Introduction
The insulin-like growth factor (IGF) pathway is fundamental in cell proliferation, differentiation and transformation across all stages of growth and development.Reference Le Roith1, Reference Monzavi and Cohen2 IGFs are part of a complex system promoting cellular communication with the physiologic environment often denoted the IGF ‘axis’.Reference Rajaram, Baylink and Mohan3 The IGF-axis comprises two cell-surface receptors (IGF-1R, IGF-2R), two ligands (IGF-1, IGF-2), a family of 10 IGF-binding proteins (IGFBP 1–10) and associated IGFBP degrading enzymes known collectively as proteases. The ligands interact with IGF-1R more readily than IGF-2R.Reference Rother and Accili4 The IGFBPs and proteases play a vital role in controlling and modulating the effects of IGFs.Reference Monzavi and Cohen2, Reference Delafontaine, Song and Li5–Reference Wolf, Lahm, Wu, Wanke and Hoeflich7
The regulation of fetal and postnatal growth depends on multiple hormones including insulin, IGF-1, IGF-2 and growth hormone (GH).Reference Ahmad, Beharry and Valencia8 IGF-1 and IGF-2 are essential for fetal growth.Reference Gohlke, Fahnenstich, Dame and Albers9–Reference Westwood13 IGF-1 is the predominant regulator of postnatal growth,Reference Le Roith, Werner, Beitner-Johnson and Roberts14 acting as an important mediator between GH and growth during childhood.Reference Smith, Gunnell and Holly15 IGF-2 has been shown to be primarily responsible for early development, particularly intra-uterine growth.Reference Constancia, Hemberger and Hughes16–Reference Sara and Hall18
The interaction of IGFs and their receptors play a critical role in promoting and regulating growth from mid-gestation onwards optimizing pre- and postnatal growth and development.Reference Rother and Accili4, Reference Robertson17 Although 10 IGFBPs are known to exist, most studies (including this one) have focussed on IGFBPs 1 to 5, which have a high affinity towards the ligands and tightly regulate IGF function.Reference Rajaram, Baylink and Mohan3, Reference Delafontaine, Song and Li5, Reference Firth and Baxter19, Reference Salih, Tripathi and Holding20 Overall IGFBPs are known to affect cell motility and adhesion, apoptosis, survival and cell cycle by aiding IGF-1 and IGF-2 transport and modulating their interactions with IGF receptorsReference Westwood13, Reference Firth and Baxter19 (Fig. 1).

Fig. 1 Elements of the insulin-like growth factor (IGF)-axis [adapted from multiple sources (1–4, 7, 9–13, 21–24, 27–31)]. The factors are IGF-1 and IGF-2. IGF-1 expression is required for achieving maximal growth. IGF-1 is the predominant regulator of postnatal growth,Reference Le Roith, Werner, Beitner-Johnson and Roberts14 acting as an important mediator between growth hormone (GH) and growth during childhood.Reference Smith, Gunnell and Holly15 IGF-2 is thought to be the primary growth factor required for early development.Reference Tisi, Liu, Wykes, Skinner and Koskl49 Receptors (IGF-1R, IGF-2R): The IGFs are known to bind with receptors including IGF-1R, IGF-2R and the insulin receptor.Reference Rother and Accili4, Reference Massague and Czech30 IGF-1R is a transmembrane tyrosine kinase receptor, which dominates IGF-2R, acting as the primary receptor to both IGF-1 and IGF-2.Reference Rother and Accili4, Reference Massague and Czech30 Its principal role mediates mitogenic responses of the cell but can also affect metabolic responses from tissues.Reference Le Roith, Werner, Beitner-Johnson and Roberts14 IGF-2R binds only IGF-2. IGF2R may indirectly affect downstream signalling via its interaction with other proteinsReference El-Shewy and Luttrell50 and clearance of IGF-2; however, its major function is to maintain the correct IGF-2 levels in tissues and in the circulation. Binding proteins (IGFBP1–5): Generally, it is accepted that IGFBPs 1–5 have similar affinities for IGF-1 and IGF-2.Reference Kalus, Zweckstetter and Renner51–Reference Forbes, McCarthy and Norton56 IGFBP-1 has the ability to either inhibit or potentiate the effects of IGFs, depending upon its state of phosphorylation.Reference Rajaram, Baylink and Mohan3, Reference Firth and Baxter19, Reference Jones and Clemmons31, Reference Busby, Klapper and Clemmons57, Reference Yu, Iwashita, Kudo and Takeda58 In contrast, the main function of IGFBP-2, is to inhibit IGF-2 function.Reference Wolf, Lahm, Wu, Wanke and Hoeflich7, Reference Hoflich, Lahm, Blum, Kolb and Wolf59 IGFBP-3 like IGFBP-1 has the ability to either inhibit or augment IGF activity by controlling the binding of IGF-1 and IGF-1R.Reference Mohseni-Zadeh and Binoux60 IGFBP-4 levels have been associated with an inhibitory effect caused by preventing the binding of IGF-1 with IGF-1R.Reference Jones and Clemmons31, Reference Rechler32 IGFBP-5 is thought to have a stimulatory rather than an inhibitory effect on IGFs.Reference Rajaram, Baylink and Mohan3
The aim of the current study was to investigate the association between SNPs in genes within the IGF-axis and antenatal and postnatal growth from birth to adolescence in the Raine Cohort adjusting for common environmental influences on IGF levels. Analyses were replicated where possible in the Generation R Pregnancy Cohort.
