Introduction
Nematodes include an enormously various group of creatures with the most members in marine and terrestrial habitats that show a significant portion of the soil community (Kaya & Gaugler, Reference Kaya and Gaugler1993). Entomopathogenic nematodes (EPNs) and other parasitic nematodes can cause the death of insects and slugs. In the last decade, they have been utilized as biological approaches to control a range of pests (Poinar, Reference Poinar1979; Hazir et al., Reference Hazir, Kaya, Stock and Keskin2003; Nguyen et al., Reference Nguyen, Půža and Mracek2008; Stock et al., Reference Stock, Rivera-Orduno and Flores-Lara2009; Tamura et al., Reference Tamura, Peterson, Peterson and Stecher2011; Kanzaki et al., Reference Kanzaki, Ragsdale, Herrmann, Roseler and Sommer2013; San-Blas et al., Reference San-Blas, Morales-Montero, Portillo, Nermut and Půža2016; Malan & Ferreira, Reference Malan, Ferreira, Fourie, Spaull, Jones, Daneel and De Waele2017; Koppenhöfer et al., Reference Koppenhöfer, Shapiro-Ilan and Hiltpold2020; Sivaramakrishnan & Razia, Reference Sivaramakrishnan and Razia2021). Two families of nematodes, Steinernematidae and Heterorhabditidae (Rhabditida), are the most known nematodes for the utilization as biocontrol agents. These nematodes can control insect pests which are widely dispersed in the soils across the globe and have a benign effect on vertebrates and plants since they are mutually associated with pathogenic bacteria (Kaya & Gaugler, Reference Kaya and Gaugler1993; Gaugler, Reference Gaugler2002; Grewal et al., Reference Grewal, Ehlers and Shapiro-Ilan2005; Ye et al., Reference Ye, Torres-Barragan and Cardoza2010; Torres-Barragan et al., Reference Torres-Barragan, Suazo, Buhler and Cardoza2011). Furthermore, other members of the Rhabditida indicated potential to infect insects and are promising as new candidates for biological control of pests (Sudhaus, Reference Sudhaus2011; Liu et al., Reference Liu, Mrácěk, Zhang, Půža and Dong2012; Zhang et al., Reference Zhang, Liu and Tan2012). The genus Oscheius belongs to the Rhabditidae family and contains free-living nematodes (Andrássy, Reference Andrássy1976) that feed on bacteria, but some are reported as entomopathogenic (Tabassum & Shahina, Reference Tabassum and Shahina2010; Ye et al., Reference Ye, Torres-Barragan and Cardoza2010; Liu et al., Reference Liu, Mrácěk, Zhang, Půža and Dong2012; Pervez et al., Reference Pervez, Eapen, Devasahayam and Jacob2013; Torrini et al., Reference Torrini, Mazza, Carletti, Benvenuti, Roversi, Fanelli, de Luca, Troccoli and Tarasco2015; Castro-Ortega et al., Reference Castro-Ortega, Caspeta-Mandujano, Suárez-Rodríguez, Peña-Chora, Ramírez-Trujillo, Cruz-Pérez, Sosa and Hernández-Velázquezl2020) or scavenging behaviour (Campos-Herrera et al., Reference Campos-Herrera, Půža, Jaffuel, Blanco-Pérez, Čepulyte-Rakauskiene and Turlings2015; Zhang et al., Reference Zhang, Baiocchi, Lu, Chang and Dillman2019). Due to this, Oscheius has received increased attention lately. However, heretofore the number of valid species of Oscheius has been expressed as 45 species (Zhou et al., Reference Zhou, Yang, Wang, Bao, Wang, Hou, Lin, Yedid and Zhang2017; Serepa-Dlamini & Gray, Reference Serepa-Dlamini and Gray2018; Kumar et al., Reference Kumar, Jamal, Somvanshi, Chauhan and Mumtaz2019), but recently the verifiable species of this genus have been reported as 39. The revised list includes two subgenera, Oscheius Andrássy, Reference Andrássy1976, (including 16 species), and Dolichorhabditis Andrássy, 1983, (containing 23 species) (Abolafia & Peña-Santiago, Reference Abolafia and Peña-Santiago2019; Castro-Ortega et al., Reference Castro-Ortega, Caspeta-Mandujano, Suárez-Rodríguez, Peña-Chora, Ramírez-Trujillo, Cruz-Pérez, Sosa and Hernández-Velázquezl2020). The species belonging to the insectivore-group are characterized by leptoderan bursa, crochet needle-shaped spicules and normal rectum, while the dolichura-group has peloderan bursa, probe head spicule tips and expandable rectum (Sudhaus, Reference Sudhaus1976). In Iran, few studies have been done on the identification, ecology, and biology of Oscheius spp., and few species including, Oscheius chongmingensis (Darsouei et al., Reference Darsouei, Karimi and Shokoohi2014) (Oscheius rugaoensis newly was proposed as a junior synonym of O. chongmingensis by Bhat et al., Reference Bhat, Gautum, Rana, Chaubey, Abolafia and Půža2021), Oscheius necromenus, Oscheius onirici, O. Oscheius tipulae (Valizadeh et al., Reference Valizadeh, Goldasteh, Rafiei-Karahroodi and Pedram2017; Karimi et al., Reference Karimi, Rezaei and Shokoohi2018) and Oscheius myriophilus (Gholami Ghavamabad et al., Reference Gholami Ghavamabad, Talebi, Mehrabadi, Farashiani and Pedram2021) have been reported until now.
Recent evidence has supported the pathogenicity of Oscheius spp. on insects. This genus has mutually associated with 19 bacterial genera (Fu & Liu, Reference Fu and Liu2019), mainly with the bacteria belonging to genera Serratia, Enterococcus, Ochrobactrum and some genera dependent on the order of Rhizobiales (Liu et al., Reference Liu, Mrácěk, Zhang, Půža and Dong2012, Reference Liu, Zhang and Zeng2016; Zhang et al., Reference Zhang, Liu and Tan2012; Lephoto et al., Reference Lephoto, Featherston and Gray2015; Zhou et al., Reference Zhou, Yang, Wang, Bao, Wang, Hou, Lin, Yedid and Zhang2017; Serepa-Dlamini & Gray, Reference Serepa-Dlamini and Gray2018; Fu & Liu, Reference Fu and Liu2019; Lephoto & Gray, Reference Lephoto and Gray2019; Castro-Ortega et al., Reference Castro-Ortega, Caspeta-Mandujano, Suárez-Rodríguez, Peña-Chora, Ramírez-Trujillo, Cruz-Pérez, Sosa and Hernández-Velázquezl2020).
The genus Ochrobactrum belongs to the family Brucellaceae (Rhizobiales: Alphaproteobacteria) and contains pervasive and multifunction bacteria with extensive ecological niches.
