Introduction
The genus Angiostrongylus is represented by 21 valid named species and an unnamed species (Spratt, Reference Spratt2015). Rodents serve as the definitive host for 15 species of this genus of nematode parasites (Eamsobhana, Reference Eamsobhana2014). Two species, A. cantonensis (Chen, 1935) and A. costaricensis (Morera & Céspedes, 1971) are zoonotic parasites of public health importance (Cross & Chen Reference Cross, Chen, Murrel and Fried2007; Eamsobhana, Reference Eamsobhana2014; Spratt, Reference Spratt2015). Two other species of the A. cantonensis species complex, A. mackerrasae (Bhaibulaya, 1968) and A. malaysiensis (Bhaibulaya & Cross, 1971), have not been unequivocally shown to be involved in human infections, but their potential needs to be investigated as they share a similar life cycle with A. cantonensis.
When first documented in Malaysia, A. malaysiensis was referred to as A. cantonensis (Schacher & Cheong, Reference Schacher and Cheong1960; Lim et al., Reference Lim, Ow-Yang and Lie1965). Both mitochondrial and nuclear genes have been used to differentiate A. cantonensis and A. malaysiensis. The mitochondrial genes cytochrome c oxidase subunit I (cox1) (Eamsobhana et al., Reference Eamsobhana, Lim, Solano, Zhang, Gan and Yong2010b, Reference Eamsobhana, Yong, Song, Prasartvit, Boonyong and Tungtrongchitr2017a, Reference Eamsobhana, Song, Yong, Prasartvit, Boonyong and Tangtrongchitrb) and cytochrome b (cob) (Yong et al., Reference Yong, Eamsobhana, Song, Prasartvit and Lim2015a) as well as the complete mitochondrial genome (Yong et al., Reference Yong, Song, Eamsobhana and Lim2016) unequivocally separated A. cantonensis and A. malaysiensis. Likewise, the nuclear genes, 66-kDa protein gene (Eamsobhana et al., Reference Eamsobhana, Lim, Zhang, Gan and Yong2010a) and ribosomal RNA small subunit (18S rRNA) gene (Eamsobhana et al., Reference Eamsobhana, Lim and Yong2015) clearly separated A. cantonensis and A. malaysiensis.
As for A. malaysiensis, A. mackerrasae, when first documented in Australia, was referred to as A. cantonensis (Mackerras & Sandars, Reference Mackerras and Sandars1954, Reference Mackerras and Sandars1955). Recent studies based on molecular markers indicate that it is highly similar to A. cantonensis (Aghazadeh, Reference Aghazadeh2015; Aghazadeh et al., Reference Aghazadeh, Trau, Mohandas, Aland, Reid, McCarthy and Jones2015; Chan et al., Reference Chan, Barratt, Roberts, Lee, Shea, Marriott, Harkness, Malik, Jones, Aghazadeh, Ellis and Stark2015). These studies did not include A. malaysiensis, another member of the A. cantonensis species complex, for comparison. Furthermore, only a single mitogenome of A. cantonensis from China (GQ398121) was included for comparison based on amino acid sequence (Aghazadeh et al., Reference Aghazadeh, Trau, Mohandas, Aland, Reid, McCarthy and Jones2015).
In the present study, we examined the genetic distance and phylogenetic relationships between the component taxa (A. cantonensis, A. mackerrasae and A. malaysiensis) within the A. cantonensis species complex, based on the 12 protein-coding genes from their mitochondrial genomes. Both the nucleotide and amino acid sequences were analysed. The results indicate that A. mackerrasae and A. cantonensis are members of the same genetic lineage and both are genetically distinct from A. malaysiensis.
Materials and methods
The complete mitochondrial genome of A cantonensis (KT947978 from Thailand; NC_013065 from China) and A. malaysiensis (NC_030332 from Malaysia) were downloaded from GenBank (National Center for Biotechnology Information, NCBI). The nucleotide sequences of the 12 protein-coding genes (PCGs) of A. mackerrasae from Australia were from Aghazadeh (Reference Aghazadeh2015). Other mitogenomes of Angiostrongylus available in NCBI's GenBank were included for comparison – A. costaricensis Costa Rica (KR827449), A. costaricensis Brazil (NC_013067) and A. vasorum (NC_018602). The mitogenomes of Metastrongylus pudendotectus (NC_013813) and M. salmi (NC_013815) were included as outgroup taxa.
