Introduction
Entomopathogenic nematodes (EPN) belonging to the genera Steinernema and Heterorhabditis (Nematoda: Steinernematidae, Heterorhabditidae) are lethal, obligate parasites of insects. They have been known to science for almost 100 years. These organisms can be produced commercially in vitro and are used in biological control of insect pests (Lacey & Georgis, Reference Lacey and Georgis2012). For this reason, they arouse the interest of scientists searching for new species that are potentially useful in insect population control, and more than 100 species have been described.
Steinernematids have a worldwide distribution and occur on all continents except Antarctica (Hominick, Reference Hominick and Gaugler2002; Spiridonov & Subbotin, Reference Spiridonov, Subbotin, Hunt and Nguyen2016). According to molecular data, the family Steinernematidae is divided into five clades: carpocapsae, affine-intermedium, bicornutum, feltiae–kraussei and glaseri (Nadler et al., Reference Nadler, Bolotin and Stock2006; Spiridonov & Subbotin, Reference Spiridonov, Subbotin, Hunt and Nguyen2016). Of all described species of the genus Steinernema, the most frequently isolated are those from the feltiae–kraussei group (Mráček et al., Reference Mráček, Nguyen and Hunt2007; Hunt et al., Reference Hunt, Nguyen, Spiridonov, Hunt and Nguyen2016). This clade constitutes ~35% of all described Steinernema species. Infective juveniles (IJs) from the feltiae–kraussei group exhibit some common features: body length ≤1000 μm, a characteristic elliptical shape of the bacterial pouch, and 6–8 lateral fields. Males and females of this genus are generally similar in size and morphology.
In Europe, six species of the feltiae–kraussei group have been identified: S. kraussei, S. feltiae, S. silvaticum, S. weiseri, S. ichnusae (Mráček et al., Reference Mráček, Nguyen and Hunt2007) and S. schliemanni (Spiridonov et. al., Reference Spiridonov, Waeyenberge and Moens2010). Steinernema silvaticum was isolated for the first time from woodland soil near Berlin (Germany) and described by Sturhan et al. (Reference Sturhan, Spiridonov and Mráček2005). This EPN species is symbiotically associated with Xenorhabdus bovienii (Kazimierczak et al., Reference Kazimierczak2016). According to Sturhan et al. (Reference Sturhan, Spiridonov and Mráček2005), S. silvaticum is common in Europe and has been found in soil samples from Germany, the Netherlands, Sweden and the UK. This species is also prevalent in Polish soil, constituting c. 10% of isolated Steinernematidae nematodes (Kazimierczak et al., unpubl. data).
The original description of S. silvaticum was based on material obtained by direct isolation of nematodes from the soil. However, no stable laboratory cultures of this species were established and all isolates were lost (Sturhan et al., Reference Sturhan, Spiridonov and Mráček2005). Consequently, the original description of S. silvaticum contains no description of first-generation females or males. Additionally, there are no available data that compare the sequence variation of the internal transcribed spacer (ITS)-rDNA with those of other molecular markers of S. silvaticum. Our aim was to complete the description of this species, based on morphological, morphometric and molecular studies of four isolates from Poland.
Materials and methods
Isolates of S. silvaticum were recovered from soil samples taken at different localities in 2016 using a modified live trap method (Bedding & Akhurst, Reference Bedding and Akhurst1975). Soil samples (~1 dm3 volume) from the south-east of Poland were collected in coniferous forests from 0–20 cm depth, as suggested by Sturhan et al. (Reference Sturhan, Spiridonov and Mráček2005). After isolation, straight lines of all nematode strains were obtained (offspring of two IJs) and initially microscopically identified. The species identification was confirmed by ITS sequence analysis. Four isolates of S. silvaticum, designated S16/019, S16/056, S16/082 and S16/090, respectively, were chosen for detailed morphological and molecular studies. All these strains were able to reproduce normally in Galleria mellonella (Lepidoptera: Pyralidae) larvae and were maintained successfully in our laboratory. Infective juveniles, the first- and second-generation males and females of the S16/082 strain, constituted the main material for this research. In addition, morphological and morphometric studies were performed for IJs of the other S. silvaticum isolates.
Light (LM) and scanning electron microscopy (SEM)
To make observations and measurements using LM and SEM, different life stages of S. silvaticum were obtained from infected G. mellonella larvae exposed to c. 100 IJs per insect in a 7 cm diameter Petri dish lined with moistened filter paper at 17.5°C. Male and female nematodes of the first and second generations were retrieved during dissections of insect cadavers in Ringer's solution after five and ten days, respectively. IJs were harvested using a modified White trap method (Stock & Goodrich-Blair, Reference Stock, Goodrich-Blair and Lacey2012), and collected in tap water one week after initial migration.
For LM, all developmental stages of the nematodes were heat-killed and fixed in a 2% (v/v) formaldehyde solution in 0.9% (w/v) aqueous NaCl. After 48 h fixation at room temperature, the nematodes were dehydrated in a graded ethanol + glycerol series [10% (v/v) ethanol + 1% (v/v) glycerol + 89% (v/v) deionized water, 20% ethanol + 2% glycerol + 78% deionized water, …, 90% ethanol + 10% glycerol], evaporated, transferred to pure glycerin and mounted on permanent slides. Observations and measurements were made using a Leica 5500B microscope (Leica Camera AG, Wetzlar, Germany) equipped with differential interference contrast (DIC) optics and the Leica Application Suite LAS 3.8 software (live measurements, montage). For SEM of IJs, first-generation male and female nematodes were prepared as described previously (Skrzypek et al., Reference Skrzypek2011) and examined using LEO 1430VP scanning electron microscope at 15 kV accelerating voltage.
Hybridization tests
Reproductive isolation of the Polish strains of S. silvaticum studied as well as S. kraussei, S. feltiae, S. weiseri and S. ichnusae from the collection of Dr Z. Mràček was tested using a modified Nguyen & Duncan (Reference Nguyen and Duncan2002) method. A drop of G. mellonella haemolymph was placed at the bottom of a 2 ml sterile polypropylene test tube and one IJ of S. silvaticum and one IJ of another nematode species were added. Crosses between IJs of the same species (including crosses between each S. silvaticum isolate) were performed in control tubes. All the treatments were replicated 20 times for each combination of the nematode isolates and observed for 15 consecutive days at 17.5°C.
