Introduction
Hemicycliophora spp., also known as ‘sheath nematodes’, are obligate ectoparasites of various agricultural crops, fruit and nut trees, and ornamental plants. These nematodes are temporarily sedentary during their feeding process and can induce the formation of root galls, causing stunted growth and the thickening of root terminuses (Chitambar & Subbotin, Reference Chitambar and Subbotin2014). This nematode group is widely distributed over all the continents and can even be found in areas of extreme conditions such as at 13 m depth in Lake Untersee, Antarctica, and in brackish coastal environments (Loos, Reference Loos1948; van den Berg, Reference van den Berg1988; Chitambar & Subbotin, Reference Chitambar and Subbotin2014). Chitambar & Subbotin (Reference Chitambar and Subbotin2014) listed 132 valid species of the genus Hemicycliophora. Subsequently, Van den Berg et al. (Reference Van den Berg, Tiedt, Liébanas, Chitambar, Stanley, Inserra, Castillo and Subbotin2018) described two new species, namely H. onubensis and H. robbinsi, and the authors also proposed H. wyei and H. ripa as junior synonyms of H. parvana Tarjan, 1952 and H. poranga Monteiro & Lordello, 1978, respectively. Recently, H. subbotini has been described by Maria et al. (Reference Maria, Cai, Qu, Castillo and Zheng2018), bringing the total number of valid species to 133. However, only a limited number of species, including H. arenaria Raski, 1958, H. conida Thorne, 1955, H. parvana, H. poranga, H. similis Thorne, 1955, H. typica de Man, 1921 and H. ripa Van den Berg, 1981 (H. poranga), have been reported as causing damage to agriculture crops. This is a major reason why accurate identification to species level of this genus is of crucial importance for pest management strategies.
Thanks to the valuable work of Chitambar & Subbotin (Reference Chitambar and Subbotin2014), comprehensive descriptions of 132 valid Hemicycliophora species combined with both a dichotomous and a tabular key are available. However, the use of either dichotomous key or tabular key both have drawbacks, including the dependence on starting entry (each choice leads to another certain choice), and an increasing error load for larger datasets (Nguyen et al., Reference Nguyen, Trinh, Couvreur, Singh, Decraemer and Bert2019a, Reference Nguyen, Trinh, Couvreur, Singh, Decraemer and Bertb). A web-based key is therefore recommended to facilitate the accuracy and speed of identification process. Nonetheless, given that the morphological identification of species in the genus Hemicycliophora is greatly hampered by a large number of valid species with not always well-delineated traits and a high number of diagnostic features that need to be considered, an integrated approach that includes molecular data is vitally needed.
In Vietnam, the genus Hemicycliophora has been poorly investigated. Nguyen (Reference Nguyen1983) reported the presence of Hemicycliophora spp. in association with eight different host plants from three provinces, but only H. vietnamensis Nguyen & Nguyen, 2001 found in the rhizosphere of bamboo was characterized on species level. During our survey in the growing area of the medicinal plant, Amomum longiligulare T.L. Wu, in the Central Highlands of Vietnam, a new plant-parasitic species was discovered. A. longiligulare, also known as Malabar cardamom or Tavoy cardamom, is one of precious medicinal plants that contain essential oils bearing many valuable chemical compounds such as camphen, β-pinen, limonen, camphor, borneol and saponin, which can be used to treat different diseases including indigestion, cold, diarrhoea, vomiting, miscarriage, dysentery, toothache, and oedema (Dang et al., Reference Dang, Nguyen, Duong and Truong2011; Lim, Reference Lim2013; Chau et al., Reference Chau, Thang, Huong and Ogunwande2015). However, to the best of our knowledge, no plant-parasitic nematodes associated with A. longiligulare have so far been recorded.
This paper reports on a new Hemicycliophora species associated with A. longiligulare encountered during surveys in the Central Highlands of Vietnam. Morphological and molecular characteristics (D2-D3 expansion segment of 28S rRNA, ITS and COI mtDNA gene sequences) of this new species, which is described herein as Hemicycliophora cardamomi sp. n., differentiate it from all other nominal species. Additionally, a web-based key for species identification of the genus Hemicycliophora is proposed.
Materials and methods
Sampling and nematode extraction
Soil and root samples were collected from the upper 30-cm soil layer in the rhizosphere of A. longiligulare in the Central Highlands of Vietnam (GPS coordinates: 14°26′10.2″N, 107°43′14.9″E). Subsequently, nematodes were extracted from soil and roots using a modified Baermann tray method (Whitehead & Hemming, Reference Whitehead and Hemming1965).