Only the 12 PCGs were used for phylogenetic analysis as sequences for rRNA and tRNA genes were not available for A. mackerrasae. Phylogenetic analysis based on nucleotide sequences was performed as described in Yong et al. (Reference Yong, Song, Eamsobhana and Lim2016). Evolutionary analyses based on amino acid sequences were conducted in MEGA7 (Kumar et al., Reference Kumar, Stecher and Tamura2016). The median joining (MJ) network (Bandelt et al., Reference Bandelt, Forster and Röhl1999) was used to estimate the genealogical relationships of the haplotypes. The MJ network was calculated using NETWORK v5.0.0.1 (http://www.fluxus-engineering.com).
Results and discussion
Figure 1 depicts the molecular phylogeny of A. mackerrasae in relation to other members (A. cantonensis and A. malaysiensis) of the A. cantonensis species complex and other taxa of the Angiostrongylidae, as well as Metastrongylus taxa, based on 12 mt-PCGs. Angiostrongylus mackerrasae formed a sister lineage with A. cantonensis and both were distinctly different from A. malaysiensis. The A. cantonensis species complex formed a distinct clade from that comprising A. costaricensis and A. vasorum.
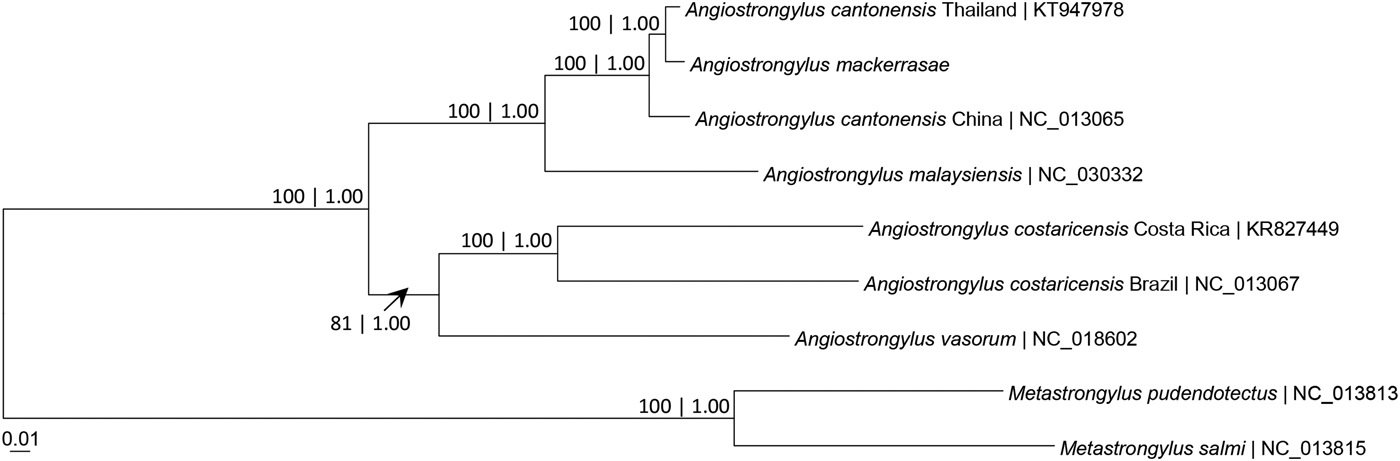
Fig. 1. Maximum likelihood and Bayesian inference tree based on 12 protein-coding genes of the whole mitogenomes of Angiostrongylus and Metastrongylus taxa. The total nucleotide sequence was 10,429 bp, AIC model = GTR + Gamma and BIC model = SYM + Gamma. Numeric values at the nodes are ML bootstrap/Bayesian posterior probabilities.The A. cantonensis species complex formed a distinct clade, with A. mackerrasae and A. cantonensis forming a sister lineage.