Molecular characterization
DNA was extracted from single, virgin first-generation females of S. silvaticum using a DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). Amplification of the molecular markers was performed using primers 18S and 26S for the ITS of the rDNA (Vrain et al., Reference Vrain1992), D2F and 536F for the D2-D3 domain of 28S rRNA (Stock et al., Reference Stock, Campbell and Nadler2001; Nguyen, Reference Nguyen, Nguyen and Hunt2007), and 507F and 588R for the mitochondrial cytochrome c oxidase subunit 1 (cox1) gene (Nadler et al., Reference Nadler, Bolotin and Stock2006). Polymerase chain reaction (PCR) amplifications were performed in a Bio-Rad C1000 thermal cycler (Bio-Rad Laboratories, Hercules, USA) programmed for an initial denaturation at 95°C for 3 minutes, followed by 35 cycles of 94°C for 1 minute, 30 s at 50°C for rDNA or 42°C for cox1, 1 minute at 72°C, and a final extension of 7 minutes at 72°C. The PCR reaction was performed in a total volume of 25 μl containing 0.2 mM of each dNTP, 0.2 μl of each starter, 1.5 U of Taq DNA polymerase, and 1.5–2.5 mM of MgCl2 as required to optimize the amplification. The PCR products were purified using standard methods and sent to Genomed (Poland) for direct sequencing in both directions.
The ITS and D2-D3 as well as cox1 gene sequences of S. silvaticum obtained in this study were analysed and compared with those deposited in GenBank using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple sequence alignments were produced by default ClustalW configuration included in MEGA 6.06 (Tamura et al., Reference Tamura2013). The resulting alignments were corrected manually; the longer sequences were truncated to obtain the same number of analysed positions. The phylogenetic trees were constructed with the Maximum Likelihood method using MEGA 6.06. The best fit model was identified as the general time reversible (GTR) model (Nei & Kumar, Reference Nei and Kumar2000). The percentage of trees in which the associated taxa clustered together is shown next to the branches. Sequence identity values and the number of substitutions between sequences of different taxa were calculated using BioEdit and MEGA 6.0, respectively.
All the gene sequences of the four S. silvaticum isolates S16/019, S16/056, S16/082 and S16/090 were deposited in the GenBank database under accession numbers MG543845–48 for ITS rDNA, MG547576–79 for D2-D3 rDNA, and MG547572–75 for the cox1 gene. Accession numbers of the other sequences of nematode species are shown on the phylogenetic trees.
Results
Measurements
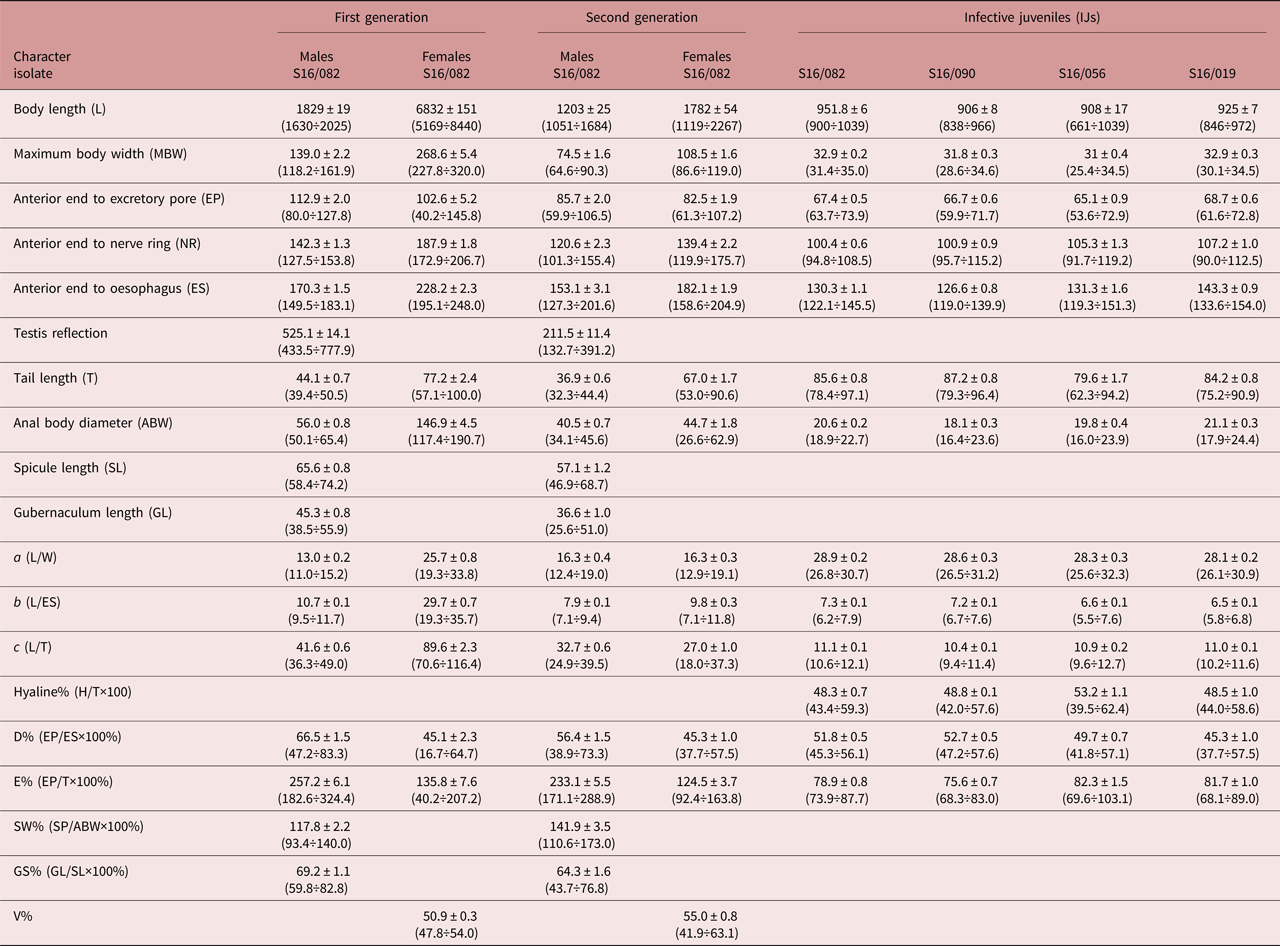
Morphometrics data of infective juveniles (isolates S16/019, S16/056, S16/82, S16/090) are presented in table 1. Measurements of the first- and second-generation males and females were performed for S16/082 isolate only. All developmental stages of nematodes were reared in G. mellonella larvae.