Morphological characterization
For morphometric and morphological examination, permanent slides were prepared following Nguyen et al. (Reference Nguyen, Trinh, Couvreur, Singh, Decraemer and Bert2019a). Slides were examined and photographed using an Olympus BX51 DIC Microscope (Olympus Optical, Tokyo, Japan), equipped with a drawing tube and Olympus C5060Wz camera. Measurements from the obtained pictures were calculated using ImageJ 1.51. For scanning electron microscope (SEM), specimens preserved on permanent slides were transferred to a drop of glycerine (50 μl) in an embryo dish. Two drops of 95% ethanol (100–200 μl) were added every 10 min, for six times. Subsequently, the specimens were dehydrated by passing them through a graded ethanol concentration series of 95, 98, 100% (1 h each), 100% (overnight) and 100% (20 min, next morning). In the last step, the specimens were critical point-dried with liquid CO2, mounted on stubs with carbon discs and coated with gold (25 nm, JEOL 1200jfc) before observation with a JSM-840 EM (JEOL) at 12 kV. Digital drawings were made using Adobe Illustrator CS 3 based on pencil drawings and SEM pictures.
Molecular characterization
Nematode morphological vouchers (photomicrographs and videos) were prepared prior to DNA extraction. These vouchers were made based on temporary mounts, and these are available online at http://nematodes.myspecies.info/node/789. DNA from a single vouchered nematode was extracted following Nguyen et al. (Reference Nguyen, Trinh, Couvreur, Singh, Decraemer and Bert2019b). The D2-D3 expansion segment of 28S rRNA, ITS region, and cytochrome c oxidase subunit 1 (COI) gene fragments were amplified with respectively D2A/D3B, Vrain2 F/Vrain2R, and JB3/JB4 primers (Vrain et al., Reference Vrain, Wakarchuk, Levesque and Hamilton1992; Subbotin et al., Reference Subbotin, Sturhan, Chizhov, Vovlas and Baldwin2006; Derycke et al., Reference Derycke, Vanaverbeke, Rigaux, Backeljau and Moens2010). The thermal profile to amplify 28S rRNA and ITS regions was one cycle of 94°C for 4 min, followed by five cycles of 94°C for 30 s, 56°C for 30 s, 72°C for 2 min, and 45 cycles of 94°C for 30 s, 54°C for 30 s, 72°C for 1 min and finished at 10°C for 10 min. The same thermal profile was set to amplify COI mtDNA gene but annealing temperature was set to 45°C.
Successful PCR reactions were purified and sequenced commercially by Macrogen Europe (Amsterdam, Nederland). Geneious R11 (www.geneious.com) was used to assemble forward and reverse sequences. The BLAST search was used to check for closely related sequences of other species on GenBank (Altschul et al., Reference Altschul, Madden, Schäffer, Zhang, Zhang, Miller and Lipman1997). Multiple alignments for all genes were made using MUSCLE in Geneious R11. The best fit models were chosen based on the BIC 150 criterion in MEGA 7 (Barry, Reference Barry2011). The HKY + G model was selected for the ITS dataset; the GTR + G model was selected for the 28S rRNA and COI mtDNA datasets. Bayesian phylogenetic analysis was carried out for all genes using MrBayes 3.2.6 add-in in Geneious R11 (Huelsenbeck & Ronquist, Reference Huelsenbeck and Ronquist2001). Analyses were run under 1 × 106 generations (two independent runs with four chains); Markov chains were sampled every 200 generations and 20% of the converged runs were regarded as burn-in.
Web-based key
To facilitate the morphological identification of Hemicycliophora spp. based on the tabular key of Chitambar & Subbotin (Reference Chitambar and Subbotin2014), a web-based key was created based both on Bray-Curtis similarity (Bray & Curtis, Reference Bray and Curtis1957) and a newly developed similarity formula. The web-based key was designed using Notepad++ v7.5.6 and debugged using Google Chrome browser version 81.0.4044.129. This web-based key is freely available at http://nematodeidentification.mypressonline.com/news/identification-key-for-hemicycliophora-spp/.
The Bray–Curtis similarity formula used in our web-based key was
where
B AB is the Bray–Curtis similarity between species A and B
i = 1 to n. where n is the number of characters used in the tabular key of Chitambar & Subsbotin (Reference Chitambar and Subbotin2014)
Ai is the ranked value for the character i of species A
Bi is the ranked value for the character i of species B.
