked eye. Positive samples turned green in colour and the negative samples remaining orange in colour.
Evaluation of the LAMP assay for specificity and sensitivity in comparison with the Kato–Katz technique
The performance of the LAMP assay for detection of T. trichiura was evaluated for specificity and sensitivity against the microscopy-based Kato–Katz technique using 137 faecal samples collected from schoolchildren from Mwea.
To determine the specificity of the LAMP assay in detecting T. trichiura DNA, the LAMP primers designed for this purpose were tested in the assay using DNA extracted from the other common, faecal-based helminth parasites – namely, A. lumbricoides, hookworm (N. americanus) and S. mansoni – and the amplification products analysed by agarose gel electrophoresis and SYBR green staining alongside the amplified T. trichiura DNA.
To determine the analytical sensitivity of the assay, 1:10 serial dilutions of T. trichiura DNA were prepared from an initial solution at 98.6 ng/μl, and the DNA samples from each dilution were then amplified separately in the assay. The amplified products from each dilution were visualized on stained agarose gel after electrophoresis.
Data analysis
For the entire 137 human faecal samples, parasite detection was done using both the Kato–Katz procedure and the developed LAMP assay, and the results recorded as either positive or negative. The data were then analysed for sensitivity and specificity using the following formula developed by Cohen (Reference J1960):