Table 1. Morphometrics (in μm) of different developmental stages of Steinernema silvaticum. Mean ± SE (range); N = 25.
Description
First-generation male
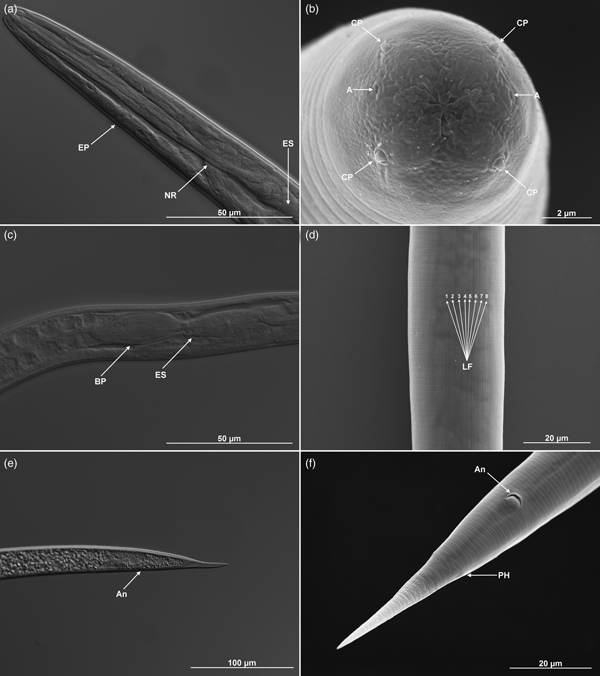
Body C- or J-shaped posteriorly when heat-killed. Cuticle with fine, annular, irregular striation under SEM, but smooth under the light microscope (figs 1b, e and 2). Lateral fields absent. Anterior end rounded, with six labial and four cephalic papillae. Amphidial apertures slit-like, slightly above the middle pair of labial papillae. Labial papillae longer than cephalic papillae, but with a smaller diameter (fig. 1b). Stoma triangular to rounded, narrow. Cheilorhabdions prominent, well cuticularized. Pharynx muscular, narrower isthmus region surrounded by nerve ring and rounded basal bulb. Excretory pore in centre of ventral side and located at ~2/3 of oesophagus distance from anterior end (fig. 2a, b; table 1). Nerve ring surrounding isthmus, basal bulb distinct. Anterior deirids located laterally, slightly posteriorly to the oesophagus (fig. 1a). Gonad reflexed. Testis with ventral reflexion. Spicules paired and brown in colour. Spicules curved in shape and tapered to the end, and velum prominent. Each spicule with two internal ribs. Gubernaculum boat-shaped in lateral view, with narrowed and flattened end and wide space in the centre (fig. 2c, d). Males usually have a single precloacal papilla and 11 pairs of genital papillae arranged in a normal position for Steinernema (fig. 1c). Nine pairs of genital papillae are located subventrally in a single lane, running from the tail end towards the centre of the body. The sixth pair of papillae is located laterally and the eighth and ninth pair subdorsally. Pairs 8, 9 and 10 lie behind the anus, and pair 11 in front of it. Twin post-deirids visible usually before the first pair of papillae. Tail terminus without (fig. 2c) or with tiny mucron, visible only in SEM (fig. 1f).
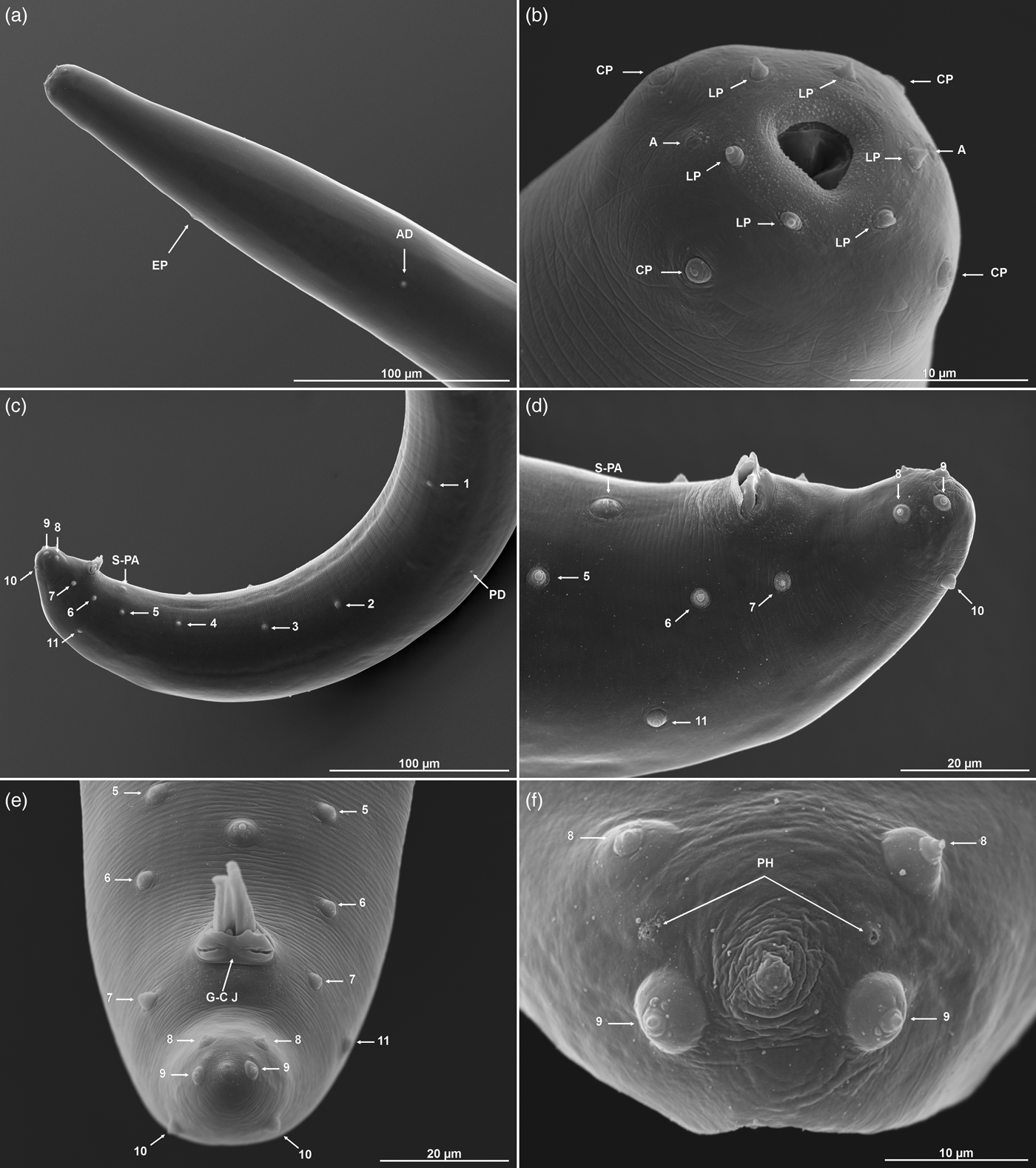
Fig. 1. Steinernema silvaticum. Scanning electron microscopy of first-generation males. (a, b) Head region with excretory pore (EP), anterior deirid (AD), amphids (A), cephalic (CP) and labial papillae (LP). (c–f) Posterior region with a typical complete set of genital papillae (11 pairs + single preanal papilla, S-PA), posterior deirid (PD), gubernaculum–cuticle junction (G–C J), and phasmid openings (PH).
