Introduction
The disruption and clearing of forested areas by activities such as residential construction, agriculture, industrial operations and public utility development are occurring worldwide. This disturbance of wildlife habitats has led to an increase in various socio-economic problems, including emerging and re-emerging zoonoses. In Thailand, elephants (Elephas maximus) have been seriously affected by increasing fragmentation of their forest habitat and decreasing habitat quality, forcing them to appear frequently at forest edges and to trespass in agricultural and residential areas. In addition to these threatening and destructive activities, wild elephants are reservoirs of diseases, such as helminthiasis, that cause health problems in both humans and domestic animals (Horak et al., Reference Horak, Boomker, De Vos and Potgieter1988). Parasitic nematodes that have been frequently observed in previous studies of Thai and African elephants include Murshidia falcifera, M. neveulemairei, Murshidia sp., Leiperenia leiperi, Khalilia buta, K. sameera, Quilonia spp., Parabronema africanum, P. rhodesiense, Parabronema spp., Oesophagostomum mocambiquei and O. mwanzae (Greve, Reference Greve1969; Basson et al., Reference Basson, McCully, de Vos, Young and Kruger1971; Sikes, Reference Sikes1971; Soulsby, Reference Soulsby1982; Horak et al., Reference Horak, Boomker, De Vos and Potgieter1988; Boomker et al., Reference Boomker, Horak and Flamand1991; Carreno et al., Reference Carreno, Neimanis, Lindsjo, Thongnoppakun, Barta and Peregrine2001; du Toit, Reference du Toit2001; Boomker, Reference Boomker2014). Interestingly, no eggs or larvae of Oesophagostomum spp. have been reported in Thai wild elephants.
Oesophagostomum, or nodule worm, is a parasitic nematode belonging to the family Chabertiidae. Infection with these helminths can cause granulomas in the walls of the small and large intestines. Oesophagostomum bifurcum is a common species that has been detected in livestock, non-human primates and humans (Gasser et al., Reference Gasser, de Gruijter and Polderman2006), while O. aculeatum has been reported in the Japanese macaque and is also potentially transmitted to humans (Arizono et al., Reference Arizono, Yamada, Tegoshi and Onishi2012; Ghai et al., Reference Ghai, Chapman, Omeja, Davies and Goldberg2014). We thus screened faecal samples of wild elephants for Oesophagostomum, using the small subunit ribosomal DNA gene (SSU rDNA) as a genetic marker. This marker has been widely used to clarify the taxonomy and molecular relationships of many parasite groups (Dorris et al., Reference Dorris, Viney and Blaxter2002; Marigo et al., Reference Marigo, Thompson, Santos and Iñiguez2011; Anderson et al., Reference Anderson, Upadhayay, Sudimack, Nair, Leland, Williams and Anderson2012). The present results indicate that Oesophagostomum found in wild elephants may pose a potential health risk to local inhabitants and domestic animals.
Materials and methods
Collection and examination of faecal samples
Forty-five faecal samples from wild elephants were collected from the grounds of the Salakpra Wildlife Sanctuary, Kanchanaburi Province, Thailand during January through November 2013. All samples were observed, characterized and graded for freshness by veterinarians. Geographic coordinates of each sample were recorded (table 1).
Table 1 Collection sites, with coordinates, of elephant faecal samples from Kanjanaburi Province, Thailand.

To identify helminth eggs and larvae, samples were subjected to faecal concentration techniques, including sedimentation and flotation methods (Setasuban, Reference Setasuban1989). All samples were examined for helminth eggs and larvae using a light microscope (Carl Zeiss, Jena, Germany). To prepare faecal samples for extraction of copro-DNA, 2 g of each sample was dissolved in 200 ml of distilled water, filtered through two layers of wet cotton gauze, and left undisturbed for 1–2 h to obtain precipitates. After two additional cycles of dissolution and filtration, the precipitates were combined and stored at − 70°C for future molecular analysis.