Some species in the Ochrobactrum genus, Ochrobactrum anthropi and Ochrobactrum intermedium, almost have been recognized as opportunistic human pathogens. Until now, there had been reported only one Ochrobactrum species as an associated bacterium from Rhabditis (Oscheius) sp.; named Ochrobactrum tritici isolated from Oscheius chongmingensis (Fu & Liu, Reference Fu and Liu2019). Ochrobactrum pseudogrignonense is discovered as the dominant bacterium for the first time.
On the other hand, the combination of related bacteria and their potential attendant in this nematode species have been insufficiently described (Fu & Liu, Reference Fu and Liu2019). So, it is necessary to obtain a better insight into bacteria–Oscheius relationships (Aujoulat et al., Reference Aujoulat, Pagès, Masnou, Emboulé, Teyssier, Marchandin, Gaudriault, Givaudan and Jumas-Bilak2019).
Moreover, the new species, Oscheius cyrus n. sp., and its dominant attendant bacterium were described for the first time globally.
So, this study aims to introduce and identify the new species of collected nematodes belonging to the Oscheius genus with comprehensive details, including molecular and morphological traits of the nematode and the introduction of its dominant bacterium.
Furthermore, the efficiency of the new nematode species and its dominant bacterium was evaluated in suppressing the Galleria mellonella (Linnaeus, 1758) larvae as an insect model.
Materials and methods
Nematode isolation from soil
During the summer of 2018 and 2019, a survey was conducted from 10 plots of the heights of Dalkhani forest, Ramsar city, Mazandaran Province (36.3822° N, 48.5550° E) from Iran. Samples were individually collected in the moist soils from the surface to a depth of 20–25 cm, by a small shovel. They were put in plastic bags and transported to the laboratory, mixed and placed in plastic bowls (300 ml) with lids. The nematodes were collected by baiting with 10 last instar larvae of G. mellonella in each bowl. Then, they were maintained at room temperature (25 ± 2°C) for 10 days (Bedding & Akhurst, Reference Bedding and Akhurst1975). The killed larvae of G. mellonella were recognizable via colour change due to infection. Mixed stages of nematodes emerged from the dead body of Galleria and were collected from the white trap 10 days post-inoculation (White, Reference White1927). Koch's postulates were conducted by about 100 infective juveniles (IJs) on G. mellonella (Kaya & Stock, Reference Kaya, Stock and Lacey1997). Then, they were stored in the refrigerator at 8–10°C for subsequent use.
Morphological observation
About 30–40 females and juveniles were randomly selected from cadavers of G. mellonella for morphological characterization. They were fixed using hot (80°C) formaldehyde 4% solution and transmitted to anhydrous glycerine for mounting (Ryss, Reference Ryss2017). Using an ocular micrometer, measurements and microphotographs of fixed nematodes were collected, and a drawing tube coupled to the microscope was used to create drawings. The ratio measurements were based on specimens put on slides.
Scanning electron microscope (SEM)
Females and juveniles of nematodes were fixed in glutaraldehyde 3% with 0.1 M sodium cacodylate at pH 7.2 for 24 h at 4°C in the dark. Then, the samples were postfixed with 2% osmium tetroxide solution for 16 h at laboratory temperature. Then, they were washed with 0.1 M sodium cacodylate three times and were dehydrated through a graded ethanol series with 10, 20, 30, 40, 50, 60, 70, 80, 90, 95 and 100%, each for 20 min, all on dried ice (dehydrated with 100% ethanol three times at room temperature). Finally, they were mounted on stubs, and gold-coated by mini sputter coater SC7620 (Quorum Technologies, UK). The SEM images of the nematodes were attained with an LEO 1450VP SEM (LEO Co. Ltd., Germany) (Ye et al., Reference Ye, Torres-Barragan and Cardoza2010; Nikdel & Niknam, Reference Nikdel and Niknam2015).
DNA extraction, polymerase chain reaction (PCR) and sequencing
The DNA was extracted from a single female nematode. The nematode was transmitted by eyelash into an Eppendorf tube. After grinding in 50 μl 5% Chelex-100 (Sigma Aldrich, Germany) and two μl proteinase K, it was incubated at 60°C for three hr. Then, placed at 95°C for 10 min. In the end, it centrifuged at 13,000 rpm for 3 min, and the extracted DNA was stored at −20°C for subsequent use. For molecular characterization of the isolate, a molecular strategy including sequencing analysis of tree genes, internal transcribed spacer (ITS), 18S, and 28S was examined. The primers for the amplification of ITS gene were 5′-GTTTCCGTAGGTGAACCTGC-3′ (TW81, forward), 5′-ATATGCTTAAGTTCAGCGGGT-3′ (AB28, reverse) (Joyce et al., Reference Joyce, Reid, Driver, Curran, Burnell, Ehlers and Masson1994). The other region comprising 18S rDNA (small subunit (SSU)) was distinguished using 5′-AAAGATTAAGCCATGCATG-3′ (18A, forward) and 5′-CATTCTTGGCAAATGCTTTCG-3′ (26R, reverse) (Blaxter et al., Reference Blaxter, de Ley, Garey, Liu, Scheldeman, Vierstraete, Vanfleteren, Mackey, Dorris, Frisse, Vida and Thomas1998). The section including the D2D3 expansion segments of 28S rDNA (large subunit (LSU)) amplification was identified using 5′-ACAAGTACCGTGAGGGAAAGT-3′ (D2A, forward) and 5′-TGCGAAGGAACCAGCTACTA-3′ (D3B, reverse) (Nunn, Reference Nunn1992). The PCR mixture was carried out in a reaction volume of 25 μl, including 12.5 μl 2X Taq PreMix, 6.5 μl sterilized water, one μl of each forward and reverse primers (10 pmol/μl) and four μl DNA. Then, all PCR products were loaded on 1% agarose gel. PCR products were electrophoresed at 100 V for 40 min and the green viewer was used for gel staining. At the end, the PCR products were sequenced by Macrogen Co. (Korea).
Multiple alignments and phylogenetic analysis
The quality of chromatograms was checked, and the sequences from this study were compared with other nematode species available in GenBank utilizing the Basic Local Alignment Search Tool (BLAST) program to confirm the sequences. Geneious prime software (version 2019.1.3; (https://www.geneious.com)) was used to assemble and trim the consensus sequence for all tree genes. According to Bhat et al. (Reference Bhat, Gautum, Rana, Chaubey, Abolafia and Půža2021), legitimate and validated sequences of the genus and outgroup sequences were chosen for phylogenetic analysis. The sequences for all three amplified locus (ITS, SSU and LSU) were aligned using the ClustalX with default parameters and the alignment was manually edited in Molecular Evolutionary Genetics Analysis (MEGA) 7 (Kumar et al., Reference Kumar, Stecher and Tamura2016). The number of base differences per site were computed using Geneious, and pairwise distances were computed using MEGA 7.0 by Kimura-2 model and 10,000 bootstraps (Kumar et al., Reference Kumar, Stecher and Tamura2016).