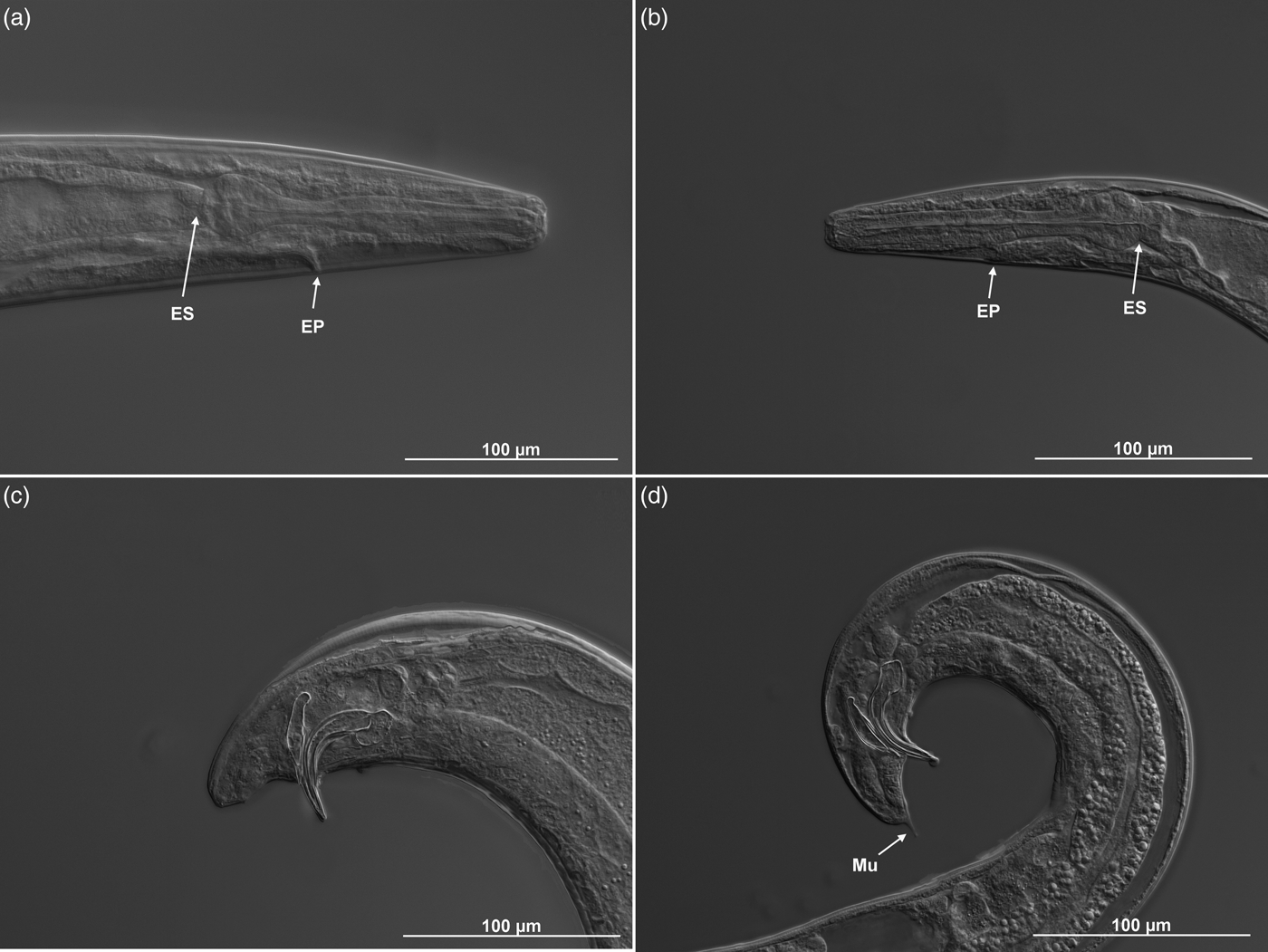
Fig. 2. Steinernema silvaticum. Differential contrast microscopy of (a, c) first- and (b, d) second-generation males. (a, b) Head region with oesophagus (ES) and excretory pore. (c, d) Tail with spicules, gubernaculum and mucron (Mu, second-generation only).
Second-generation male
Similar to the first-generation male, but more slender in body, and smaller in body length and other morphometric values. Tail terminus with a mucron, usually subterminal subventral in position (fig. 2d).
First-generation female
Body C-shaped or S-shaped when heat-relaxed and fixed with formalin. Head rounded, continuous with body. Cuticle smooth or with delicate rings that disappear on the anterior and posterior ends. Six labial papillae present and four cephalic papillae behind labial papillae. Between labial papillae, amphids visible only in SEM (fig. 3b). Pharynx with procorpus cylindrical and muscular, isthmus distinct, basal bulb enlarged. Nerve ring anterior to basal bulb, surrounding isthmus. Excretory pore position variable, near mid-pharynx (fig. 3a). Vulva as a transverse slit on a protruding area located in middle of the body (~50%) (fig. 3c, d). Tail with pointed tip, much shorter than anal body diameter. Postanal swelling present (fig. 3e, f).

Fig. 3. Steinernema silvaticum. (a, c, e) Differential contrast and (b, d, f) scanning electron microscopy of first-generation females. A, amphid openings; An, anus; CP, cephalic papillae; EP, excretory pore; ES, oesophagus; LP, labial papillae; V, vulva; T, tail ending.