The percentage of uncorrected ‘p’ genetic distances between different pairs of Angiostrongylus taxa and Metastrongylus taxa, based on nucleotide sequence and amino acid sequence of 12 PCGs, are summarized in table 1. Based on the nucleotide sequence, the genetic distance between A. mackerrasae and A. cantonensis from Thailand was p = 1.73%, while that between A. mackerrasae and A. cantonensis from China was p = 3.70%; the genetic distance between A. cantonensis from Thailand and from China was p = 3.52%. The genetic distance between A. mackerrasae and A. malaysiensis was p = 12.07% while that between A. malaysiensis and A. cantonensis was 11.89% (from Thailand) and 12.15% (from China). The genetic distance between other congeners was p > 15%.
Table 1. Percentage of uncorrected ‘p’ genetic distance between different pairs of Angiostrongylus taxa and Metastrongylus taxa based on the nucleotide sequence (below diagonal) and amino acid sequence (above diagonal) of 12 protein-coding genes (PCGs) of the mitochondrial genome.

The magnitude of genetic difference was similar based on amino acid sequence (table 1). The genetic distance between A. mackerrasae and A. cantonensis was p = 2.6% (for the Thai taxon) and p = 5.8% (for the Chinese taxon), while that between the Thai and Chinese taxa of A. cantonensis was p = 5.6%. Compared to A. malaysiensis, the genetic distance was p = 20.2% for A. mackerrasae, p = 19.6% for A. cantonensis from Thailand, and p = 20.7% for A. cantonensis from China.
Based on the nucleotide sequence of cytochrome c oxidase subunit I (cox1) gene, the genetic distance between A. mackerrasae and A. cantonensis from Thailand was p = 1.90%, while that between the Thai and Chinese taxa of A. cantonensis was p = 3.80% (table 2). The genetic distance between A. mackerrasae and A. malaysiensis was p = 9.00%, and that between A. cantonensis and A. malaysiensis was p = 8.56–9.13%. Other congeners had genetic distances of p > 11%.
Table 2. Percentage of uncorrected ‘p’ genetic distance between different pairs of Angiostrongylus taxa and Metastrongylus taxa based on the nucleotide sequence of the complete cytochrome c oxidase subunit I (cox1) gene.

Haplotype network analysis based on individual mt-PCG revealed close genealogical relationships of A. mackerrasae and A. cantonensis compared to A. malaysiensis (table 3; fig. 2). Angiostrongylus mackerrasae and A. cantonensis from Thailand had the smallest difference in the number of base pairs in 11 of the 12 mt-PCGs (ATP6, COI, COB, COII, COIII, ND2, ND3, ND4, ND4L, ND5 and ND6), while A. cantonensis from Thailand and China had a larger difference (table 3). For the ND1 gene, A. mackerrasae differed from A. cantonensis from Thailand by 37 bp (4.35%) and from A. cantonensis from China by 50 bp (5.88%), while the difference between A. cantonensis from Thailand and China was 29 bp (3.41%).

Fig. 2. Haplotype network of Angiostrongylus cantonensis species complex based on mitochondrial protein-coding gene sequences generated by NETWORK software. Abbreviations of gene names: ATP6, ATP synthase subunit 6; COB, cytochrome b; COI–III, cytochrome c oxidase subunit 1–3; ND1–6, nicotinamide-adenine-dinucleotide dehydrogenase subunit 1–6.
Table 3. Differences in number of base pairs between component taxa of Angiostrongylus cantonensis species complex as determined by haplotype network analysis of 12 mitochondrial protein-coding genes. Amac, A. mackerrasae; AcanT, A. cantonensis Thailand; AcanC, A. cantonensis China; Amal, A. malaysiensis.

ATP6, ATP synthase subunit 6; COB, cytochrome b; COI–III, cytochrome c oxidase subunit 1–3; ND1–6, nicotinamide-adenine-dinucleotide dehydrogenase subunit 1–6.
Until recently, molecular studies were lacking for A. mackerrasae compared to the other members (A. cantonensis and A. malaysiensis) of the A. cantonensis species complex. A recent study indicates that A. mackerrasae and A. cantonensis are almost identical at two genetic loci (the nuclear internal transcribed spacer (ITS)-1 and 18S rRNA genes), indicating that these two taxa are conspecific (Chan et al., Reference Chan, Barratt, Roberts, Lee, Shea, Marriott, Harkness, Malik, Jones, Aghazadeh, Ellis and Stark2015). Another study, based on restriction enzymes for the cox3 region, failed to distinguish A. mackerrasae from A. cantonensis (Aghazadeh, Reference Aghazadeh2015). Based on the amino acid sequence of the 12 mitochondrial PCGs, the genetic distance between A. mackerrasae and A. cantonensis is p = 2.4% (Aghazadeh et al., Reference Aghazadeh, Trau, Mohandas, Aland, Reid, McCarthy and Jones2015), indicating that these two taxa are genetically very similar.