The best fit model was identified under the GTR + I + G (ITS and SSU) and GTR + G (LSU) model using the MrModeltest 2 (Nylander, Reference Nylander2004). Bayesian analysis was performed using MrBayes 3.1.2 (Ronquist & Huelsenbeck, Reference Ronquist and Huelsenbeck2003) for two million generations for all datasets. To estimate the posterior probabilities of the phylogenetic trees, the Markov chain Monte Carlo chains were sampled every 100 generations (Larget & Simon, Reference Larget and Simon1999) using the 50% majority rule. Then, the burn-in step was set at 25% of the converged runs. The Dendroscope V.3.5.7 software (Huson & Scornavacca, Reference Huson and Scornavacca2012) was used to visualize the output file of the phylogenetic program, and the tree was resized in CorelDRAW software version 2020.
Laboratory bioassay
Efficacy on G. mellonella
The bioassay was conducted to determine the lethal concentration of nematodes against the last instar larvae of G. mellonella under laboratory conditions. Five concentrations of nematodes (25, 50, 100, 200 and 400 IJs/larva) were inoculated in moistened filter papers in 6-centimetre-diameter in Petri dishes. Each treatment (concentration) with the control, tree duplicates (10 larvae per replicate) were used. The total quantity of water in Petri dishes was the same (400 μl). Until one week post-infection, dead larvae were documented. The experiment was repeated three times.
Isolation of bacterial strain from nematode
After disinfecting about 100 juveniles with 10% sodium hypochlorite for 10 min, they were washed three times with sterilized water to remove any remaining sodium hypochlorite. Then, juveniles were crushed using a modified pipette in 10 μl sterile water. Finally, the obtained suspension was cultured using a nutrient agar (NA) with 0.04 g (w/v) triphenyltetrazolium chloride and 0.025% (w/v) bromothymol blue (NBTA) at 28°C for 48 h at dark.
They were cultured on NBTA medium (NA supplemented with 0.025 g bromothymol blue and 0.04 g 2,3,5-triphenyltetrazolium chloride per litre) (Woodring & Kaya, Reference Woodring and Kaya1988).
DNA extraction, PCR, DNA sequencing and phylogenetic analyses
The DNA content of the Ochrobactrum strain was acquired using PCR cloning (Mcpherson & Møller, Reference Mcpherson and Møller2006). To identify bacteria, partial sequences of the 16S ribosomal RNA (rRNA) gene were used (with universal bacterial primers: 27 F and 1492R). 35 cycles of 1 min at 94°C, 1 min at 56.5°C, and 2 min at 72°C comprised the PCR conditions. The agarose gel containing PCR results and a molecular DNA ladder from Pars Tous, Iran, was subsequently delivered to Macrogen Co. (Korea) for sequencing. The 16S rRNA sequence was edited using Geneious prime software and was compared with the sequences deposited in GenBank sequencing-genome databases by the BLAST program (http://www.ncbi.nlm.nih.gov/blast). To reconstruct the phylogenetic tree, MEGA7 was used by the neighbour-joining method with 10,000 bootstrap replications and Kimura 2-parameter model to determine the number of base compositions per site in units (Kumar et al., Reference Kumar, Stecher and Tamura2016). Mycoplana dimorpha (Gray and Thornton 1928) DSM strain (NR116128), Lutibaculum baratangense (Anil Kumar et al. 2012) AMV1 strain (FN297835) and Oligotropha carboxidovorans (Paul et al., Reference Paul, Bridges, Burgess, Dandass and Lawrence2008) OM5 strain (NR074142) were used as outgroups.
Bacteria phenotypic characterization
A few significant phenotypic characterizations of the associated bacterium, such as morphological characters (size, shape and colour), dye absorb (Akhurst, Reference Akhurst1986), antibiotic test (Kazmierczak et al., Reference Kazmierczak, Bieden ach, Hackel, Rabine, De Jonge, Bouchillon, Sahm and Bradford2016), catalase test, movement test and lipase activity were investigated (Akhurst & Boemare, Reference Akhurst and Boemare1988;Tailliez et al., Reference Tailliez, Laroui, Ginibre, Paule, Pagès and Boemare2010).
Furthermore, the last instar larvae of G. mellonella were used to evaluate the pathogenicity of the bacterium according to a method described by Peel et al. (Reference Peel, Alfredson, Gerrard, Davis, Robson, Mcdougall, Scullie and Akhurst1999).
Results
A thorough method was used to identify the isolated populations as O. cyrus n. sp. Female and juvenile morphological traits are listed in table 1. The morphological characteristics of new species were very close to O. myriophilus species from Iran and China with accession number MW430436 and KT825914, respectively.
Table 1. Morphometrics of Oscheius cyrus. n. sp. Measurements in μm and in the form: mean ± standard deviation (range).

a: total body length/greatest body diam or greatest body Width(L/W); b: total body length/pharynx length (L/ES); c: total body length/tail length(L/T); ć: Tail length/Anal body diam (T/ABD), D%: excretory pore position/ pharynx length*100 (EP/ES*100); E%: excretory pore position/tail length (EP/T*100).
Oscheius cyrus n. sp.