Because Bray–Curtis similarity between species A and B only provides the total distance between the ranked value of each character and does not reflex the number of actual differences, another new formula was also incorporated in our web-based key that measures the similarity between two species based on the number of identical character states:
where SAB is the similarity index between species A and B. How identical character states are processed in the computational program is further explained in the web-based key.
Results
Hemicycliophora cardamomi sp. n. has been found in 30% of the 20 investigated soil samples taken from the growing area of A. longiligulare with a mean density of 50 individuals/100 ml in the positive samples. Detailed information about this new species is given below.
Measurements
See table 1.
Table 1. Morphometric data of fixed specimens of Hemicycliophora cardamomi sp. n. All measurements are in μm and in the form: mean ± s.d. (range).

Description

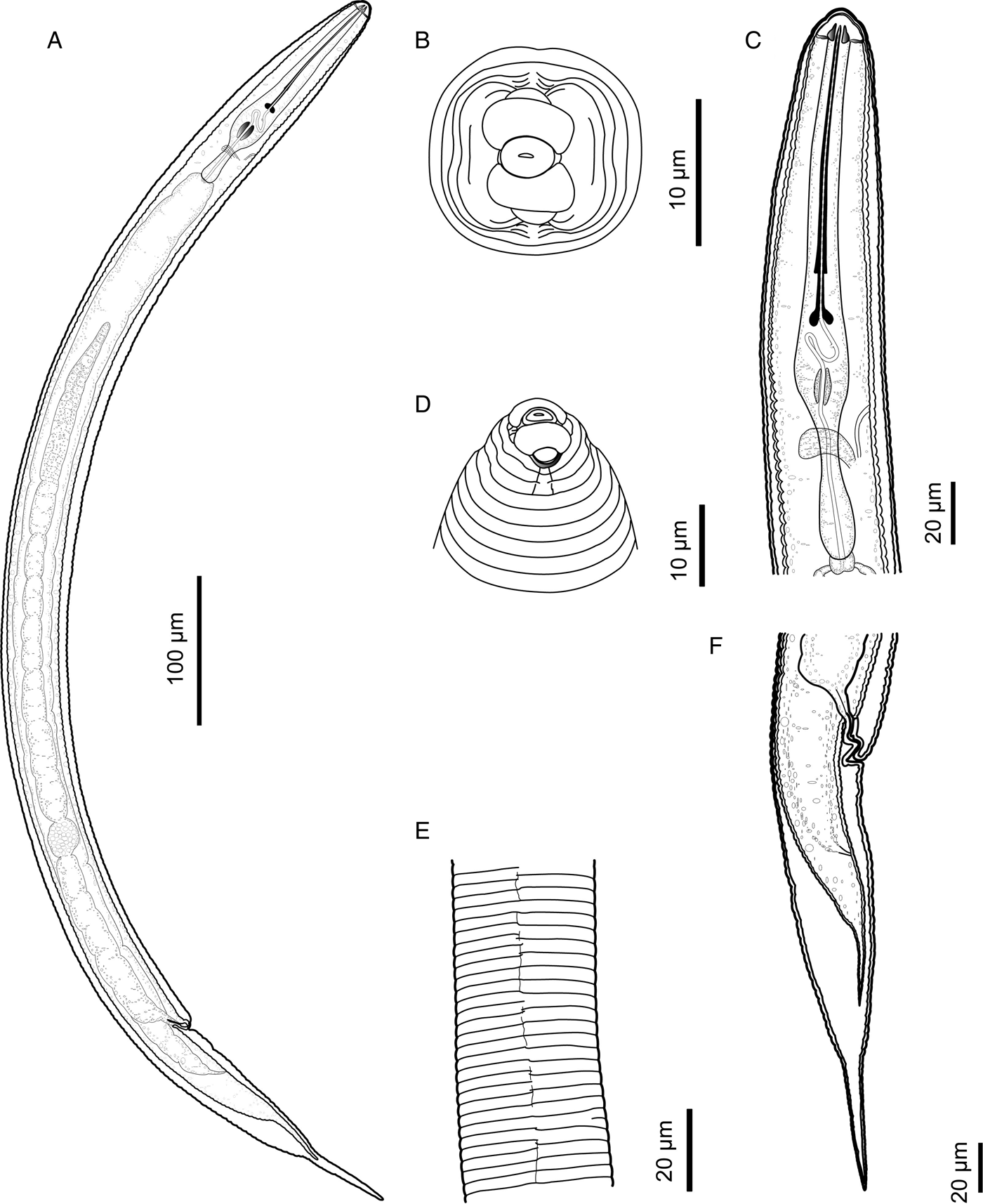
Fig. 1. Hemicycliophora cardamomi sp. n. (LM ♀). A: entire body; B: pharyngeal region; E: anterior end region; H: tail region; K: lateral field region (SEM ♀). C: anterior end region; D: enface view; F: anterior end region; G: lateral field region; I: posterior end region, lateral view; J: posterior end region, ventral view.

