Introduction
Trichinella spiralis is one of the most widespread nematodes and can infect humans and many other species of mammal. Trichinellosis remains an important food-borne parasitic zoonosis. The formation of nurse cells in host muscle cells is a key step of the infective mechanism, as it provides a suitable long-term habitat for larvae (Polvere et al., Reference Polvere, Kabbash, Capo, Kadan and Despommier1997; Capo et al., Reference Capo, Despommier and Polvere1998), comprising a constant and reliable system for the supply of nutrients and waste removal required for the worm to develop (Kang et al., Reference Kang, Jo, Cho, Yu, Ock and Cha2011). The T. spiralis surface includes a cuticle, an epicuticle and a surface coat, which are vital for physiological functioning (Maizels et al., Reference Maizels, Blaxter and Selkirk1993). During their invasion and development, parasites produce numerous molecules as part of a survival strategy against adverse changes (Dzik, Reference Dzik2006). Among the antigens of T. spiralis infective larvae, excretory–secretory products (ESP) and surface antigens are the main target of the immune response, and play an important role in the invasion and development process of Trichinella larvae (Robinson et al., Reference Robinson, Massie and Connolly2007). Trichinella antigens are specific to different developmental stages (Parkhouse & Ortega-Pierres, Reference Parkhouse and Ortega-Pierres1984), evoking a protective stage-specific host immune response, owing to the uniqueness of both the cuticular antigens and the ESP antigens of each stage (Wang & Bell, Reference Wang and Bell1992).
Briefly, the cuticle is a multilayered structure composed mainly of collagens. It is synthesized five times during development: during embryogenesis and during each of the four larval stages before each moult (Johnstone, Reference Johnstone2000). Collagen biosynthesis is a complex multistep process, involving chaperones and numerous modifying, folding and processing enzymes. The first important cotranslational modification of procollagen is prolyl 4-hydroxylation, which allows its proper folding into a thermally stable form. The α-subunit of the prolyl 4-hydroxylase enzyme associates with the β-subunit of protein disulphide isomerase (PDI) to form active soluble enzyme complexes, which are resident in the endoplasmic reticulum. Collagen trimerization is set up via PDI-catalysed disulphide bond formation. In nematodes, the presence of PDI has been described in Onchocerca volvulus (Wilson et al., Reference Wilson, Tuan, Shepley, Freedman, Greene, Awadzi and Unnasch1994), Ancylostoma caninum (Epe et al., Reference Epe, Kohlmetz and Schnieder1998, Reference Epe, Berens and Strube2007), Ostertagia ostertagi (Vercauteren et al., Reference Vercauteren, Geldhof, Peelaers, Claerebout, Berx and Vercruysse2003) and Caenorhabditis elegans (Winter & Page, Reference Winter and Page2000). The draft genome of T. spiralis has been published (Mitreva et al., Reference Mitreva, Jasmer, Zarlenga, Wang, Abubucker, Martin, Taylor, Yin, Fulton, Minx, Yang, Warren, Fulton, Bhonagiri, Zhang, Hallsworth-Pepin, Clifton, McCarter, Appleton, Mardis and Wilson2011), and different PDI family members are listed, such as PDI (PDIA2), ERp57 (PDIA3) and ERp72 (PDIA4). Endoplasmic reticulum molecular chaperones, such as calreticulin (CRT), calnexin (CNX) and the 78 kDa glucose-regulated protein precursor/immunoglobulin heavy chain-binding protein (GRP78/BIP) are also described. Both CRT and CNX are functionally coupled, establishing a calreticulin–calnexin cycle, which is physically associated with ERp57 (PDIA3). Protein disulphide isomerase is an essential catalyst of the endoplasmic reticulum in different biological systems, acting as a dithiol–disulphide oxidoreductase capable of reducing, oxidizing and isomerizing disulphide bonds, and also acting as a chaperone. It is the founding member of a family of about 20 related mammalian proteins that are mainly located and functioning in the endoplasmic reticulum. The members vary in length and domain arrangement, but share the common structural feature of having at least one domain with a thioredoxin-like structural fold. Most PDI family members (PDIs) contain both catalytic motifs and non-catalytic thioredoxin-like domains (Kozlov et al., Reference Kozlov, Määttänen, Thomas and Gehring2010). Although PDIs are endoplasmic-reticulum-resident proteins, they can be found at the plasma membrane, where they regulate the redox status of the cell surface.
No specific antibodies are available for the localization of T. spiralis PDIs. This fact prompted our group to search the T. spiralis databases of PDIs for the identity and similarity of amino acid peptide sequences used in the production of commercial antibodies for human-orthologous PDI family members. A search of the T. spiralis database (taxid 6334) revealed that some of these antibodies should cross-react with T. spiralis PDIs. This paper reports our immunocytochemical analysis of the localization of T. spiralis PDIs in muscle larvae. We demonstrate the presence of different members of the PDI family and chaperones, as well as their localization on the larvae.