Compared to A. mackerrasae, molecular markers (both mitochondrial and nuclear genes) could distinguish A. malaysiensis from A. cantonensis unambigiously (Eamsobhana et al., Reference Eamsobhana, Lim, Zhang, Gan and Yong2010a, Reference Eamsobhana, Lim, Solano, Zhang, Gan and Yongb, Reference Eamsobhana, Lim and Yong2015, Reference Eamsobhana, Yong, Song, Prasartvit, Boonyong and Tungtrongchitr2017a, Reference Eamsobhana, Song, Yong, Prasartvit, Boonyong and Tangtrongchitrb; Yong et al., Reference Yong, Eamsobhana, Song, Prasartvit and Lim2015a, Reference Yong, Song, Eamsobhana and Lim2016). The genetic distance bwteen A. cantonensis and A. malaysiensis is p = 11.9% based on 12 PCGs, p = 9.5% based on 2 rRNA genes, and p = 11.6% based on 14 mt-genes (Yong et al., Reference Yong, Song, Eamsobhana and Lim2016).
In the present study, the genetic distance based on concatenated nucleotide sequences of 12 mt-PCGs indicates that A. mackerrasae falls within the intraspecific range for A. cantonensis – p = 1.73% between A. mackerrasae and A. cantonensis from Thailand compared to p = 3.52% between the Thai and Chinese taxa of A. cantonensis. Both A. mackerrasae and A. cantonensis are genetically distinct from A. malaysiensis (p > 11%). A large genetic difference between sibling species is also found in the Costa Rican and Brazilian taxa of A. costaricensis (p = 16.17%, table 1; Yong et al., Reference Yong, Song, Eamsobhana, Goh, Lim and Chow2015b). The magnitude of the genetic difference is similar for the amino acid sequence of the 12 PCGs (table 1) as well as the complete mitochondrial cox1 nucleotide sequence (table 2).
Phylogenetic analysis based on 12 mt-PCGs indicates that A. mackerrasae is a member of the A. cantonensis genetic lineage (fig. 1). Based on haplotype network analysis, 11 of the 12 mt-PCGs also show a closer genetic relationship between A. mackerrasae and A. cantonensis from Thailand than between both of these and A. cantonensis from China (table 3; fig. 2). These results agree with the notion that A. mackerrasae may be conspecific with A. cantonensis (Chan et al., Reference Chan, Barratt, Roberts, Lee, Shea, Marriott, Harkness, Malik, Jones, Aghazadeh, Ellis and Stark2015).
In Australia, A. cantonensis occurs in introduced rat species (Rattus rattus and Rattus norvegicus) while A. mackerrasae is found mainly in native bush rats, Rattus fuscipes (Spratt, Reference Spratt2015). In mainland Asia, various species of murid rodents (both native and introduced) are the final or definitive hosts of A. cantonensis (Yong & Eamsobhana, Reference Yong and Eamsobhana2013; Eamsobhana et al., Reference Eamsobhana, Yong, Prasartvit, Wanachiwanawin and Boonyong2016). An extensive phylogeographical study of the parasites from mainland Asia, Indonesia, New Guinea, Australia and other Pacific islands will provide conclusive evidence for the taxonomic status and origin of these parasites.
In this study, we have demonstrated that A. mackerrasae is genetically very similar to A. cantonensis and that these taxa form a sister lineage. The small genetic distance between these taxa (p = 1.73–3.70%), compared to that with A. malaysiensis (p = 11.89–12.15%), indicates the need to re-examine the taxonomic status of A. mackerrasae (referred to as A. cantonensis when first documented in Australia). Further studies, particularly phylogeography, are needed to resolve whether it is conspecific with A. cantonensis or undergoing incipient speciation.
Financial support
We thank our institutions for providing various research facilities and other support. This study was funded by the University of Malaya (H-5620009).
Conflict of interest
None.