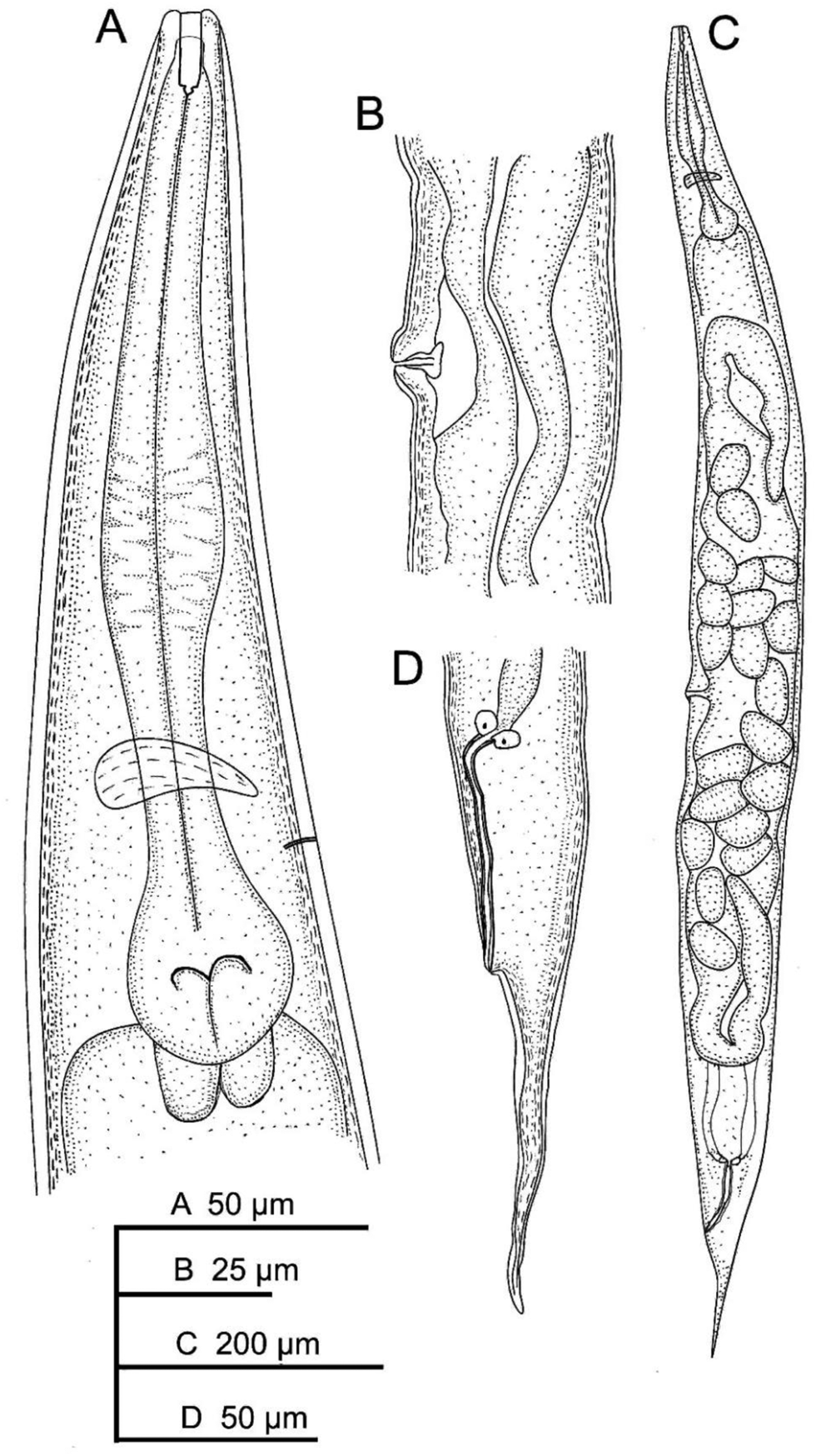
Fig. 1. Oscheius cyrus sn. sp.: (A) female anterior end; (B) vaginal region; (C) entire female; and (D) female posterior end.
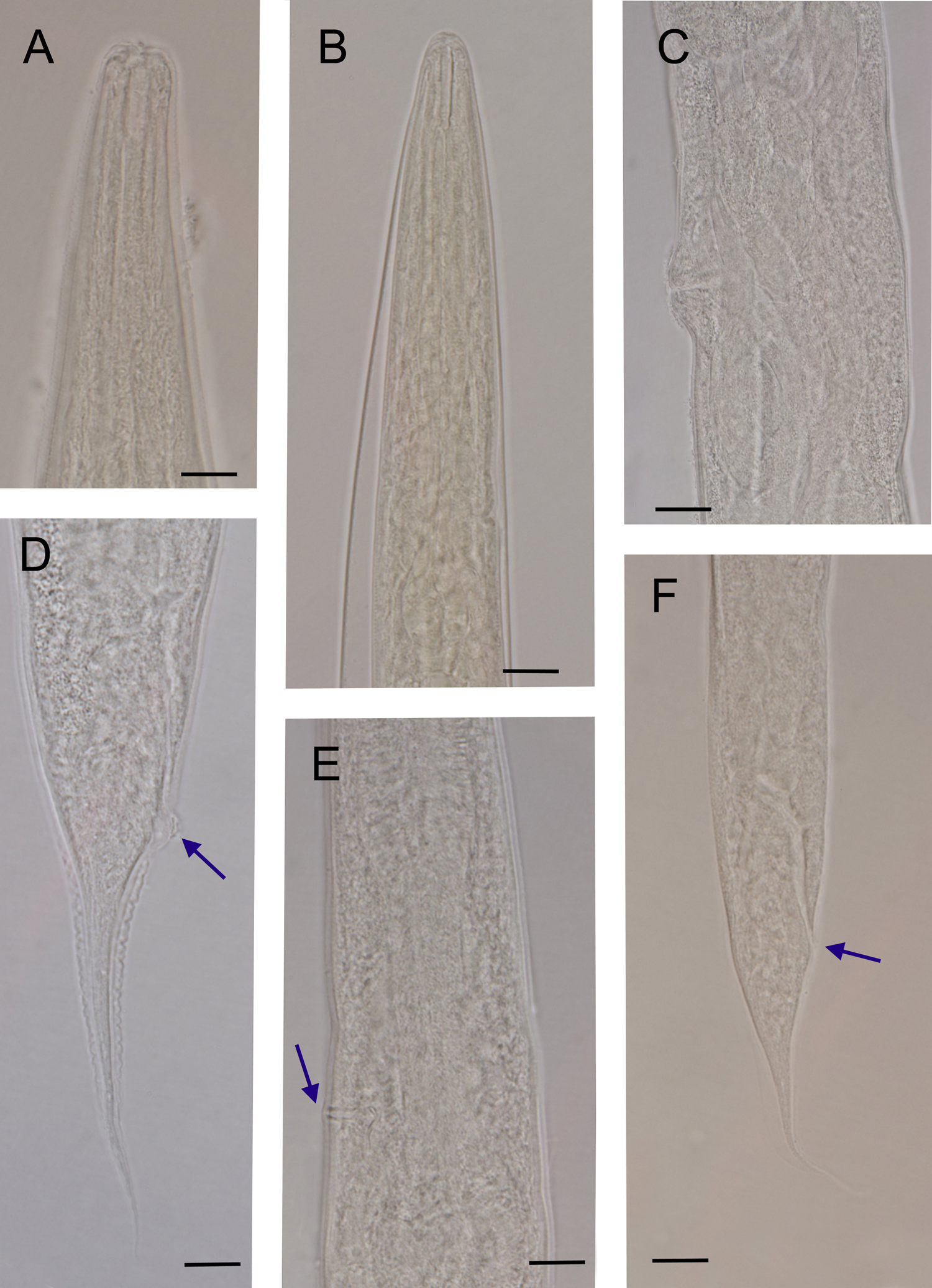
Fig. 2. Oscheius cyrus n. sp. (light microscope): (A) anterior end of female; (B) anterior end of infective juvenile; (C) vaginal region; (D) female posterior end (arrow indicates anus); (E) excretory pore (arrow); and (F) infective juvenile (arrow indicates anus) (scale bar 10 μm).