Molecular analysis
Precipitates of faecal samples were snap-frozen in liquid nitrogen and then ground with a plastic pestle. The homogenization was repeated three times to ensure complete breakage of helminth eggs and larvae. The copro-DNA was then extracted using a FavorPrep Stool DNA Isolation Mini kit (Favorgen, Ping-Tung, Taiwan) according to the manufacturer's instructions.
To confirm and determine the quality of wild elephant DNA, a portion of the cytochrome b (Cytb) gene was amplified in all faecal samples. A portion of the SSU rDNA gene was amplified for identification of intestinal nematode species. Sequences of primers used to amplify Cytb and SSU rDNA genes, which followed Fernando et al. (Reference Fernando, Pfrender, Encalada and Lande2000) and Dorris et al. (Reference Dorris, Viney and Blaxter2002), respectively, are shown in table 2. Amplifications of both genes were carried out in 50-μl reaction volumes containing 1 μl of DNA template, 0.5 μm of each primer, and 1 × TopTaq Master Mix (Qiagen, Hilden, Germany). The amplification profile used for Cytb was as follows: 95°C for 5 min; followed by 35 cycles of 95°C for 60 s, 60°C for 60 s and 72°C for 60 s; and a final extension of 72°C for 5 min. For SSU rDNA, the amplification profile consisted of 95°C for 5 min; followed by 35 cycles of 95°C for 30 s, 53°C for 30 s and 72°C for 30 s; and a final extension of 72°C for 5 min. All polymerase chain reaction (PCR) products were analysed by 1% agarose gel electrophoresis and submitted to Macrogen (Seoul, Republic of Korea) for sequencing.
Table 2 Oligonucleotide primers used for PCR amplification of tetramicrosatellite loci and Cytb and SSU rDNA genes.
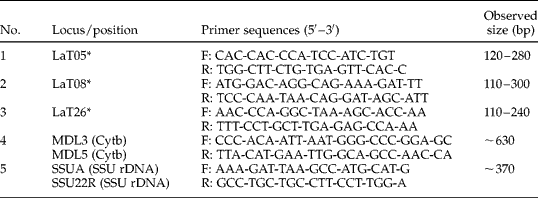
* Microsatellite markers; F, forward; R, reverse.
Tetranucleotide microsatellite markers were used to determine the number of individual elephants, according to previous studies (Archie et al., Reference Archie, Moss and Alberts2003; Suwattana et al., Reference Suwattana, Jirasupphachok, Kanchanapangka and Koykul2010). In particular, three tetramicrosatellite loci (LaT05, LaT08 and LaT26) were amplified in 50-μl reaction volumes containing 1 × TopTaq Master Mix (Qiagen), 0.5 μm of each primer (table 2), and 1 μl of DNA template. Amplification conditions for LaT05 and LaT08 consisted of 95°C for 10 min, followed by one cycle of 95°C for 30 s, 66°C for 30 s and 72°C for 30 s. Ten subsequent cycles were performed under identical conditions, except that the annealing temperature was decreased by 1°C per cycle; this was followed by 30 cycles of 95°C for 30 s, 56°C for 30 s and 72°C for 30 s; with a single extension of 72°C for 5 min. For LaT26, amplification conditions were 95°C for 10 min; followed by 40 cycles of 95°C for 30 s, 53°C for 30 s and 72°C for 30 s; and a single extension step of 72°C for 5 min. PCR products were analysed by 6% polyacrylamide gel electrophoresis.
Data analysis
The obtained partial SSU rDNA sequences were compared with related nematode sequences (table 3) to identify species and haplotypes, using the multiple alignment program Clustal X version 2.0 (Thompson et al., Reference Thompson, Gibson, Plewniak, Jeanmougin and Higgins1997). A neighbour-joining phylogram based on p-distances was constructed in MEGA version 5.0 (Tamura et al., Reference Tamura, Peterson, Peterson, Stecher, Nei and Kumar2011). Bootstrap analyses were conducted using 1000 replicates.