where K is kappa value, OA is observed agreement and AC is agreement by chance.
Results
Primer design and selection for the LAMP reaction
Five primer sets were generated by Primer Explorer software version 5 (Fujitsu Ltd., Tokyo, Japan). After trying all primer combinations, one set of primers (table 1) gave consistent and reliable amplification. This was subsequently used in all the analysis and optimization of the technique.
LAMP reaction optimization
Primer concentration ratios
When ratios of different primer concentrations of inner (FIP, BIP):outer (F3, B3) between 1:1 and 8:10, respectively, were carried out, the most optimum ratio based on the brightness of the amplicon bands on the agarose gel after electrophoresis was observed to be 4:2.5 (fig. 1).
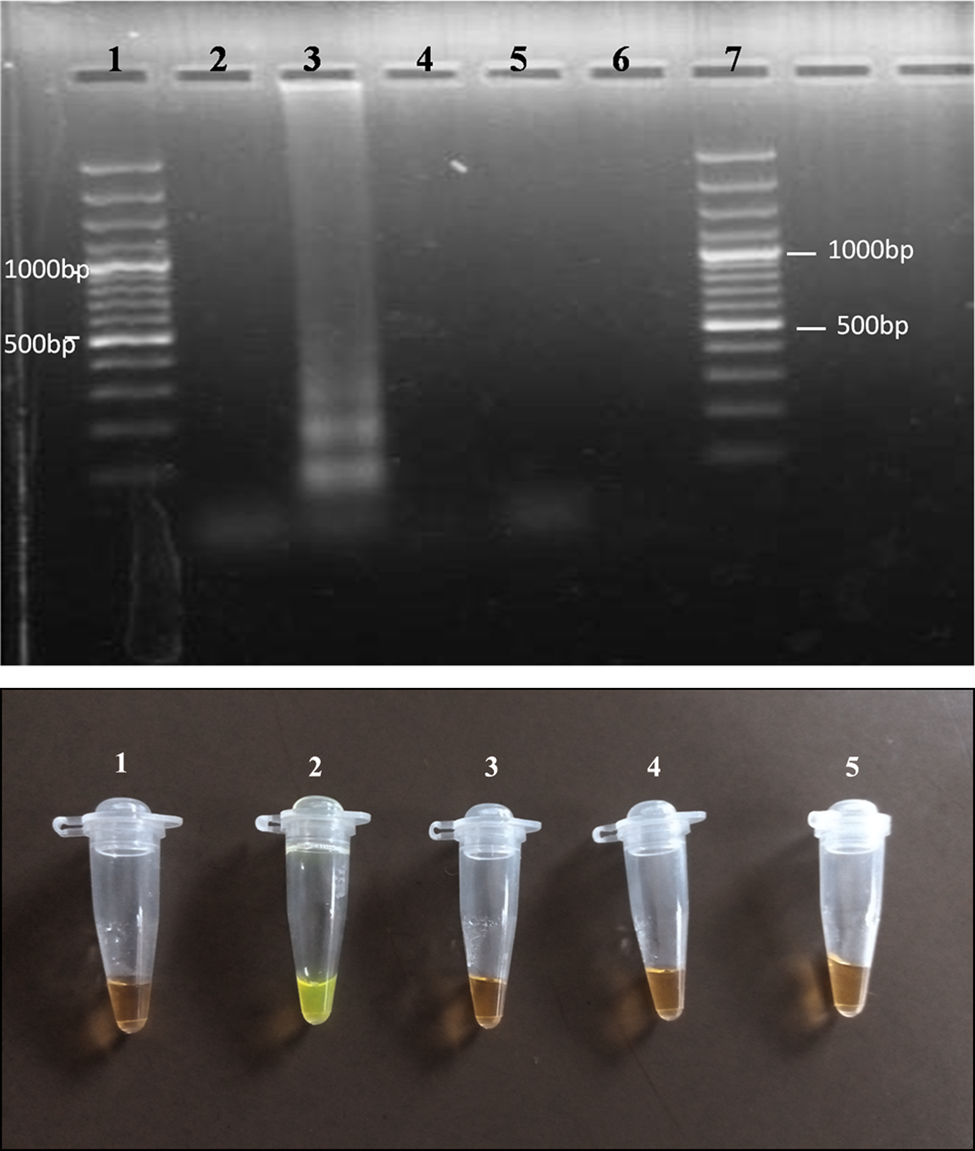
Fig. 1. (a) Agarose gel photo showing results of the LAMP assay when varying primer ratios. Lanes 2, 3, 4 and 5 represent ratios 1:1, 4:2.5, 6:5 and 8:10, respectively. Lane 6 is the negative control (reaction mix without DNA), and Lanes 1 and 7 are 100-bp molecular markers. (b) LAMP visual detection for colour change using the SYBR green dye. Tubes 1, 2, 3 and 4 represent primer ratios 1:1, 4:2.5, 6:5 and 8:10, respectively. Tube 5 is the negative control.
Reaction temperature
When different isothermal temperature conditions were tested between 60°C and 65°C, based on the brightness of DNA amplicons, the optimal temperature for reaction mix was determined to be 63°C (fig. 2)

Fig. 2. (a) Agarose gel photo showing amplified fragments at temperatures in the range of 60–65°C for Lanes 2, 3, 4 and 5 representing 60°C, 62°C, 63°C and 65°C, respectively. Lane 6 is the negative control (reaction mix without DNA), Lanes 1 and 7 are 100-bp molecular markers. (b) Detection of amplicon by the naked eye based on colour change using the SYBR green dye. Tubes 1, 2, 3 and 4 represent amplification at temperatures of 60°C, 62°C, 63°C and 65°C, respectively. Tube 5 represents the negative control (reaction mix without DNA).
Ability to detect single whipworm egg DNA
Whipworm parasite eggs were isolated from fresh faecal samples, counted and subjected to LAMP assay to determine the optimum number that can be successfully amplified; 1, 5, 10 and 15 eggs were tested, respectively. Amplification was possible even for a single egg (fig. 3).

Fig. 3. (a) Agarose gel showing amplification of isolated egg(s). Lanes 2, 3, 4 and 5 are DNA amplification from 1, 5, 10 and 15 eggs, respectively. Lane 6 is the negative control (reaction mix without DNA) and Lanes 1 and 7 are 100-bp molecular markers. (b) Detection of amplicons using SYBR green dye. Tubes 1, 2, 3 and 4 represent amplifications of 1, 5, 10 and 15, respectively. Tube 5 is the negative control (reaction mix without DNA).
Specificity of the LAMP reaction
Only samples of T. trichiura were amplified with no cross-reactivity or amplification observed with closely related parasites, illustrating the high specificity of the developed LAMP assay, as in fig. 4.