Materials and methods
Experimental infection and sample preparation
Trichinella spiralis (Spanish strain) was maintained by serial passage infections in female rats (Rattus rattus) according to the method of Pozio et al. (Reference Pozio, La Rosa, Rossi and Murrell1992). Tissue samples of the tongue, diaphragm and masseter muscle were collected from rats infected with T. spiralis for immunocytochemical analysis. Immediately after dissection, tissues were fixed by immersion in 10% formalin for 24 h, dehydrated by immersion in increasing concentrations of alcohol, incubated with xylene, infiltrated with paraffin (60°C) and embedded in paraffin blocks.
Trichinella spiralis genome databases
A string search of the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/) for previously listed proteins was undertaken. A search of the T. spiralis genome database (taxid 6334) for orthologues to human PDI family members was made using the BLAST algorithm (Altschul et al., Reference Altschul, Madden, Schaffer, Zhang, Zhang, Miller and Lipman1997). The Scratch program (http://scratch.proteomics.ics.uci.edu/; Cheng et al., Reference Cheng, Randall, Sweredoski and Baldi2005) was used for a linear B-cell epitope search of the amino acid sequences of the antigen used for commercial anti-PDI antibody production.
Immunocytochemistry
Paraffin sections, 3 μm thick, of routinely processed, formalin-fixed, paraffin-embedded material were cut and mounted on glass slides (SuperFrost Ultra Plus®, Thermo Scientific, Waltham, Massachusetts, USA), ensuring a firm electrostatic attraction. Immunostaining was performed using an automated immunostainer (Ventana BenchMark® ULTRA, Ventana Medical Systems Inc., Tucson, Arizona, USA). Briefly, sections were deparaffinized and rehydrated with EZ Prep (Ventana), and heat-induced epitope retrieval was accomplished using CC1 (Ventana). Primary antibody anti-PDIs (Sigma, St. Louis, Missouri, USA) were employed at optimized dilutions: polyclonal rabbit anti-human PDIA3 (HPA003230), 1:100; polyclonal rabbit anti-human GRP78 (G8918), 1:300; polyclonal rabbit anti-human TMX4 (HPA000399), 1:50. Reactivity was detected using the UltraView detection kit (Ventana). Slides were counterstained with haematoxylin and blue-dye solutions. Sections were dehydrated with a graded series of ethanol and cleared in xylene. A cover slip was placed over the sections and secured with mounting medium, to protect the tissue. Positive and negative controls were included with each assay.
The human appendix (from the hospital's histological archive) was used as a positive control. Negative controls were treated in the same way as samples and positive controls, except for the absence of the primary antibody. No stain was observed in negative control tissues (results not shown).
Comparative modelling and validation
A theoretical three-dimensional model for PDIA3 from T. spiralis was constructed by comparative modelling using the Swiss-Model server, version 3.7 (Arnold et al., Reference Arnold, Bordoli, Kopp and Schwede2006; Kiefer et al., Reference Kiefer, Arnold, Künzli, Bordoli and Schwede2009; Biasini et al., Reference Biasini, Bienert, Waterhouse, Arnold, Studer, Schmidt, Kiefer, Cassarino, Bertoni, Bordoli and Schwede2014; http://swissmodel.expasy.org/), which is accessible via the ExPASy web server (http://expasy.org/). The model quality was checked using the QMEAN program (Benkert et al., Reference Benkert, Biasini and Schwede2011). The Swiss-Pdb Viewer, version 4.0.4 (Guex & Peitsch, Reference Guex and Peitsch1997) was used for model visualization.
Results and discussion
Trichinella spiralis genome database
The string search performed for T. spiralis PDI found a number of annotated proteins (table 1): PDI2, PDIA3 and PDIA4, and a DnaJ protein subfamily C member 10. In addition, the chaperones CRT, CNX and heat-shock protein 70 were listed in the database.
Table 1 An exploration of the Trichinella. spiralis genome database for human-orthologous PDI family members using accession codes from databases. Query cover: % protein in analysis (T. spiralis) matching the template protein (human); identity: % sequence alignment presenting identical amino acids in both query and template sequences; E-value: statistical significance of a pairwise alignment of sequences, reflecting the size of the database and the quality of alignment (score) used, the lower the E-value the higher the congruity between the query and retrieved sequences, where a value of 0 indicates a precise match.

Reverse search on Human genome database found *TMX2 and **TMX3 as the highest homologous proteins.