Table 3 Accession numbers of sequences downloaded from GenBank for use in the present study.
Results
Microscopic examination of elephant faeces revealed helminth eggs and larvae in 40 of the 45 samples, and these stages were categorized into three nematode groups, including the strongylids, trichurids and ascarids. Strongylid eggs measuring 80–85 × 42–47 μm were found in all 40 infected samples, compared to trichurid eggs, measuring 55–62 × 27–31 μm, which were found in one sample (fig. 1A and B). Ascarid eggs measuring 58–65 × 43–47 μm were also found in one sample (fig. 1C), which was concurrently infected with strongylid eggs. Additionally, eight samples contained rhabditiform larvae with buccal canal and rhabditiform oesophagus lengths of 15–19 μm and 100–110 μm, respectively. Their tails were slightly long, 90–95 μm from the anus to the tip of the tail. Larval bodies in entirety were 590–640 × 17–21 μm (fig. 1D), with unclear genital primordia.
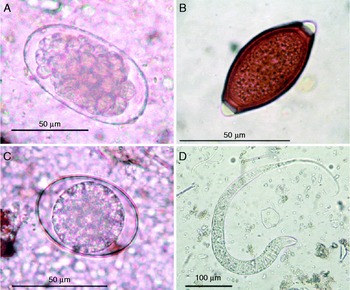
Fig. 1 Egg and larval stages identified in faecal samples of wild elephants, including eggs of (A) O. aculeatum, (B) Trichuris sp., (C) Ascaris sp. and (D) the rhabditiform larva of O. aculeatum.
An accurate determination of helminth prevalence and genetic diversity was impeded by the difficulty in identifying the source of each faecal sample. Tetranucleotide microsatellite analysis was therefore performed to identify the actual number of distinct faecal samples. The Asian elephant Cytb gene was first amplified in all samples by conventional PCR to determine the specificity and quality of extracted DNA. All samples yielded PCR products of the expected size for the genus Elephas (elephant) – approximately 630 bp long. Sequences generated from these products closely matched DNA sequences of E. maximus (Asian elephant) in GenBank (data not shown). Samples were further analysed for individual differences using three tetramicrosatellite loci. This analysis yielded 8–10 alleles per locus and 11–15 haplotypes (data not shown). The analysis results indicated that the 45 faecal samples originated from 43 individual wild elephants. Consequently, the actual distribution of nematodes in our group of samples was 93.0% strongylids, 2.3% trichurids and 2.3% ascarids.
Microscopic examination revealed that 40 faecal samples were positive for strongylid eggs or larvae but did not allow species determination. SSU rDNA was therefore used as a marker for species identification. All samples were amplified with specific primers and gave products of the expected size, 370 bp. All amplicons were subsequently sequenced, but good-quality sequences were only obtained from 27 of the 40 samples. Alignment and comparison of the 27 sequences in Clustal X revealed five distinct haplotypes (A, B, C, D and E) (table 4). Thirteen DNA sequences belonging to haplotype A showed 100% identity with O. aculeatum SSU rDNA sequences from the Japanese macaque (Macaca fuscata yakui). Another 13 sequences, consisting of two haplotype B, eight haplotype C and three haplotype D sequences, were 99% identical to O. aculeatum. The sole haplotype E sequence matched Ancylostoma caninum (dog and cat hookworm), showing 99% identity.
Table 4 Identified haplotypes and variable nucleotide positions of the partial SSU rDNA gene (370 bp) and frequencies of these haplotypes in 27 O. aculeatum samples.
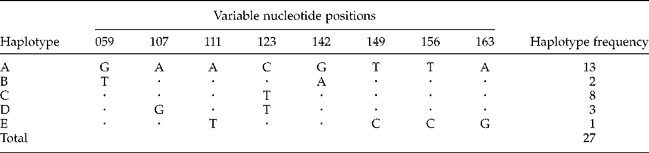
Dots correspond to nucleotides identical to the corresponding nucleotide in the haplotype A sequence. Position numbers refer to positions 1–895 of the SSU rDNA gene partial sequence (895 bp) of O. aculeatum (GenBank number AB677956).