Fig. 2. Hemicycliophora cardamomi sp. n. (drawings ♀). A. Entire body; B: En face view; C: pharyngeal region; D: Lip region, lateral view; E: lateral field; F: posterior end region.
Females
Body slightly curved ventrally (figs 1A and 2A). Cuticular sheath loosely fitting anterior and posterior end areas (figs 1A and 2A). Lateral field mostly marked by one central longitudinal line, discontinuous over short distances, indistinct additional short lines also present in some parts along body. Lip region broadly rounded, not offset, with 3 annuli and a distinctly protruding labial disc (figs 1B–F, and 2D). Labial framework moderately sclerotized (fig. 1E). Stylet long, slender, straight to slightly curved dorsally (figs 1B and 2C). Stylet knobs sloping posteriorly with a large cavity (fig. 2B). Opening of dorsal pharyngeal gland indistinct in most specimens. Hemizonid and hemizonion not seen. Excretory pore located from level of nerve ring to level of pharyngo-intestinal junction (fig. 2B). Spermatheca rounded, full of sperm (fig. 2A). Anus indistinct. Vulval lips modified, elongate, vulval sleeve 4–5 annuli long. Body posterior to vulva gradually narrowing to about last one-third then narrowing more rapidly to a distinctly offset narrower and elongate distal portion with a narrowly rounded terminus (figs 1H–J and 2F).
Male
Not found.
Tabular key code
According to the polytomous key (which is actually a tabular key or matrix key) of Chitambar & Subbotin (Reference Chitambar and Subbotin2014), the matrix code for Hemicycliophora cardamomi sp. n. is: A:4, B:3, C:1, D:1, E:2, F:1, G:2, H:1, I:3, J:4, K:4, L:3, M:2, N:1, O:3, P:1, Q:3, R:6,1, S:3, T:2, U:3, V:1, W:2, X:1, Y:-
Web-based key analysis
In this study, a web-based key has been developed, which is freely available at http://nematodeidentification.mypressonline.com/news/identification-key-for-hemicycliophora-spp/. After the matrix codes are filled in, which are explained in the key, the most similar species will be forwarded, based on both Bray–Curtis similarity and the newly developed formulae. The range of similarity can be adapted to have more or less similar species.
The analysis using the web-based key showed that H. cardamomi sp. n. is most similar to H. vitiensis Orton Williams, 1978 (90%) based on Bray–Curtis similarity measure and to H. poranga (72%) based on the formula (2) developed in this study, respectively (fig. 3).

Fig. 3. Identification based on matrix codes of Hemicycliophora cardamomi sp. n. using the web-based key developed in this study.