A search for human-orthologous accession codes on the T. spiralis genome database revealed other PDI family members and chaperones (table 1). Orthologues for the human thioredoxin-related-transmembrane protein (TMX) were found: the thioredoxin domain-containing protein 1, the thioredoxin domain-containing protein 14-like protein and the conserved hypothetical protein. In addition, human PDIA10 matched, with 45% identity, the sequence from T. spiralis thioredoxin domain-containing protein 4; both exhibited the same active site amino acid sequence (CRFS).
Immunolocalization of Trichinella spiralis protein disulphide isomerase family members
Analysis of the paraffin sections of rat muscle infected with T. spiralis larvae after staining with commercial antibodies showed positive results for CRT, CNX, GRP78, PDIA3, TMX1 and TMX4. These results suggest that, in a first approach, some commercial anti-human antibodies could be used to detect orthologous molecules in T. spiralis. To validate these results, the specificity of these antibodies was checked by searching the amino acid sequence of the peptides used for commercial production of antibodies on the T. spiralis genome database (taxid 6334). When large amino acid sequences were used as immunogens, the potential linear B-cell epitope propensity and respective amino acid sequence were also predicted using the Scratch prediction method (Cheng et al., Reference Cheng, Randall, Sweredoski and Baldi2005) and a search for homologous amino acid sequences in T. spiralis genome (taxid 6334) was performed.
Immunocytochemical analysis of binding cross-reactivity between Homo sapiens and T. spiralis of the commercial antibodies gave a positive result (not shown), suggesting that both anti-human PDIA3 antibodies could be used for the orthologous proteins in T. spiralis. A search of the T. spiralis genome database for the whole sequences used as an immunogen, in addition to the predicted amino acid sequences for the linear B-cell epitope, identified a match with T. spiralis orthologue XP_003373217.1 (EFV54589.1).
With respect to CNX, CRT and TMX1, similar searches of the T. spiralis database did not provide any results, suggesting that the binding of antibodies does not confirm their potential use in immunolocalization.
However, a database search using the amino acid sequences of the immunogens used for production of commercial human antibodies to TMX4 and GRP78 gave a different result. The peptide corresponding to human GRP78 matched the entries in the T. spiralis database for heat-shock protein 70 (AAK85149.2) and heat-shock protein C (EFV60727.1). A reverse search of these two amino acid sequences on the human database (taxid 9606) found, with the highest amino acid sequence identities, the same three entries for AAK85149.2: the GRP78 precursor entries NP_005338.1 and AAA52614.1 (82% and 81% identity, respectively), and the BIP protein entry AAF13605.1 (82% identity), validating the results obtained by immunocytochemical analysis.
In the case of the antibody to human TMX4, the amino acid sequence used as an immunogen matched the entry in the T. spiralis database for thioredoxin domain-containing protein 1 (EFV59702.1), which corresponds to the TMX1 human orthologue. This result suggests that the anti-human TMX4 binds to the TMX1 orthologue in T. spiralis.
From the constructed three-dimensional model for T. spiralis ERp57 (PDIA3) (not shown), it is possible to identify the expected four domains. It is also possible to see that the peptides predicted to be the linear B-cell epitopes for the commercial antibodies are located in domains b (127 GPSSKELKTADDFKK 141), b′ (256 DVDYERN 262; 323 KYVMKDEFS 331) and a′ (352 KSEPIPETNDNP 363; 448 PQSYTGGRTLDDFI 461) at the surface of the molecule, as expected, and accessible to the binding of commercial antibodies.