Second-generation female
Body C-shaped when heat-relaxed and fixed. Similar to the first-generation female but smaller and more slender. Body diameter greater anterior to vulva than posterior. Vulva situated at mid-body (~55%). Postanal swelling present. Tail longer than anal body diameter, tapering gently to a pointed tip. In some larger females, tail shorter than anal body diameter.
Infective juveniles
Body straight or gently curved and narrows slightly at the anterior and posterior ends (fig. 4a, e, f). Four cephalic papillae. Labial region smooth, continuous with the rest of body. Amphidial apertures prominent (fig. 4b). Cuticle transversely ribbed (fig. 4b, d, e). Excretory pore anterior to nerve ring (fig. 4a). Lateral field with eight ridges in mid-body region (fig. 4d), deirids not visible in SEM. Near phasmid, only two ridges (poorly separated) observed in lateral field. Pharynx with narrow corpus, metacorpus slightly swollen, isthmus present, nerve ring usually in the middle of isthmus (fig. 4a). Basal bulb elongated with visible valve. Bacterial pouch located posterior to basal bulb, containing bacterial cells (fig. 4c). Tail conical and hyaline part accounts for 48.3%, almost half of its length. Phasmid present near mid-tail, ventral to lateral field (fig. 4f).

Fig. 4. Steinernema silvaticum. (a, c, e) Differential contrast and (b, d, f) scanning electron microscopy of infective juveniles. A, amphid openings; An, anus; BP, bacterial pouch; CP, cephalic papillae; EP, excretory pore; ES, oesophagus; LF, lateral fields (1–8); NR, nerve ring; PH, phasmid opening.
Life cycle
Only some Polish isolates of S. silvaticum are able to complete their life cycle at a temperature over 20°C. In the experimental infections of G. mellonella with a dose of ~20 IJs at a temperature of 17.5°C, the insects were dead after 48–72 h when fourth-stage juveniles and young males and females were present in the host body cavity. Second-generation nematodes were observed 7–10 days after infection. First mature IJs started to migrate into the White trap after 18–20 days.
Cross-mating test
All the European representatives of the feltiae–kraussei group used are reproductively isolated, yielding offspring only with conspecific partners.
Molecular characterization and phylogenetic position
The ITS region of isolates S16/019, S16/056, S16/082 and S16/90 consisted of 727 bp. The sequence length of each transcribed spacer was 266 bp for ITS1 and 304 bp for ITS2, which correlates with data published by Sturhan et al. (Reference Sturhan, Spiridonov and Mráček2005), describing S. silvaticum as a new species. The ITS rDNA of the tested nematodes showed 99.6–100% sequence identity to each other (0–3 nucleotide substitutions) and 99.3–99.9% sequence identity (1–4 nucleotide substitutions) to the corresponding sequences of German and British S. silvaticum strains available in GenBank. Compared to the other species of the genus Steinernema, the isolates showed the highest ITS sequence identity with S. kraussei (95–96%, 27–34 bp differences), S. cholashanense (94%, 42–43 bp differences), and S. oregonense (92%, 49–51 bp differences). Phylogenetic analysis based on the sequences of the ITS region showed that the studied isolates formed a cluster at bootstrap support of 96% along with S. silvaticum strains. In the ITS phylogram, S. silvaticum and S. kraussei formed sister branches with 87% bootstrap support. The phylogenetic relationships between the tested isolates and the selected species of the genus Steinernema based on ITS rDNA sequences are presented in fig. 5.
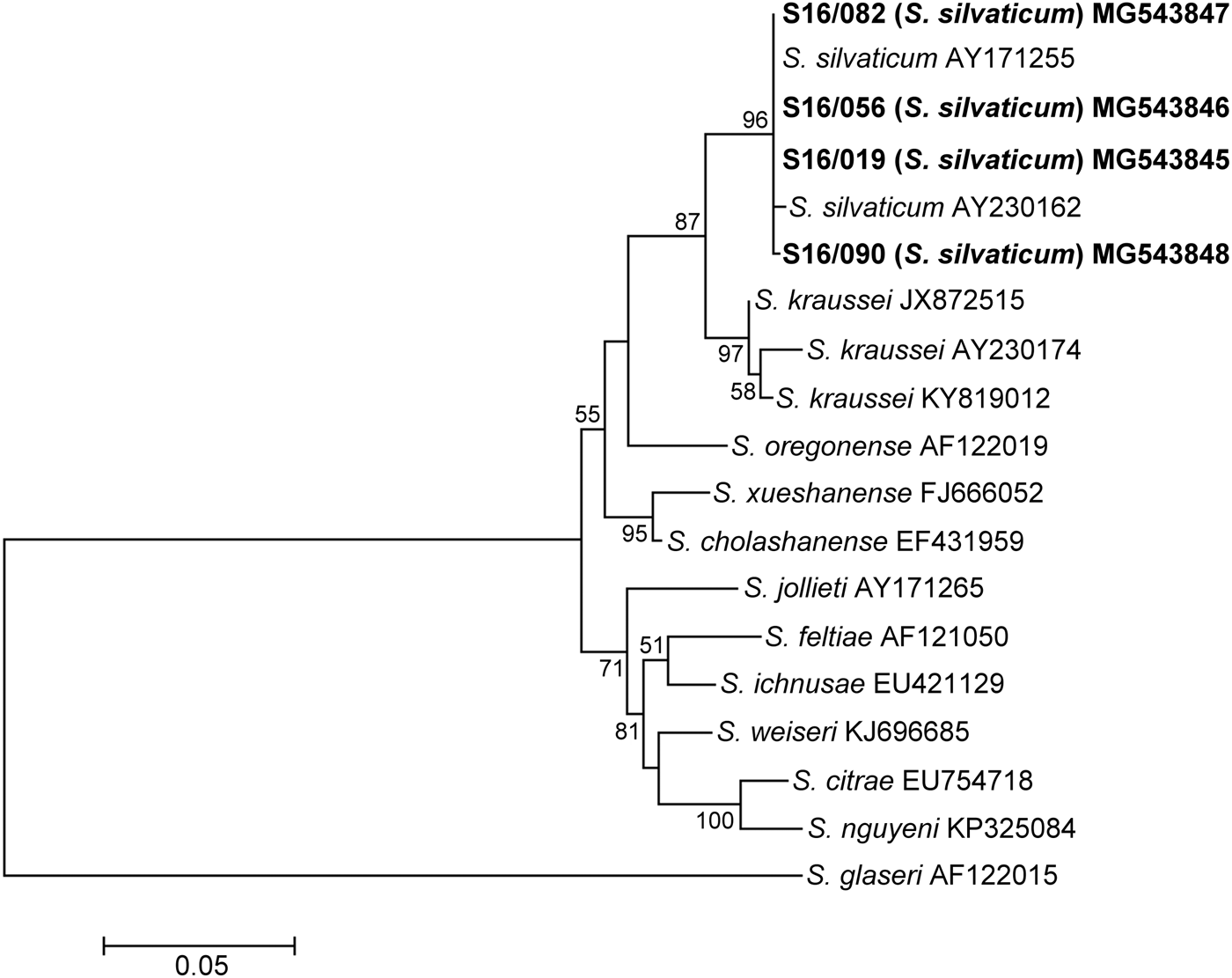
Fig. 5. Maximum Likelihood tree showing the phylogenetic relationships of the S. silvaticum isolates studied with the reference species of the genus Steinernema based on ITS rDNA sequences. Bootstrap values >50% are indicated at the branching points. The scale bar indicates the number of nucleotide substitutions per site.