Nematode identification
Characterization
Description
Female: Cuticle annulated; annuli 0.9–1.1 μm. Lateral field bearing five incisures, visible under SEM (fig. 3E), starting at 3–4 annuli posterior to lip region, ending at phasmid (fig. 3H). Lip region continuous with body contour, 9–11 μm width, having six rounded lips (fig. 3A-D), bearing small papillae. Stoma rhabditoid, 13–18 μm long, with distinct cheilo-, gymno- and stegostom. Cheilostom finely cuticularized. Gymnostom longer than cheilostom, having well cuticularized lumen. Stegostom has isomorphic glottoid apparatus without denticles (fig. 2A). The pharyngeal collar is present and partially covers the stoma. The length of pharyngeal corpus is between 1.6 and 1.9 times length of the isthmus, with the procorpus being longer than the metacorpus. Metacorpus is distinct and bloated. Strong isthmus, clearly separated from metacorpus and basal bulb ovoid with valvular apparatus. Cardia conoid, surrounded by intestinal tissue. Nerve ring at isthmus level, at 60–69% of neck length. Excretory pore opening at posterior isthmus level, very clear with sclerotized pore, at 74–79% of neck length. Deirid not visible. Intestine without distinct specialization. Reproductive system didelphic–amphidelphic. Vulva protruded, located posterior to middle part of body. Oviduct short. Uterus tubular, with swollen lumen, about 1.5–2 times as long as corresponding body diameter. Vagina with fine walls, extending about 0.2 of the body width. Rectum 1.7–1.9 times anal body diameter. Tail conical, with acute end (fig. 3G, I). Phasmid at 42–45% of the tail length from the anus.

Fig. 3. Oscheius cyrus. n. sp. (scanning electron microscope): (A, B) lip region (frontal view); (C, D) anterior end of female (sub-ventral and sub-lateral view, respectively); (E) vulval region (arrow indicates lateral field incisures); (F) female anal region (arrow); (G, H) female posterior end (in H, arrow indicates phasmid); and (I) entire female.
Infective juveniles: The specimens have similar morphology to the adult females; however, they have shorter body length (318–540 μm), more slender body (a = 16.8–25.1 vs. 10.5–19.6), narrower stoma (fig. 2B) and shorter tail length (49–67 μm). They also differ in c (total body length/tail length(L/T)) (5.2–9.2 vs. 12.0–14.5) and D% (excretory pore position/ pharynx length*100 (EP/ES*100))(122.6–136.4 vs. 73.6–111.0).
Male:
Not found.
Relationship with other Oscheius species
The last update for the genus Oscheius has been provided by Abolafia & Peña-Santiago (Reference Abolafia and Peña-Santiago2019). They have indicated 22 species under Oscheius subgenus with Oscheius (Oscheius) insectivorus (Körner, 1954) Andrássy, Reference Andrássy1976 as type species, and 15 species under the Dolichorhabditis Andrássy, 1983 with subgenus, with Oscheius (Dolichorhabditis) dolichura (Schneider, 1866) Sudhaus & Hooper, Reference Sudhaus and Hooper1994 as a type species. The new species, O. cyrus , falls under the subgenus Oscheius as it has tubular stoma without teeth. Therefore, the new species resembles O. myriophilus. However, compared with the material studied by Poinar (Reference Poinar1986), they differ in the upper range for female body length (1097–1293 vs. 1200–1500 μm), female tail (45–100 vs. 108–135 μm) and infective juvenile tail length (40–67 vs. 75–80 μm. Compared with the material examined by Sudhaus & Schulte (Reference Sudhaus and Schulte1989), they differ in the lower range of the female body length (1097–1293 vs. 792–1530 μm). Compared with the Iranian population of O. myriophilus (Gholami Ghavamabad et al., Reference Gholami Ghavamabad, Talebi, Mehrabadi, Farashiani and Pedram2021), they differ in female body length (vs. 862–1247 μm) and the lower range of the tail length (45–100 vs. 90–126 μm). Moreover, SEM observation revealed six incisures for the lateral field, with four incisures observed in O. myriophilus. Furthermore, the new species resembles O. myriophilus (presented by Zhou et al., Reference Zhou, Yang, Wang, Bao, Wang, Hou, Lin, Yedid and Zhang2017 as Oscheius microvilli). However, the new species, in comparison with the specimens studied by Zhou et al. (Reference Zhou, Yang, Wang, Bao, Wang, Hou, Lin, Yedid and Zhang2017), differs in female body length (1097–1293 vs. 846–1446 μm), female tail length (89–191 μm), median bulb (oval shaped vs. swollen), epiptygma (absent vs. present) and lateral field (5 vs. 4). Compared with O. insectivorus (Körner, 1954) Andrássy, Reference Andrássy1976, they differ in cheilostom (cheilostom shorter than stomatal tube length vs. cheilostom as long as stomatal tube length) and have shorter body length (1097–1293 vs. 2200–2912 μm). Compared with Oscheius caulleryi Maupas, 1919, they differ in body length (vs. 1910–2364 μm) and stoma length (13–18 vs. 16–20μm). Compared with Oscheius niazii Tabassum & Shahina, Reference Tabassum and Shahina2010, they differ in body length (vs. 715–1020 μm) and longer neck length (177–225 vs. 160–175 μm). Compared with Oscheius cobbi Tabassum, Shahina, Nasira and Erum, 2016, they differ in male (absent vs. present), and female tail (filiform at posterior vs. not filiform at posterior) and tail length (45–100 vs. 80–170 μm). Compared with Oscheius siddiqii Tabassum & Shahina, Reference Tabassum and Shahina2010, they differ in rectum length (39–59 vs. 45–70 μm), lips (six separate lips vs. lips fused to form three doublets), female tail length (45–100 vs. 100–332 μm) and male (absent vs. present). Compared with Oscheius andrassyi Tabassum and Shahina, 2008, they differ in neck length (177–225 vs. 158–178 μm), and tail length (45––100 vs. 45–63 μm). Compared with Oscheius citrii Tabassum, Shahina, Nasira and Erum, 2016, they differ in tail length (45–100 vs. 35-50 μm). Compared with Oscheius lucianii (Maupas, 1919) Sudhaus & Hooper, Reference Sudhaus and Hooper1994, they differ in body length (1097–1293 vs. 1601–2871 μm) and c value (12.0–14.5 vs. 14.5–17.8). Compared with Oscheius chongmingensis (Zhang, Liu, Xu, Sun, Yang, An, Gao, Lin, Lai, He, Wu, and Zhang, 2008) Ye et al., Reference Ye, Torres-Barragan and Cardoza2010, they differ in body length (vs. 809–2220 μm) and stoma (13–18 vs. 9.8–12 μm). Liu et