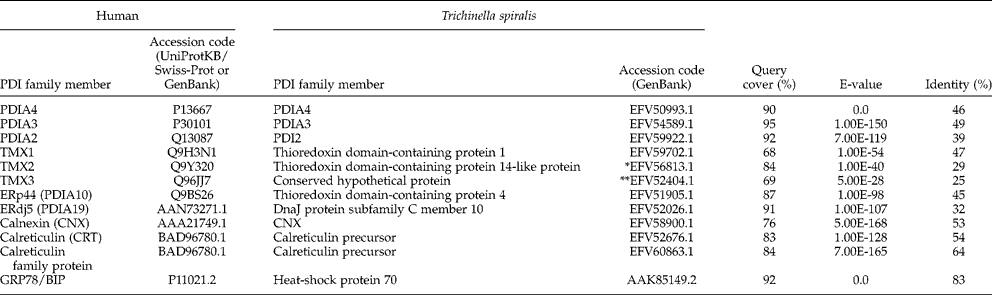
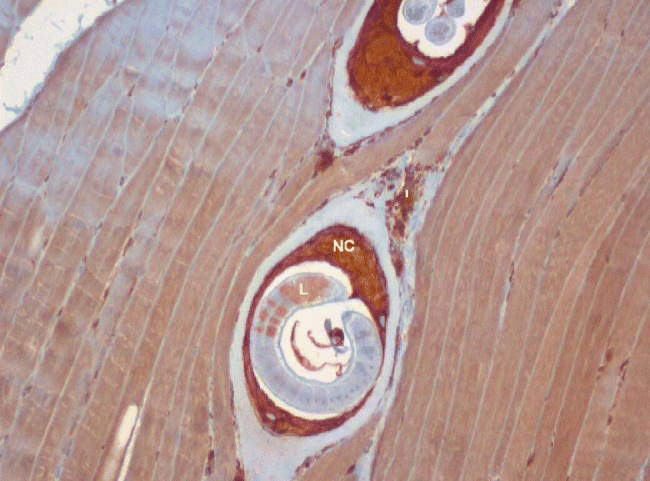
Paraffin sections stained using the antibodies whose predicted specificity suggested that they could be used for orthologous identification in T. spiralis larvae are shown in fig. 1 (for PDIA3; those for TMX1 and GRP78 are not shown). Analysing fig. 1, we can see intense immunostaining for PDIA3 in the developing nurse cell cytoplasm, moderate staining for PDIA3 in the surrounding inflammatory cell infiltrate and weak focal staining for PDIA3 in the larvae. The intense expression of PDIA3 in the nurse cell cytoplasm is suggestive of its role in the formation of the nurse cell. An intense immunostaining of TMX1 (cross-reacting with the anti-TMX4 antibody used) was observed in the nurse cell cytoplasm and in particular detail in the larvae. For GRP78, moderate immunostaining was noted in the developing nurse cell cytoplasm, while intense granular nuclear expression was observed in the larvae.
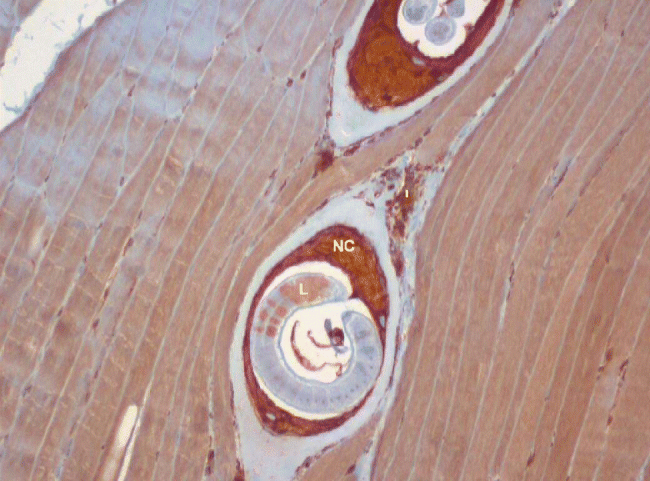
Fig. 1 Immunocytochemistry analysis. Rat muscle infected with T. spiralis larvae stained with commercial anti-human PDIA3 (ERp57) antibody. Magnification: × 100. NC, nurse cell cytoplasm; I, inflammatory infiltrate; L, larva.
The immunocytochemical results suggest the localization of three important molecules belonging to the PDI family and chaperones, which were predicted by computational genome analysis. Unfortunately, the immunostaining results obtained with other anti-human antibodies did not agree with the computational cross-reaction prediction; these results are not shown. In particular, it is important to localize the CRT/CNX chaperone system, as it is intimately related with the activity of PDIA3. Moreover, the relationships between these molecules and induced endoplasmic reticulum stress (Yu et al., Reference Yu, Deng, Lu, Zhang, Jia, Huang and Qi2014) and their role in the induced protective immunity from infection in the host (Fang et al., Reference Fang, Sun, Yang, Gu, Zhan, Huang and Zhu2014) suggests the importance of knowing the localization of these molecules during nurse cell formation.
Acknowledgements
We thank Dr Dinora Ferreira, Vitor Domingos and Maria Rosário Tito for assistance with animals in the IHMT_UNL animal room.
Financial support
This research received no specific grant from any funding agency, commercial interest or not-for-profit sector.
Conflict of interest
None.
Ethical standards
Work dealing with pathogens and drugs was performed under controlled conditions, and all safety measures were taken. If needed, the material used for laboratory practices was sterilized before being discarded appropriately. Good animal handling practices as well as good laboratory practices were used and all EU Directives were followed, namely: 2000/54/CE Exposure to biological agents, 88/320/CE Good laboratory practices, 90/679/CE, 405/98/PT and 1036/98/PT. Workers were protected against exposure to dangerous biological agents during work time. In no known way did this project contravene the social, ethical and environmental laws or principles accepted in Portugal and in the European Union. The person developing the project was certified by the national body for conducting experimental animal manipulations.