The sequences of the D2-D3 expansion fragment of the 28S rRNA gene of strains S16/019, S16/056, S16/082 and S16/090 were 867 bp long and displayed 99.2–99.7% sequence identity to each other (3–7 nucleotide substitutions). However, their sequence similarity to the only three full-length D2-D3 rDNA sequences marked as derived from LN30k isolates of S. silvaticum available in GenBank was relatively low: 96–97% (21–30 bp differences). The highest sequence identity of the D2-D3 rDNA region of the studied isolates to corresponding sequences of other species from the feltiae–kraussei group was 97% in respect to S. kraussei, S. cholashanense and S. oregonense, corresponding to 23–27, 23–26 and 26–29 nucleotide substitutions, respectively. The phylogenetic analysis of the D2-D3 dataset yielded a tree with branches separating the tested S. silvaticum isolates from the other strains of the Steinernema species (100% bootstrap). Steinernema silvaticum was a part of the clade comprising S. kraussei, S. cholashanense and S. oregonense supported by 74% bootstrap. The phylogenetic relationships between the tested isolates and the selected species of the feltiae–kraussei group based on the D2-D3 region of 28S rDNA are presented in fig 6.
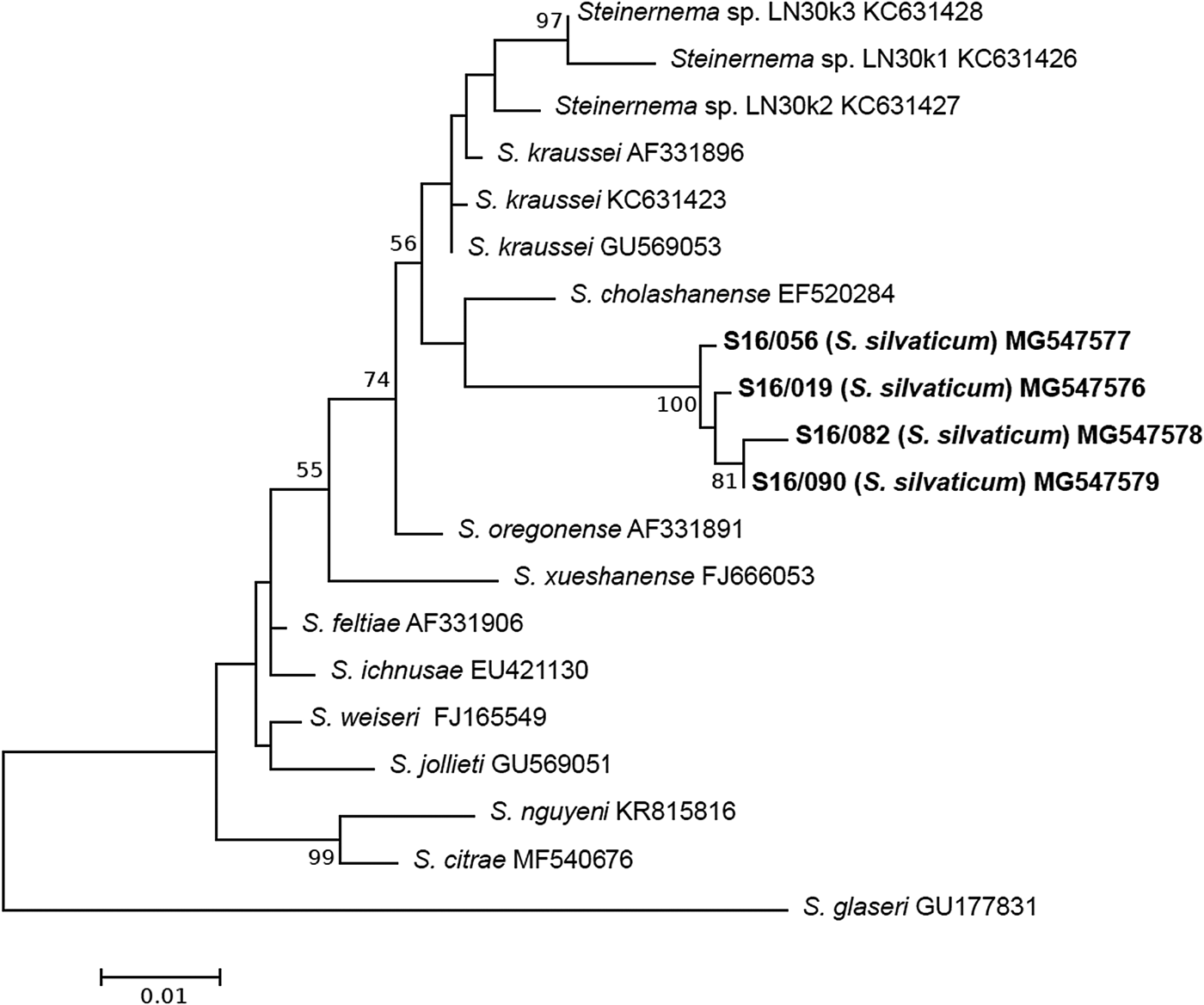
Fig. 6. Maximum Likelihood tree showing the phylogenetic relationships of the S. silvaticum isolates studied with the reference species of the genus Steinernema based on D2-D3 rDNA sequences. Bootstrap values >50% are indicated at the branching points. The scale bar indicates the number of nucleotide substitutions per site.