Fig. 4. (a) Agarose gel photo showing results for the specificity of the LAMP assay for Trichuris trichiura. Lanes 2 and 3 are T. trichiura DNA at 82.8 ng/μl and 99.2 ng/μl, respectively. Lane 4 is Schistosoma mansoni DNA at 98.6 ng/μl, Lane 5 is Necator americanus DNA at 90.6 ng/μl and Lane 6 is Ascaris lumbricoides at 50.2 ng/μl. Lane 7 is a negative control (master mix without DNA). Lanes 1 and 8 represent 100-bp molecular markers. (b) Detection using the SYBR green dye. Tubes 1 and 2 represent amplification of T. trichiura DNA, and 3, 4 and 5 are S. mansoni DNA, N. americanus DNA and A. lumbricoides DNA, respectively. Tube 6 represents a negative control (master mix without DNA).
Sensitivity of the LAMP reaction
When 1:10 serial dilutions of total genomic DNA from T. trichiura were used to test the sensitivity of the developed LAMP assay from an initial concentration of 98.6 ng/μl, the LAMP assay was able to amplify DNA concentration as low as 98.6 × 10−9 ng/μl (fig. 5).

Fig. 5. Agarose gel photo of the LAMP assay amplified products of different concentrations of Trichuris trichiura DNA, following 1:10 serial dilutions, starting with a 98.6 ng/μl DNA concentration (Lane 2) to 98.6 × 10−9 ng/μl (Lane 11). Lane 12 is the negative control (master mix with no DNA), and Lanes 1 and 13 are 100-bp molecular markers.
Comparison of LAMP against Kato–Katz
When the performance of the developed LAMP technique was compared to the Kato–Katz technique, results displayed in a 2 × 2 contingency table, and sensitivity, specificity and predictive values determined, our results indicated that the LAMP technique had a sensitivity of 77% compared to that of Kato–Katz. The specificity was 88% (table 2).



Table 2. Contingency table comparing LAMP assay with the Kato–Katz technique.

showing substantial agreement.