al. (Reference Liu, Mrácěk, Zhang, Půža and Dong2012) redescribed O. chongmingensis with higher upper range of the female body length (1293 vs. 2363 μm). Compared with O. chongmingensis (described as Oscheius rugaoensis by Zhang et al., Reference Zhang, Liu and Tan2012; Darsouei et al., Reference Darsouei, Karimi and Shokoohi2014), they differ in the lateral field (5 vs. 1 wart) and tail length (45–100 vs. 113–155 μm). Compared with Oscheius indicus Kumar et al., Reference Kumar, Jamal, Somvanshi, Chauhan and Mumtaz2019, they differ in tail length (131–156 μm) and lateral field (5 vs. 3 warts). Compared with Oscheius shamimi Tahseen and Nisa, 2006; they differ in lateral field (5 vs. 2 warts), stoma length (13–18 vs. 19–23 μm) and tail length (45–100 vs. 110–132 μm). Compared with Oscheius nadarajani Ali, Asif, and Shaheen, 2011, they differ in body length (vs. 1358–1606 μm) and neck length (177–225 vs. 256–267 μm). Compared with Oscheius colombianus Stock, Caicedo, and Calatayud, 2005, they differ in smaller stoma in females (13–18 μm vs. 21–28 μm) and female tail (45–100 vs. 110–167 μm). Compared with Oscheius carolinensis Ye et al., Reference Ye, Torres-Barragan and Cardoza2010, they differ in body length (1097–1293 vs. 1360–2420 μm) and female tail length (45–100 vs. 108–206 μm). Compared with Oscheius safricana Serepa-Dlamini & Gray, Reference Serepa-Dlamini and Gray2018, they differ in the lateral field (5 vs. 3 warts), stoma length (13–18 vs. 6.9–9.9 μm) and male (absent vs. present).). Compared with the Iranian population of O. necromenus (Sudhaus & Schulte, Reference Sudhaus and Schulte1989) Sudhaus & Hooper, Reference Sudhaus and Hooper1994, studied by Valizadeh et al. (Reference Valizadeh, Goldasteh, Rafiei-Karahroodi and Pedram2017), they differ in V (49–50 vs. 41.4–52.3), lower range of the female tail length (45–100 vs. 91–127 μm) and lateral field (5 vs. 3 warts).
Diagnosis
Rhabditidae (Oscheius subgenus). Small-sized to medium-sized nematodes, 1097–1293 μm long. Lateral field with five warts (six incisures). Lip region continuous. Stoma tubular, bearing glottoid apparatus without teeth. Pharynx consists of a cylindrical corpus gradually enlarging posteriorly, with an oval-shaped median bulb, and basal bulb bearing a haustrulum. Secretory–excretory pore sclerotized. Female genital system didelphic–amphidelphic, with equatorial vulva. Vulva without epiptygma. Female rectum conspicuously longer than the anal body diameter. Female tail conical elongate with fine tip. Phasmid posterior to anus, about 42–45% of tail length.
Type locality and habitat
Iran, Mazandaran, Guilan and Khorasan Razavi province, in association with forest soil.
Type material
Twenty females (holotype and paratypes) were deposited in the Department of Plant Protection of the Ferdowsi University of Mashhad, Iran. 10 paratype females and 10 dauer juveniles were deposited in the Zoological Museum of Ferdowsi University of Mashhad, Iran. In addition, slides from both females and dauer juveniles will be sent for deposition in the American Museum of Natural History, New York, USA.
Etymology
This species was named respecting king, Cyrus the Great, the founder of the Achaemenid dynasty. The first declaration of human rights, Cyrus Cylinder, has been attributed to him.
Molecular analysis and identification of nematodes
Molecular approaches were conducted to confirm the morphological identification of the nematode isolate. The partial sequence of the ITS, 18S, and 28S were determined and deposited in the GenBank under the accession numbers OM502415 (ITS), OM502416 (18S) and ON695780 (28S) as O. cyrus n. s. A BLAST search by GenBank showed that the new species, O. cyrus n. sp., has highest identity with unknown species of Oscheius, KX068706 and MT702996, (ITS), HQ332390 (18S), and KY366264, KT884891, and MW584618 (28S).
Aligned files including O. cyrus n. sp. and 36 taxa (for ITS), 27 taxa (for I8S) and 39 taxa (for 28S) contained 1013, 1725 and 623 base pairs (bp) segments, respectively (figs 4–6). Also, analysis on ITS, 18S, and 28S genes indicated that 166, 1209 and 280 sites were conserved, 786, 475 and 341 sites were variable, 688, 331, and 254 sites were parsimony informative, and 89, 143 and 87 sites were singletons, respectively. The overall average pairwise distance of ITS, 18S and 28S sequences between O. cyrus with different species of Oscheius were 0.62 (range 0.00–1.34), 0.15 (range 0.01–0.27) and 0.34 (range 0.00–0.69), which were calculated by the Kimura 2-parameter model (tables 2–4).

Fig. 4. Phylogenetic relationships of the Iranian strain of Oscheius cyrus and other closely related species as inferred from Bayesian analysis of sequences of the internal transcribed spacer (ITS) rDNA region under GTR + I + G model. Bayesian posterior probability values equal to or more than 0.50 are given for appropriate clades. The scale bar shows the number of substitutions per site.

Fig. 5. Phylogenetic relationships of the Iranian strain of Oscheius cyrus and other closely related species as inferred from Bayesian analysis of sequences of the small subunit rDNA region under GTR + I + G model. Bayesian posterior probability values equal to or more than 0.50 are given for appropriate clades. The scale bar shows the number of substitutions per site.

Fig. 6. Phylogenetic relationships of the Iranian strain of Oscheius cyrus and other closely related species as inferred from Bayesian analysis of sequences of the large subunit rDNA region under GTR + G model. Bayesian posterior probability values equal to or more than 0.50 are given for appropriate clades. The scale bar shows the number of substitutions per site.
Table 2. The number of bases which are not identical (upper triangle), and pairwise comparison on the number of nucleotide differences (lower triangle) among some of Oscheius species and Oscheius isolates FUM222 based on ITS rDNA sequences.

The data set in bold showing the difference between the new species and others.
Table 3. The number of bases which are not identical (upper triangle), and pairwise comparison on the number of nucleotide differences (lower triangle) among some of Oscheius species and Oscheius isolates FUM222 based on 18S rDNA sequences.