In this investigation, we sequenced for the first time the mitochondrial cox1 gene of S. silvaticum. The cox1 gene sequences of S16/019, S16/056, S16/082 and S16/090 were 655 bp long. They were separated from each other by 3–6 nucleotide substitutions, displaying 99.1–99.5% sequence identity. These sequences showed relatively low identity to the other sequences of Steinerema spp.: 87–88% to S. kraussei (84–72 nucleotide substitution), 85% to S. oregonense (86–87 nucleotide substitutions) and <84% to the other nematodes from the feltiae–kraussei group. The phylogenetic analysis of the cox1 gene sequences showed a clade grouping S. silvaticum and S. kraussei supported by a moderate bootstrap value (81%). The phylogenetic tree inferred for the cox1 gene is presented in fig. 7.
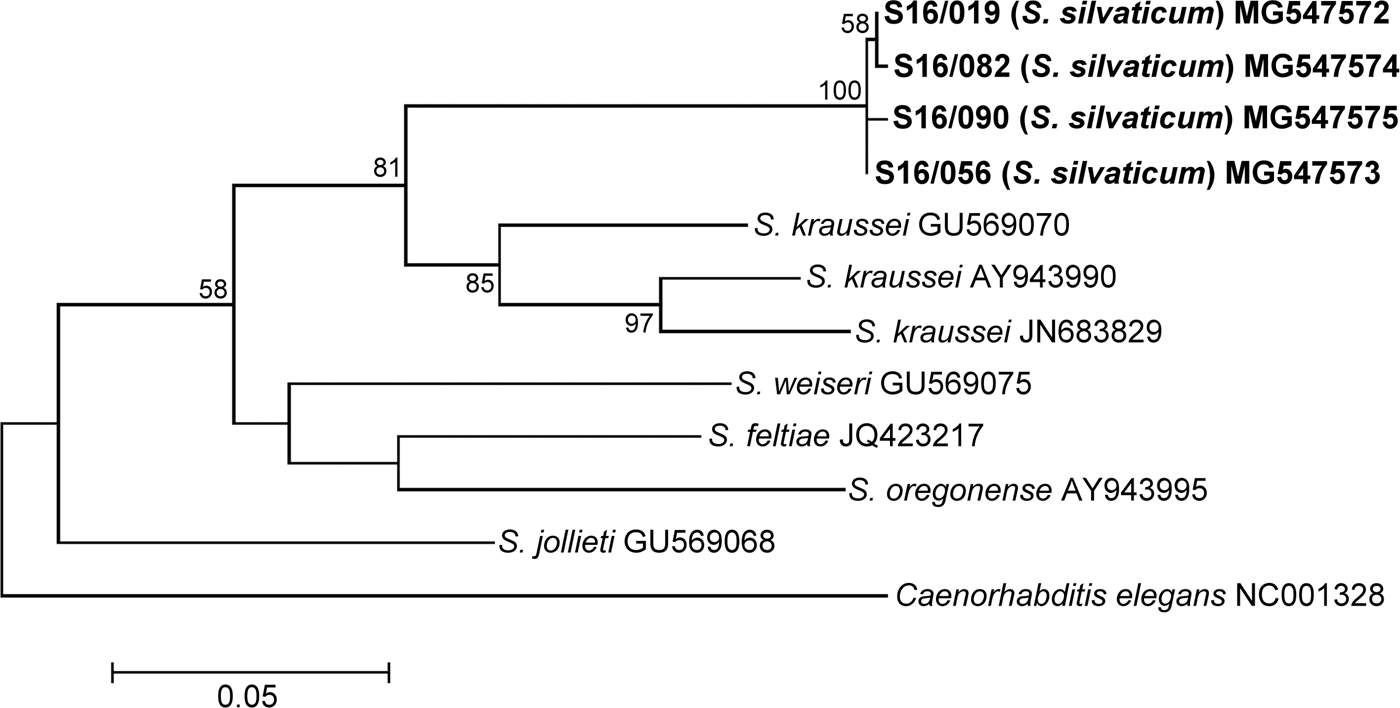
Fig. 7. Maximum Likelihood tree showing the phylogenetic relationships of the S. silvaticum isolates studied with the reference species of the genus Steinernema based on cox1 gene sequences. Bootstrap values >50% are indicated at the branching points. The scale bar indicates the number of nucleotide substitutions per site.
Discussion
Isolates of S. silvaticum can be distinguished from the other European feltiae–kraussei group representatives by means of a combination of morphological and morphometric characteristics of IJs and males. The IJs of Polish isolates are longer (>900 μm) than those of S. weiseri (740 μm), S. ichnusae (866 μm) and S. feltiae (879 μm). The total body length of the Polish strains of S. silvaticum is also slightly to distinctly greater than that of most isolates used in the original species description (917, 860, 785, 780, 775 and 705 μm, respectively) (Sturhan et al., Reference Sturhan, Spiridonov and Mráček2005). This may be a result of the use of different sample preparation methods. It should also be remembered that the isolates used in the original species description were isolated directly from the soil (not propagated on G. mellonella larvae), which can influence the size of the nematodes. Infective juveniles of S. silvaticum and S. kraussei differ also by the hyaline tail portion (43–59% vs 35–40%).
First-generation males of S. silvaticum can be distinguished from the males of S. kraussei, S. weiseri and S. ichnusae by the body length of 1829 (1630–2025) vs 1400 (1200–1600), 1180 (990–1395) and 1341 (1151–1494) μm, respectively. Males of S. silvaticum and S. kraussei differ also in the distance from the anterior end to the nerve ring (142 vs 105 μm), spicule (66 vs 49 μm) and gubernaculum length (45 vs 33 μm). The males of S. silvaticum differ from S. weiseri and S. feltiae in the tail length of 44 (39–51) vs 25 (19–32) and 31 (24–38) μm, respectively. First-generation males of S. silvaticum can be distinguished from the males of S. weiseri by the anal body diameter of 56 (50–65) vs 38 (29–43) μm, respectively, and by the distance from the anterior end to the nerve ring of 142 (128–154) vs 99 (94–115) μm, respectively. Ratios a [13 (11–15)] and b [11 (10–12)] of S. silvaticum and S. ichnusae [22 (20–29) and 8 (7–9), respectively] also differ. First-generation males of S. silvaticum do not possess a prominent mucron at the distal body end, which distinguishes this species from S. kraussei and S. feltiae.