Molecular characterization and phylogenetic relationship
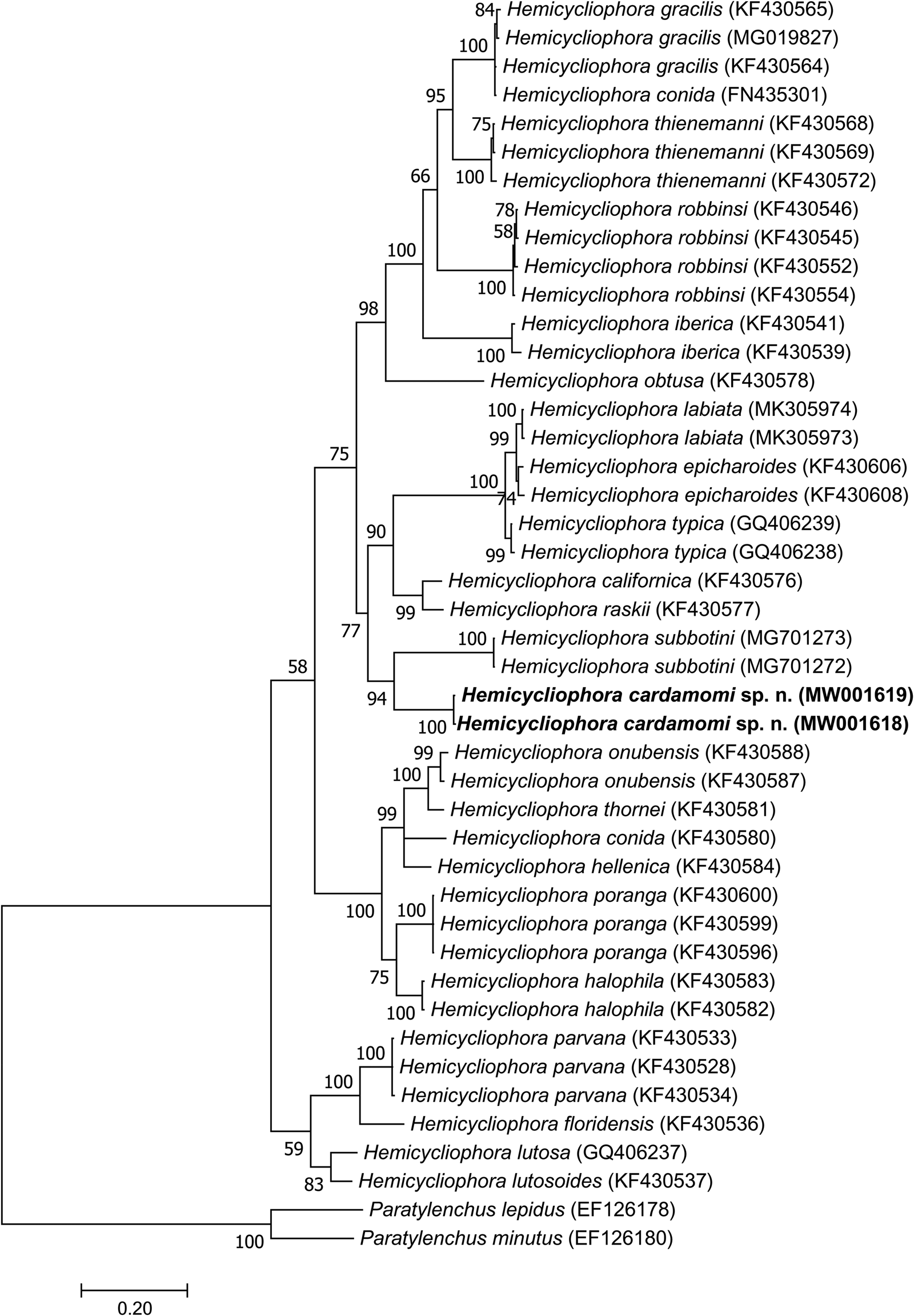
Two 743 bp long D2-D3 expansion segment of 28S rRNA sequences of H. cardamomi sp. n. were obtained with 0.02% intraspecific variation (1 bp difference). These sequences are most similar to the sequences of H. subbotini Maria, Cai, Qu, Castillo, Zheng, 2018 (43–54 bp difference). The resulting Bayesian interference phylogenetic tree showed that the sequences of Hemicycliophora cardamomi sp. n. have a sister relation to H. californica Brzeski, 1974 and H. raskii Brzeski, 1974 (68% Posterior Probability (PP) support), and these species have a sister relation to H. subbotini (95% PP support) (fig. 4). For ITS sequences, the two obtained 909 bp long sequences of H. cardamomi sp. n. are 99% similar (6 bp difference). These sequences are closest to the sequences of H. subbotini, albeit only 79% similar (168 bp difference). In the phylogenetic tree based on ITS sequences, the sequences of H. cardamomi sp. n. formed a poorly supported clade (57% PP support) with H. subbotini (fig. 5). Finally, also two virtually identical COI mtDNA, 446 bp long, sequences of H. cardamomi sp. n. were obtained (1 bp difference), which are most similar to those of H. parvana (89–90% similar; 22–45 bp difference). The phylogenetic tree based on COI mtDNA sequences showed a well-supported sister relationship (97% PP support) of Hemicycliophora cardamomi sp. n. with H. parvana and H. floridensis (Chitwood & Birchfield, Reference Chitwood and Birchfield1957; Goodey, Reference Goodey1963 (fig. 6).

Fig. 4. BI phylogenetic tree generated from the D2-D3 of 28S rDNA region dataset under GTR + G model. Bayesian posterior probabilities (in percentage) are given next to each node. Sequences of Hemicycliophora cardamomi sp. n. are in bold.