The data set in bold showing the difference between the new species and others.
Table 4. The number of bases which are not identical (upper triangle), and pairwise comparison on the number of nucleotide differences (lower triangle) among some of Oscheius species and Oscheius isolates FUM222 based on 28S rDNA sequences.

The data set in bold showing the difference between the new species and others.
Phylogenetic analyses of symbiont bacteria
The partial sequence of the 16S region was determined and deposited in the GenBank under the accession number OM491243. Regarding the 16S rRNA gene, the associated bacterium isolate includes 911 bp. With BLAST analysis utilizing the 16S rRNA sequences of associated bacterium of FUM222 isolate the present study showed the most similarity to Ochrobactrum pseudogrignonense with accession numbers MH669291 (from South Korea) and MK517588 (from Indica) for 16S rRNA, respectively, with 99.89% similarity and 100% of query coverage for both of them.
Multiple alignments of the 911 bp section of this gene across 31 species revealed that 674 sites were conserved, 210 sites were variable, 83 sites were parsimony informative and 126 sites were unique. The reconstruction of a phylogenetic tree based on 16S rRNA sequences using neighbour-joining and 10,000 bootstrap replicates revealed that the bacterial isolate (FUM222) forms a monophyletic group with other Ochrobactrum strains (fig. 7). The main inter-specific distance of 16S rRNA sequences was 0.04% (range 0.00–0.12), which was calculated using the Kimura 2-parameter model (table 5).

Fig. 7. Phylogenetic relationship of Ochrobactrum pseudogrignonense isolate FUM222 with other Ochrobactrum species based on 16S rRNA region. Oligotropha carboxidovorans as outgroup, using neighbour-joining analysis in the bootstrap test (10,000 replicates) under Kimura 2-model.
Table 5. The number of bases which are not identical (upper triangle), and pairwise comparison on the number of nucleotide differences (lower triangle) among some of Ochrobactrum species and Ochrobactrum isolate FUM222 based on 16S rRNA sequences.

The data set in bold showing the difference between the bacterium studied and others.
For the 16S rRNA gene, the isolate shares a common ancestor with O. pseudogrignonense with a bootstrap value of 99%.
Laboratory bioassay
All the applied nematode concentrations caused mortality in the fifth instar of G. mellonella larvae. After 48 h, the larvae showed mortality based on concentration. The lowest and highest mortality percentage (20% and 82.5%) were observed in concentrations of 25 and 400 IJs/larvae, respectively. There were significant differences among the concentrations (F = 31.85, df = 1, P < 0.001) but there were no significant differences among the days (F = 1.86, df = 1, P < 0.174), and interaction of concentrations and days (F = 1.935, df = 1, P < 0.166).
Phenotypic characterization of bacterium
Table 6 and fig. 8 show the biochemical characteristics’ results of the associated bacterium.
Table 6. General biochemical characteristics of associated bacteria isolated from Oscheius cyrus FUM222.


Fig. 8. Biochemical tests: (A) mortality test; (B) lipase; (C) antibiotic; (D) dye absorption (on nutrient agar); (E) dye absorption (on bromothymol blue); (F) motility test; (G) mortality test by Xenorhabdus bovienii FUM221; and (H) control mortality test.
Discussion
During the present study, a new nematode species was recovered from the natural forests of northern Iran. The isolated nematode was collected via the Galleria trap from soil samples taken from the Mazandaran Province of Iran.
The collected isolate of Rhabditid belonged to the Oscheius genus. There are many species in the genus of Oscheius that are morphologically very close together (Abolafia & Peña-Santiago, Reference Abolafia and Peña-Santiago2019; Kumar et al., Reference Kumar, Jamal, Somvanshi, Chauhan and Mumtaz2019; Rana et al., Reference Rana, Bhat, Chaubey, Půža and Abolafia2021). Oscheius cyrus n. sp. was isolated from the soil habitat, while some species of this genus were initially recovered from Tipula larvae, such as Oscheius tipulae (Félix et al., Reference Félix, Vierstraete and Vanfleteren2001). As stated in the introduction, the Oscheius species is divided into two groups (insectivorus and dolichura) distinguished by their bursa, spicules and rectum features. Accordingly, the new species, Oscheius cyrus n. sp., belongs to the insectivorus groups (Abolafia & Peña-Santiago, Reference Abolafia and Peña-Santiago2019).
The general lip region shows morphological differences with O. myriophilus (KT825914), synonymized of O. myriophilus, from China and O. myriophilus (MW430436) from Iran. The phylogenetic tree supported the taxonomic identification of the nematode. Hence, the molecular analysis of the new species based on ITS, 18S and 28S rDNA sequences are described. The results are presented in the trees (figs 4–6). Also, table 2 clearly shows that the ITS region of two undescribed species of South Africa (MT702996 and KX068706) and O. cyrus are identical, which means they are the same. So, based on the findings from Iran and South Africa, the distribution of O. cyrus is broad.
The absence of a vital life stage is a significant obstacle in most Rhabditid identifications. In some Rhabditids, including the genus Oscheius, the males are the primary diagnostic trait used to identify and differentiate species. The form and size of spicules and gubernaculums, the shape and location of the bursa, and the number and arrangement of genital papillae are examples of these defining characteristics (Ye et al., Reference Ye, Foye, MacGuidwin and Steffan2018). Some previous works showed a lack of male stage in the collected samples (Ye et al., Reference Ye, Foye, MacGuidwin and Steffan2018; Abolafia & Peña-Santiago, Reference Abolafia and Peña-Santiago2019; Félix et al., Reference Félix, Vierstraete and Vanfleteren2001), and in some cases, the male number was rare (Torrini et al., Reference Torrini, Mazza, Carletti, Benvenuti, Roversi, Fanelli, de Luca, Troccoli and Tarasco2015). Félix et al. (Reference Félix, Vierstraete and Vanfleteren2001) found that three populations of Oscheius, which closely resembled O. pseudodolichura, were self-fertilizing hermaphrodites with facultative males that were too infrequent. They reported that the existence of the male stage was so limited. According to Ye et al. (Reference Ye, Foye, MacGuidwin and Steffan2018), as indicated in the present work, many tries were done using changes in the temperature, moisture, number of inoculated nematodes and nutriment regime in the experiments to find males in culture; nonetheless, these were not effective.