Phylogenetic analysis of the SSU rDNA sequences yielded a tree composed of two main clades (fig. 2): clade I containing all helminths sequenced in this study as well as O. aculeatum and the hookworm species N. americanus, A. caninum and A. ceylanicum, and clade II comprising various Strongyloides species. Among the five helminth haplogroups, p-distances ranged from 0.004 to 0.024 (data not shown). Haplotype A was the same haplotype as O. aculeatum and was very closely related to N. americanus (0.004). Haplotypes C and D were sister to each other; these two haplotypes were in turn sister to haplotype A, N. americanus and O. aculeatum (0.004–0.012). Haplotype B was in the same group as haplotypes A, C, D, N. americanus and O. aculeatum, but was genetically the most distinct of these members (0.008–0.016). Haplotype E was distinct from the other four haplotypes (genetic distance ≥ 0.016) and was more closely related to A. ceylanicum and A. caninum. Strongyloides species were clearly separate, being restricted to clade II. Genetic distances between the two clades ranged from 0.311 to 0.452, with an average genetic distance of 0.333.

Fig. 2 Phylogenetic tree of O. aculeatum and related species based on partial SSU rDNA gene sequences; genetic relationships were inferred by the neighbour-joining method using Ascaris lumbricoides as an outgroup, and the scale bar indicates the number of nucleotide substitutions per site. Clade I comprises O. aculeatum, hookworm species and all haplotypes in the present study; see also table 4 for haplotypes A–E. Clade II comprises Strongyloides species.
Discussion
In this study, we investigated the prevalence of helminth infections in wild elephants in Thailand. Our microscopic examination detected three groups of helminths – strongylids, trichurids and ascarids – in the faeces of wild elephants. These groups have been found frequently in both Asian and African elephants (Greve, Reference Greve1969; Basson et al., Reference Basson, McCully, de Vos, Young and Kruger1971; Sikes, Reference Sikes1971; Soulsby, Reference Soulsby1982; Carreno et al., Reference Carreno, Neimanis, Lindsjo, Thongnoppakun, Barta and Peregrine2001; du Toit, Reference du Toit2001). Strongylids were the most common, being present in more than 90% of collected samples. The taxonomic characteristics used to identify this group – morphology of eggs and rhabditiform larvae (buccal canal and genital primordium) – were insufficient to allow species-level differentiation, with one exception – the tails of rhabditiform larvae of Oesophagostomum were slightly longer (from anus to the tip of the tail) than those of hookworms and Strongyloides (Blotkamp et al., Reference Blotkamp, Krepel, Kumar, Baeta, van't Noordende and Polderman1993; Bowman et al., Reference Bowman, Lynn, Georgi and Eberhard2003).
To identify nematode species, we used an alternative DNA-based approach involving SSU rDNA. Previous research has found that SSU rDNA can resolve deep phylogenetic relationships and shed light on the evolution of nematodes (Holterman et al., Reference Holterman, van der Wurff, van den Elsen, van Megen, Bongers, Holovachov, Bakker and Helder2006). SSU rDNA markers have also been used to identify successfully nematode species in genera such as Strongyloides (Dorris et al., Reference Dorris, Viney and Blaxter2002), Mexiconema (Mejia-Madrid & Aguirre-Macedo, Reference Mejia-Madrid and Aguirre-Macedo2011) and Angiostrongylus (Constantino-Santos et al., Reference Constantino-Santos, Basiao, Wade, Santos and Fontanilla2014). Comparison of SSU rDNA sequences of nematodes infecting elephants with their orthologues indicated that 26 sequences matched O. aculeatum from the Japanese macaque (AB677956). Faecal examination of long-tailed macaques in Thailand has also uncovered Oesophagostomum spp. infections (Malaivijitnond et al., Reference Malaivijitnond, Chaiyabutr, Urasopon and Hamada2006). The monkeys inhabiting the same area as the elephants in this study, however, have neither been identified to species nor examined. A future investigation at this study site addressing these issues might provide evidence of O. aculeatum transmission between monkeys and elephants.