Fig. 5. BI phylogenetic tree generated from ITS sequences under HKY + G model. Bayesian posterior probabilities are given next to each node. Sequences of Hemicycliophora cardamomi sp. n. are in bold.

Fig. 6. BI phylogenetic tree generated from COI sequences under the GTR + G model. Bayesian posterior probabilities are given next to each node. Sequences of Hemicycliophora cardamomi sp. n. are in bold.
Diagnosis and relationships
The females of H. cardamomi sp. n. are characterized by the following traits: cuticular sheath loosely fitting body; labial region continuous with body contour bearing 3 annuli; lateral field marked by one central longitudinal line, discontinuous over short distances; vulval sleeve 4–5 annuli long; spermatheca rounded and full of sperms; the body part posterior to vulva tapering slightly to about last one-third of post-vulva body, then narrowing abruptly to a distinctly offset narrower and elongate distal portion with a narrowly rounded terminus.
Hemicycliophora cardamomi sp. n. is different from all other species listed by Chitambar & Subbotin (Reference Chitambar and Subbotin2014) and species recently described by Van den Berg et al. (Reference Van den Berg, Tiedt, Liébanas, Chitambar, Stanley, Inserra, Castillo and Subbotin2018) and Maria et al. (Reference Maria, Cai, Qu, Castillo and Zheng2018). A comparison between H. cardamomi sp. n. and 133 other species based on the matrix codes sensu Chitambar & Subbotin (Reference Chitambar and Subbotin2014) with the support of our web-based key showed that H. cardamomi sp. n. is most similar to H. andrassyi Brzeski, Reference Brzeski1974, H. aquatica (Micoletzky, 1913) Loof, 1948, H. robbinsi Van Den Berg, Tiedt, Liébanas, Chitambar, Stanley, Inserra, Castillo, Subbotin, 2018, H. subbotini and H. vitiensis based on Bray–Curtis similarity (> 88% similar) and most similar to H. andrassyi, H. filicauda Doucet, 1982, H. fluvialis Bird, 1999, H. parvana, H. poranga, H. ritteri, H. robbinsi and H. similis Brizuela, 1963 based on the similarity in formula (2) developed in this study (> 65% similar) (fig. 2, table 2).
Table 2. Matrix codes comparison of Hemicycliophora cardamomi sp. n. (in bold) and similar Hemicycliophora species.

*Note: for H. aquatica, code W1 should be changed to W2 following the description in Chitambar & Subbotin (Reference Chitambar and Subbotin2014)
Hemicycliophora cardamomi sp. n. can be distinguished from H. fluvialis by five main characters used in the tabular key of Chitambar & Subbotin (Reference Chitambar and Subbotin2014) (table 2): (B) lateral field (marked by one line, discontinuous over short distances, indistinct additional short lines also present in some parts along body vs. no lateral lines apparent); (D) stylet length (95 (90–98) μm vs. 114 (107–118) μm); (I) vulval lips (anterior longer than or differently shaped than posterior vs. anterior and posterior lips about equal length); (J) vulval sleeve length (very elongate, 4–5 annuli long vs. less than 2 annuli long); (W) labial annuli (distinctly separated vs. indistinctly separated). Furthermore, Hemicycliophora cardamomi sp. n. has a smaller body length (L value) and smaller R value than H. fluvialis (L = 890 (843–931) μm vs. L = 1096 (974–1278) μm; R = 285 (276–291) vs. R = 307 (279–352)). Molecular data of H. fluvialis are not available.
Hemicycliophora cardamomi sp. n. can be distinguished from H. poranga by six out of 25 characters (table 2). Additionally, D2-D3 expansion segment of 28S rRNA, ITS, and COI mtDNA sequences of Hemicycliophora cardamomi sp. n. differ from those of H. poranga by 10–11% (64–65 bp), 14–15% (82–84 bp), and 10–11% (44–47 bp), respectively.
Hemicycliophora cardamomi sp. n. can be distinguished from H. filicauda by six main characters (table 2). In addition, H. cardamomi sp. n. has a shorter stylet length (95 (90–98) μm vs. 85 (80–90) μm) than H. filicauda. Molecular data of H. filicauda are not available.
Hemicycliophora cardamomi sp. n. can be distinguished from H. andrassyi by six main characters (table 2). Molecular data of H. andrassyi are not available.
Hemicycliophora cardamomi sp. n. can be distinguished from H. ritteri by six main characters (table 2). Molecular data of H. ritteri are not available.