This shows that 34 samples of 137 were positive for T. trichiura on both LAMP and Kato–Katz, 11 samples were negative on Kato–Katz, which were positive on LAMP, ten samples were positive on Kato–Katz, which were negative on LAMP, whereas both Kato–Katz and LAMP agreed 82 samples were negative.
Discussion
In this paper, we present a LAMP method for the diagnosis of T. trichiura (whipworm) infection in faecal samples, with the ability to detect as low as 98.6 × 10−9 ng/μl parasite DNA, suggesting that it has the ability to detect very low levels of T. trichiura infections, which are typical of endemic areas where mass drug administration (MDA) is intensely used for control or elimination efforts against T. trichiura infections and other STH infections that often co-occur. Sensitive and accurate diagnostic tools for whipworm infections are critical for the detection of low-intensity infections such as those likely to be encountered where MDA is applied intensely or where low-level transmission of whipworm occurs (Webster et al., Reference Webster, Molyneux, Hotez and Fenwick2014).
Although this assay was optimized using a thermocycler, all subsequent assays were done using a temperature controlled water bath to reliably amplify parasite DNA in the developed LAMP assay, suggesting its potential value for use in low-resource disease-endemic areas, where costly and sophisticated equipment may not be affordable for routine purposes or at the point of care (Amoah et al., Reference Amoah, Singh, Stenstrom and Reddy2017).
In previous studies, it was shown that five of 12 (41.6%) parasitological negative cases by Kato–Katz gave positive results with PCR, indicating a clear case of misdiagnosis by Kato–Katz examination (Pontes et al., Reference Pontes, Oliveira, Katz, Dias-Neto and Rabello2003). Lower sensitivities of Kato–Katz (between 46.0 and 71.8) have also been observed (Knopp et al., Reference Knopp, Rinaldi, Khamis, Stothard, Rollinson, Maurelli and Utzinger2009). The low sensitivities of a single Kato–Katz thick smear (8.0% to 22.4%) have been attributed to low infection intensities and day-to-day variation in egg output (Booth et al., Reference Booth, Vounatsou, N'goran, Tanner and Utzinger2003). Our findings indicate that LAMP has a sensitivity of 77% and a specificity of 88%, with the ability of detecting single egg DNA, showing that it is more reliable compared to the Kato–Katz technique.
The generated primers were based on the ITS-2 of the ribosomal DNA, a region chosen due to the fact that it is conserved and has low mutation rates, and, also, is known to provide species-specific markers for differentiating even closely related species. In the present study, the primers designed were highly specific to T. trichiura, and had no cross-reactivity with other commonly occurring faecal-based helminth parasites such as A. lumbricoides, S. mansoni and hookworm (N. americanus). This is an important consideration since in many endemic localities, multiple helminth infections do frequently occur, and very often in one individual (Harris et al., Reference Harris, Worrell, Davis, Odero, Mogeni, Deming and Addiss2015).
LAMP has characteristics that give it an advantage over conventional PCR, making it ideal for use in a field set-up. For example, it does not require expensive equipment, as a simple water bath or heat block are used for isothermal amplification. Also, the simple detection system based on the staining of the amplicons with SYBR green nucleic acid stain, which is visualized by the naked eye, further simplifies the technique, giving it the potential to be used at the point of care in low-resource disease-endemic regions. The LAMP technique has already been used in rural laboratories in developing areas for the diagnosis of tuberculosis (Boehme et al., Reference Boehme, Nabeta, Henostroza, Raqib, Rahim, Gerhardt and Perkins2007), trypanosomiasis (Njiru et al., Reference Mwangi, Agola, Mugambi, Shiraho and Mkoji2008) and asymptomatic malaria (Cook et al., Reference Cook, Aydin-Schmidt, González, Bell, Edlund, Nassor and Björkman2015).
While the LAMP diagnostic test has proved reliable and can be useful for field application, we do acknowledge the fact that it requires further improvements to make it applicable for routine use. For example, whether one is using a thermal cycler or a water bath, electricity is still required. This causes an addition of cost that makes it not applicable yet under field settings. Other improvements needed include a simple and precise DNA extraction method that will void the samples of polyphenols found in stool samples, which inhibits DNA polymerase. Also, pre-mixed and ready-to-use LAMP reagents are needed to minimize pipetting, which may result in contamination.
In conclusion, the developed LAMP has the advantage in specificity and sensitivity over the Kato–Katz method, and the assay described and developed in this study may constitute a turning point in the management of T. trichiura. The LAMP technique remains a ‘Gold Standard’ by which other isothermal DNA/RNA amplification methods are compared. It has the potential for use in the rapid diagnosis of T. trichiura infection, both in the laboratory and in a field set-up.
Acknowledgements
We are indebted to the parents/guardians of the children who provided the stool samples and the teachers and pupils of primary schools in Mwea for providing samples. We also acknowledge the contribution of Stephen Kamau, Martin Mutuku, Joseph Kinuthia, Geoffrey Maina and Ruth Wachira for their assistance with fieldwork. Special thanks to the staff of Kimbimbi Sub-County Hospital, for allowing the authors to use their laboratory for sample processing. This manuscript has been published with the approval of the Director General of KEMRI.
Financial support
This study was supported by a KEMRI Internal Research Grant (grant number 0219).
Conflicts of interest
None.
Ethical standards
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008. The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional guides on care and use of laboratory animals.