When we compared SSU rDNA sequences of our samples with sequences available in GenBank, we found that 13 were 100% identical to the SSU rDNA sequence of O. aculeatum (AB677956). Another 13 were 99% identical. The high similarity of these SSU rDNA sequences may not be sufficient for confirm the identity of the isolated rhabditiform larvae. This identification should be verified by morphological observation and sequencing of adult worms. To obtain worms at the adult stage, however, wild elephants must be treated with an anthelmintic drug – a logistically impossible situation. Future application of additional genetic markers may be a good option to verify the species identification.
Our phylogenetic analysis of the order Strongylida placed O. aculeatum from various hosts, including wild elephants, into the same clade as human and domestic animal hookworms (N. americanus and Ancylostoma spp.). The close relationship of these species suggests that O. aculeatum can potentially infect humans and domestic animals. The fact that this parasite infects non-human primates (Ross et al., Reference Ross, Gibson and Harris1989; Karim & Yang, Reference Karim and Yang1992; Kamel et al., Reference Kamel, Yang and Norazah1995) in addition to elephants also supports this hypothesis. Nevertheless, information about O. aculeatum host specificity and adaptation is still limited and this issue needs clarification.
In the phylogenetic tree, several Strongyloides spp. were included to compare with our obtained sequences to avoid misidentification. As mentioned elsewhere, SSU-rDNA sequences differ significantly in the genus Strongyloides – an advantage for species identification (Dorris et al., Reference Dorris, Viney and Blaxter2002). Thus, we used several Strongyloides spp. to ensure that our obtained DNA sequences were not any other Strongyloides spp.
Using tetramicrosatellites, we were able to distinguish faecal samples from individual elephants and thus could determine the frequency of helminth infection accurately. The tetramicrosatellite technique has been demonstrated to be effective for identification of individual Asian and African elephants (Archie et al., Reference Archie, Moss and Alberts2003; Suwattana et al., Reference Suwattana, Koykul, Jirasupphachok and Kanchanapangka2007, Reference Suwattana, Jirasupphachok, Kanchanapangka and Koykul2010), as well as individuals of other organisms (Kaul et al., Reference Kaul, Singh, Vijh, Tantia and Behl2001; Behl et al., Reference Behl, Kaul, Sheoran, Behl, Tantia and Vijh2002; Itoh et al., Reference Itoh, Sato, Mano and Iwata2009; Costa et al., Reference Costa, Pérez-González, Santos, Fernández-Llario, Carranza, Zsolnai, Anton, Buzgó, Varga, Monteiro and Beja-Pereira2012). In addition to their use for determination of actual sample numbers and thus helminth frequency, microsatellites can reveal the relationships and diversity of local wild elephants, which may be useful for their control and management.
In conclusion, faecal examination combined with copro-PCR amplification and sequencing of SSU rDNA identified O. aculeatum in wild elephants. Although no evidence of transmission of this helminth between elephants and humans or domestic animals has been observed to date, further investigation is warranted to determine the likelihood of risk. In the meantime, healthcare workers, veterinarians and other persons living in the elephant-incursive area should be aware of the possibility of health problems caused by O. aculeatum infection.
Acknowledgements
We express our gratitude to the wildlife rangers of the Salakpra Wildlife Sanctuary, Kanchanaburi, Thailand for their help during sample collection. We thank Miss Arissara Pongsiri, Department of Entomology, Armed Forces Research Institute of Medical Sciences for geographic information system (GIS) training. We also thank the Faculty of Tropical Medicine, Mahidol University, for funding the proofreading and editing of this manuscript.
Financial support
This work was supported by the Faculty of Tropical Medicine Research Fund for fiscal year 2012 through the Tropical Medicine One Health team.
Conflict of interest
None.