Hemicycliophora cardamomi sp. n. can be distinguished from H. robbinsi by six main characters (table 2). Furthermore, D2-D3 expansion segment of 28S rRNA, ITS, and COI mtDNA sequences of Hemicycliophora cardamomi sp. n. differ from those of H. robbinsi by 9–10% (56–59 bp), 27–28% (211–213 bp), and 16–17% (61–64 bp), respectively.
Hemicycliophora cardamomi sp. n. can be distinguished from H. similis by seven main characters (table 2). Additionally, D2-D3 expansion segment of 28S rRNA and COI mtDNA sequences of H. cardamomi sp. n. differ from those of H. similis by 11–12% (72–73 bp) and 13–14% (53–55 bp), respectively.
Hemicycliophora cardamomi sp. n. can be distinguished from H. parvana by seven main characters (table 2). Moreover, D2-D3 expansion segment of 28S rRNA, ITS, and COI mtDNA sequences of Hemicycliophora cardamomi sp. n. differ from those of H. parvana by 10–11% (66–67 bp), 16–18% (91–101 bp), and 10–11% (22–45 bp), respectively.
Hemicycliophora cardamomi sp. n. can be distinguished from H. aquatica by eight main characters (table 2). Besides, COI mtDNA sequences of H. cardamomi sp. n. differ from those of H. aquatica by 13–14% (55–56 bp).
Hemicycliophora cardamomi sp. n. can be distinguished from H. vitiensis by nine main characters (table 2). Additionally, H. cardamomi sp. n. also differs from H. vitiensis by hemizonid (not seen vs. distinct). Molecular data of H. vitiensis are not available.
Hemicycliophora cardamomi sp. n. can be distinguished from H. subbotini by eleven main characters (table 2). Furthermore, D2-D3 expansion segment of 28S rRNA and ITS sequences of H. cardamomi sp. n. differ from those of H. subbotini by 7–8% (43–54 bp) and 21–22% (163–165 bp), respectively.
Etymology
The new species is named after its common name (Malabar Cardamom or Tavoy Cardamom).
Type host and locality
The new species was recovered from the rhizosphere of A. longiligulare in the Central Highlands of Vietnam (GPS coordinates N: 14°26′10.2″, E: 107°43′14.9″).
Type material
Holotype female and three female paratypes deposited at the Ghent University Museum, Zoology Collections (slide number UGMD 104416), eight female paratypes (slides number 6001TN) are available in the Department of Nematology, Institute of Ecology and Biological Resources, Vietnam, and one slide (slide number WT3793) is deposited at the National Plant Protection Organization, Wageningen Nematode Collection (WaNeCo), the Netherlands. Additionally, four female paratypes are available in the UGent Nematode Collection (slide number UGnem-279) of the Nematology Research Unit, Department of Biology, Ghent University, Belgium.
Discussion
The identification of species in the genus Hemicycliophora is often problematic due to a huge number of valid species and many potential diagnostic features that must be considered. Chitambar & Subbotin (Reference Chitambar and Subbotin2014) have already provided comprehensive and useful dichotomous and tabular keys (note that the authors used the word ‘polytomous key’ for their tabular key, however, polytomous keys are a special case of dichotomous keys with three or more options at each choice instead of two options; see Hagedorn et al. (Reference Hagedorn, Rambold, Martellos, Nimis and Vignes Lebbe2010)). Both dichotomous and polytomous keys are single-access keys where one decision leads to a result or a further choice, and thus, a single wrong decision at any juncture will result in erroneous identification. An alternative to a single-access key is the free-access key, including the tabular key, where the sequence of choices is up to the user. In every step, the user can select from the list of characters offered, and choose a matching state or value. However, the manual utilization of such tabular keys is both time-consuming and complicated for large datasets. The developed web-based key has the advantage of being a free-access key, and hence overcomes the drawback of requiring a fixed sequence of identification steps. Furthermore, the use of developed web-based key can speed up the identification process, eliminate mistakes made in manual comparisons (especially for the large dataset in tabular key provided by Chitambar & Subbotin (Reference Chitambar and Subbotin2014)), avoid biases in the selection of closely related species to compare with (Nguyen et al., Reference Nguyen, Trinh, Couvreur, Singh, Decraemer and Bert2019a, Reference Nguyen, Trinh, Couvreur, Singh, Decraemer and Bertb), and crucially able to indicate the most closely related species along with the level of similarity after a single click of the mouse.