Thus, two hypotheses can be presented: (1) the nematodes probably can display geographical parthenogenesis (Ye et al., Reference Ye, Foye, MacGuidwin and Steffan2018). Oscheius cyru, a new species of rhabditid, is applicable to such a reproductive method without male individuals; and (2) male deficiency may suggest a form of hermaphrodite in which too few cells in the uterus indicate the presence of hermaphrodite sperm (LaMunyon & Ward, Reference LaMunyon and Ward1998; Woodruff et al., Reference Woodruff, Eke, Baird, Félix and Haag2010; Ellis & Schärer, Reference Ellis and Schärer2014; Ellis & Wei, Reference Ellis and Wei2015; Abolafia & Peña-Santiago, Reference Abolafia and Peña-Santiago2019).
Based on criteria recently suggested to define the entomopathogenicity of a nematode, it must show some traits, especially to be able to kill 50% of treated host insects within five days (Dillman et al., Reference Dillman, Chaston, Adams, Ciche, Goodrich-Blair, Stock and Sternberg2012; Torrini et al., Reference Torrini, Mazza, Carletti, Benvenuti, Roversi, Fanelli, de Luca, Troccoli and Tarasco2015). Considering this definition, the new nematode species can be presented as a reliable entomopathogen in terms of its potency to induce insect death after 48 hr depending on nematodes’ concentration. In the study by Zhou et al. (Reference Zhou, Yang, Wang, Bao, Wang, Hou, Lin, Yedid and Zhang2017), the mortality of G. mellonella larval by O. myriophilus was recorded 48 hr after inoculation. The maximum mortality (from 83 to 100%) on fifth larval instars of the box tree moth, Cydalima perspectalis (Walker, 1859), occurred via O. myriophilus in 24–144 h post-treatment (Gholami Ghavamabad et al., Reference Gholami Ghavamabad, Talebi, Mehrabadi, Farashiani and Pedram2021). Torrini et al. (Reference Torrini, Mazza, Carletti, Benvenuti, Roversi, Fanelli, de Luca, Troccoli and Tarasco2015), reported 46% of G. mellonella and 58% of Tenebrio molitor (L.) mortality by O. onirici during 7 days after infection. The host mortality of G. mellonella via O. myriophilus varied from 54% to 60% five days after inoculation (Goda et al., Reference Goda, Mirzaei and Brunetti2020). The time needed for mortality is shorter when compared to the time required for necromenic, phoretic and other kinds of strategies. However, complementary and comprehensive studies are necessary to determine if the infection is happening and effective in the field environment.
Some Oscheius species which have been recognized as insect pathogens are shown to have the type of relationship with some bacterial genera, mostly Serratia (Dillman et al., Reference Dillman, Chaston, Adams, Ciche, Goodrich-Blair, Stock and Sternberg2012; Torrini et al., Reference Torrini, Mazza, Carletti, Benvenuti, Roversi, Fanelli, de Luca, Troccoli and Tarasco2015; Aujoulat et al., Reference Aujoulat, Pagès, Masnou, Emboulé, Teyssier, Marchandin, Gaudriault, Givaudan and Jumas-Bilak2019; Fu & Liu, Reference Fu and Liu2019; Goda et al., Reference Goda, Mirzaei and Brunetti2020). Several introductory tries indicated that the O. tipulae is associated with the Serratia genus (Karimi et al., Reference Karimi, Rezaei and Shokoohi2018). Hence, the complementary attempts using molecular analysis and phenetic characterization indicated the occurrence of another bacterium, Ochrobactrum pseudogrignonense, associated with O. cyrus n. sp. This bacterium species was detected in all populations of the Rhabditid nematode. The nBLAST analysis and phylogenetic inference (16S rRNA) specified the bacterium to Ochrobactrum pseudogrignonense.
The Ochrobactrum genus includes omnipresent bacteria that can do many different functions and activities. They are alphaproteobacterial Gram-negative bacteria (Rhizobiales, Brucellaceae). The Ochrobactrum spp. is receiving more attention as a species that can adapt to a wide range of environmental niches, including soil, wastewater, plants, nematodes and other animals (Aujoulat et al., Reference Aujoulat, Pagès, Masnou, Emboulé, Teyssier, Marchandin, Gaudriault, Givaudan and Jumas-Bilak2019; Fu & Liu, Reference Fu and Liu2019; Krzyżanowska et al., Reference Krzyżanowska, Maciąg, Ossowicki, Rajewska, Kaczyński, Czerwicka, Rąbalski, Czaplewska and Jafra2019; Goda et al., Reference Goda, Mirzaei and Brunetti2020). Previously Ochrobactrum species have been reported as opportunistic pathogens in humans (Holmes et al., Reference Holmes, Popoff, Kiredjian and Kersters1988). Now, it is clear that not only they are associated with EPNs, such as Steinernema scapterisci (Nguyen & Smart, 1990) (Aguillera & Smart, Reference Aguillera and Smart1993), Steinernema siamkayai (Stock et al., 1998) (Razia et al., Reference Razia, Karthikraja, Padmanaban, Chellapandi and Sivaramakrishnan2011), Heterorhabditis sp. (Abouelhag & El-Sadawy, Reference Abouelhag and El-Sadawy2012) and Heterorhabditis indica (Poinar et al., 1992) (Aujoulat et al., Reference Aujoulat, Pagès, Masnou, Emboulé, Teyssier, Marchandin, Gaudriault, Givaudan and Jumas-Bilak2019), but also they are associated with the free-living nematodes, Acrobeloides maximus (Thorne, 1925) Thorne, 1937, (Baquiran et al., Reference Baquiran, Thater, Sedky, De Ley, Crowley and Orwin2013) and Caenorhabditis elegans (Maupas, 1900) (Dirksen et al., Reference Dirksen, Marsh and Braker2016).
The new Oscheius species was collected from the land that includes a rich fauna and flora biological resource due to diverse geography, weather and climate that causes a variety of national genetic resources, phytophagous insects, and their natural enemies. Northern Iran is an ideal area for a nematode niche because of its high yearly rainfall, wet temperate climate, excellent soils and high development potential (Marvie Mohajer, Reference Marvie Mohajer2004). Only a small number of entomoparasitic and EPN species from this region of the nation have been documented so far. So, increasing the information about their diversity in terms of species richness will increase our knowledge about this rhabditid group. Besides, the recovery of the new species of nematodes from the location site of forests in the northern part Iran suggests that it is possible to have more species diversity of the genus Oscheius and other EPNs.
Data availability
All data generated or analysed during this study are included in this published article.
Acknowledgements
The authors appreciate research assistance from the deputy of the Ferdowsi University of Mashhad. Also, the authors thank Dr Majid Pedram for assisting in providing the light microscope images and for his consultation on phylogenetic analyses.
Financial support
This study was funded by the Ferdowsi University of Mashhad (p3/48695) and grant of Iran National Science Foundation (INSF) for project 97024982.
Conflicts of interest
None.
Ethical standards
Not applicable.
