A number of studies on nematodes from the genus Steinernema have made use of the ITS region sequence analysis to help classify them and estimate their evolutionary relationships (Nguyen et al., Reference Nguyen, Maruniak and Adams2001; Stock et al., Reference Stock, Campbell and Nadler2001; Spiridonov et al., Reference Spiridonov2004). However, species distinction based on sequencing of only one molecular marker is being questioned. The most common limitations of the ITS region are intra-individual sequence variability as well as variability in nucleotide composition and length among taxa, making estimation of positional homology dubious (Peat et al., Reference Peat, Adams, Dillman, Stock, Vandenberg, Glazer and Boemare2009; Stock, Reference Stock, Stock, Vandenberg, Glazer and Boemare2009; Půža et. al., Reference Půža2015). Unfortunately, there is no universal threshold sequence identity of the ITS region for species estimation among EPN. Nguyen (Reference Nguyen, Nguyen and Hunt2007) suggested that, in the case of the new steinernematid species, the sequence similarity of the ITS region to the other strains of this genus should be 95% or lower. However, some Steinernema species were separated by less than 5% difference. Steinernema silvaticum was described as a new species on the basis of 3.7% of the ITS region sequence variation (27 bp difference), compared to its closest taxon, S. kraussei (Sturhan et al., Reference Sturhan, Spiridonov and Mráček2005). The ITS region sequence of the tested Polish S. silvaticum isolates shows 3–5% difference from S. kraussei strains, corresponding to 27–34 bp difference.
The other molecular markers proven useful for identification and studying phylogenetic relationships among entomopathogenic nematodes include 18S rDNA (SSU), 28S rDNA (LSU) and mitochondrial DNA loci such as cox1, cox2, ND4 and 12S rDNA (Adams et al., Reference Adams, Peat, Dillman, Nguyen and Hunt2007). However, they were not applied in the description of S. silvaticum. To address the potential limitations of single-locus molecular hypotheses, phylogenetic relationships among S. silvaticum and the other Steinernema taxa were estimated using 28S rRNA and cox1 gene sequences in this study.
The phylogenetic analysis of ITS rDNA has shown that S. silvaticum is a sister species to S. kraussei but this relationship was not indicated by the D2-D3 tree. Overall, the D2-D3 phylogenetic analysis yielded a tree with weaker clade support than ITS data, providing evidence that the D2-D3 region is too conservative to resolve the relationships among closely related species of Steinernema. In fact, S. silvaticum, S. kraussei, S. cholashanense and S. oregonense displayed similar nucleotide identity of the D2-D3 region: 97% (differences in the range of 23–27 bp). These species were grouped together in both the ITS and D2-D3 phylograms, which implies that they are phylogenetically closest of all the taxa tested.
Unexpected results of the phylogenetic analysis of D2-D3 sequences (KC631426-28) were observed for the LN30k1, LN30k2 and LN30k3 nematode isolates (fig. 6). Dzięgielewska et al. (Reference Dzięgielewska, Berdzik and Myśków2015) classified these isolates as S. silvaticum and proved very high identity (98–99%) between them and S. kraussei for D2-D3 and ITS1 regions. Our results indicate that the LN30k isolates, showing no more than 97.6% D2-D3 sequence identity (21–30 nucleotide substitutions) to the Polish isolates of S. silvaticum, are distantly positioned from each other on the D2-D3 phylogenetic tree. In contrast, LN30k nematode isolates showing at least 98.3% D2-D3 region identity (6–15 nucleotide substitutions) to S. kraussei strains form a common cluster with them within the feltiae–kraussei clade (fig 6). Given that taxonomic affiliation of the nematode isolates from the present study as S. silvaticum was confirmed by ITS sequences corresponding to the original ITS sequence (AY230162) for this species, the results of D2-D3 region sequencing demonstrate that the LN30k isolates are more closely related to S. kraussei than S. silvaticum. Similarly, regarding ITS1 sequences (KC631432–34), the LN30k isolates are the most similar (98% identity, 5–7% nucleotide substitutions) to the corresponding sequences of S. kraussei, not to the original ITS1 sequence of S. silvaticum (97%, 13–14 nucleotide substitutions) or the ITS1 sequences of the Polish S. silvaticum (97%, 11–13 nucleotide substitutions). In summary, these data support the argument that sequences KC631426–28 and KC631432–34 for D2-D3 and ITS1, respectively, have been mislabelled. We have also found another discordant result in relation to S. silvaticum species assignation for the 549 bp long D2-D3 sequence DQ399663. This sequence shows 98% (7–9 nucleotide substitutions) identity to the D2-D3 region of S. kraussei and only 96% identity (11–13 nucleotide substitutions) to the D2-D3 region of the Polish isolates of S. silvaticum. These observations should be an alert that many of the large number of Steinernema sequences deposited in the GenBank database do not correspond correctly to their designated taxon. In addition, this shows that the systematics of steinernematids closely related to S. kraussei are still unclear and their identification, especially on the basis of morphological parameters, is difficult and requires a lot of experience. Thus, we hope that the detailed characterization of S. silvaticum, which is the subject of this work, will help to clarify the situation and avoid further mistakes in labelling sequences.
The mtDNA cox1 sequences revealed the highest level of genetic divergence among the studied Steinernema taxa, compared to the other two genes; the level of cox1 nucleotide divergence between our S. silvaticum isolates and closely related species reached over 12%. This shows that the mitochondrial cox1 gene undergoes a rapid rate of evolution within the feltiae–kraussei clade of the genus Steinernema, performing well at revealing relationships among closely related species and even populations. The phylogenetic analysis of the cox1 gene showed the same grouping of S. silvaticum with S. kraussei as the analysis of ITS rDNA. On the other hand, the number of available cox1 sequences is limited, and direct comparison of results of clustering based on cox1 with these based on ITS and D2D3 is not feasible. Our results show that multi-locus molecular screening of additional taxa is essential to provide a reliable evolutionary history of S. silvaticum.
Overall, the morphological and molecular data validate the Polish nematode strains as S. silvaticum and confirm the status of S. silvaticum as a separate species according to the phylogenetic and evolutionary species concept (Adams, Reference Adams1998).
Acknowledgements
We thank the anonymous referees for their helpful comments on the manuscript.
Financial support
This work was supported by a Polish Ministry of Science and Higher Education grant for statutory activity. The authors gratefully acknowledge the use of the services and facilities of the Center for Interdisciplinary Research at the John Paul II Catholic University of Lublin, Poland, co-funded by the European Union from the European Regional Development Fund under the Operational Program Development of Eastern Poland 2007–2013 (POPW.01.03.00-06-003/09-00).
Conflict of interest
None.