Several studies provided computer programs or web-based keys to identify selected nematode genera (Fortuner & Wong, Reference Fortuner and Wong1984; Zullini & Manganelli, Reference Zullini and Manganelli1989; Nguyen et al., Reference Nguyen, Trinh, Couvreur, Singh, Decraemer and Bert2019a, Reference Nguyen, Trinh, Couvreur, Singh, Decraemer and Bertb). However, some of these computer programs, such as the one made by Fortuner & Wong (Reference Fortuner and Wong1984), are not freely available and can be relatively complicated for scientists without sufficient experience in the computational field (for example, they require an installation process). Furthermore, the above keys do not use the actual number of identical character states in a comparison (i.e. they compare the relative similarity, see equation (1) in our web-based key). For example, according to equation (1), H. cardamomi sp. n. is more similar to H. vitiensis (although nine characters are different) than H. poranga (only five characters are different). The new algorithm was incorporated into our web-based key (formula (2); see material and methods) to compare the absolute similarity based on the number of identical characters between two species, i.e. according to the equation (2), H. cardamomi sp. n. is most similar to the species with the fewest different characters, i.e. H. poranga (http://nematodeidentification.mypressonline.com/news/identification-key-for-hemicycliophora-spp/).
This study describes a new species, H. cardamomi sp. n. with the combination of morphological characterization and molecular data of D2-D3 expansion segment of 28S rRNA, ITS and COI mtDNA regions. Although this new species is morphologically clearly different from all known species, the support of molecular characterization is essential given the omnipresence of cryptic species in plant-parasitic nematodes (Blouin, Reference Blouin2002; Bickford et al., Reference Bickford, Lohman, Sodhi, Ng, Meier, Winker, Ingram and Das2006; Gutiérrez-Gutiérrez et al., Reference Gutiérrez-Gutiérrez, Palomares-Rius, Cantalapiedra-Navarrete, Landa, Esmenjaud and Castillo2010; Cantalapiedra-Navarrete et al., Reference Cantalapiedra-Navarrete, Navas-Cortés, Liébanas, Vovlas, Subbotin, Palomares-Rius and Castillo2013; Palomares-Rius et al., Reference Palomares-Rius, Cantalapiedra-Navarrete and Castillo2014). On the other hand, small variations of some morphological characteristics in the genus Hemicycliophora, such as the number of lip annuli and lateral field structures, are sometimes considered intraspecific variations. For example, Van den Berg et al. (Reference Van den Berg, Tiedt, Liébanas, Chitambar, Stanley, Inserra, Castillo and Subbotin2018) considered H. wyei and H. ripa to be junior synonyms of H. parvana and H. poranga respectively. Descriptions of new species using the combination of morphological and molecular data enable users to both interpret morphological variation and to evaluate the presence of cryptic species in future discoveries.
Although Vietnamese nematology has been studied since the 1970s (Andrássy, Reference Andrássy1970) with numerous investigated habitats (Nguyen & Nguyen, Reference Nguyen and Nguyen2000), this is, next to H. vietnamensis, only the second species of Hemicycliophora reported in Vietnam. Remarkably, so far only endemic Hemicycliophora species have been found in the country. However, it can be expected that other known species will be recorded in Vietnam and that H. vietnamensis and H. cardamomi sp. n. are also distributed outside Vietnam, especially given the high prevalence of the latter in the current study (30% of the total samples).
A number of studies have reported Hemicycliophora spp. as severely damaging plant's pests with experiments on their pathogenicity, such as the reports on damage of H. typica on carrot, H. arenaria on citrus, H. ripa on Swiss chard, and H. poranga on tomato (Thorne, Reference Thorne1961; Franklin & Stone, Reference Franklin and Stone1974; Chitambar, Reference Chitambar1993; Malan & Meyer, Reference Malan and Meyer1993). The relatively high density of H. cardamomi sp. n. (average density of 50 individuals/100 ml) in the rhizosphere of A. longiligulare indicates its potential importance as a plant-parasite of cardamom. However, the interaction of this potentially harmful pest with its host and other members of the ginger family needs further investigation.
Financial support
This research was supported by the National Foundation for Science & Technology Development (NAFOSTED) of Vietnam (Code: 106.05-2019.323). Huu Tien Nguyen was supported by a special research fund from Ghent University (BOF-DOS 01W02619) during his time spent studying in Belgium.
Ethical standards
The result of this work has not been published previously and is not under consideration elsewhere.
Conflict of interest
The authors declare that they have no conflict of interest.