Methods
Study cohorts
Discovery cohort
Recruitment of the Western Australian Pregnancy (Raine) Cohort has previously been described in detail.Reference Williams, Evans and Newnham21 In brief, between 1989 and 1991 2900 pregnant women were recruited before 18 weeks gestation into a randomized controlled trial to evaluate the effects of repeated ultrasound in pregnancy. Recruitment predominantly took place at King Edward Memorial Hospital (Perth, Western Australia). Ninety percent of eligible women agreed to participate in the study.Reference Newnham, Evans, Michael, Stanley and Landau22 Women were randomized to either intensive ultrasound assessment (ultrasound biometry in addition to umbilico-placental and utero-placental Doppler flow velocity waveforms measurements at 18, 24, 28, 34 and 38 weeks gestation) or to a regular ultrasound assessment at 18 weeks with subsequent scans at the clinicians discretion. The study was conducted with appropriate institutional ethics approval, and written informed consent was obtained from all mothers. The cohort has been comprehensively phenotyped through pregnancy, childhood and adolescence. In this study, we focus on a subset of 1162 individuals within the Raine Cohort who were Caucasian, singleton pregnancies with at least one ultrasound measure during pregnancy.
Gestational age (GA) was based on the date of the last menstrual period unless there was discordance of more than 7 days with ultrasound measurements at <18 weeks; in those cases (29.7%), the estimate was based on ultrasound biometry at 18 weeks gestation.Reference Williams, Evans and Newnham21 Maternal and paternal characteristics were self-reported by questionnaire. Research midwives recorded concurrent maternal medical conditions during pregnancy at recruitment and 34 weeks gestation. Fetal head circumference (HC), abdominal circumference (AC), femur length (FL) and umbilico-placental and utero-placental Doppler flow velocity waveforms were measured in triplicate using standard techniques. More than 95% of day-2 measures were taken by a single observer, the remainder by a consultant pediatrician using standardized protocols.Reference Williams, Evans and Newnham21 Postnatal weight and height were measured by trained researchers to the nearest 100 g and 0.1 cm, respectively.Reference Newnham, Evans, Michael, Stanley and Landau22 From here on we refer to the Raine Study as the discovery cohort.
Replication cohort
Recruitment of the Generation R Cohort has previously been described in detail.Reference Jaddoe, Bakker and van Duijn23, Reference Jaddoe, van Duijn and van der Heijden24 In brief, following local ethics board approval, 9778 mothers with a delivery date between April 2002 and January 2006 were recruited from two hospitals, eight midwifery practices and 16 child health centers in Rotterdam (The Netherlands) to identify early environmental and genetic causes of normal and abnormal growth, development and health. Written informed consent was obtained from all participants. Repeat ultrasound measurements were made at 12, 20 and 30 weeks gestation. Maternal and paternal characteristics and breast-feeding duration were collected by questionnaire. Children were assessed up to 4 years of age at routine child health centers. The cohort has been shown to be representative of the antenatal population in Rotterdam.Reference Jaddoe, van Duijn and van der Heijden24 From here on we refer to Generation R as the replication cohort.
Genotyping
DNA was purified from peripheral blood using standard protocols in both cohorts. Nine candidate genes were selected in the IGF-axis: two ligands, IGF-1 and IGF-2; two IGF receptors, IGF-1R and IGF-2R; and five IGFBP, IGFBP1 to IGFBP5. IGFBP6–10 were not selecting for genotyping as this stage. A multistep process was used to select SNPs in these genes. In brief, this gene-based approach entails (a) the selection of common tagging SNPs that best describe the haplotype diversity of the human genome in our population and (b) the selection of SNPs that are functionally relevant in the IGF-axis pathway. Computational programs such as SIFT, PolyPhen and SWISSProt were then utilized to characterize and model the consequences of SNPs to ensure sensitivity of evolutionary conservation, protein alignment and function. The Promoalign tool was used to identify upstream regulatory regions of genes and NetPhos to find SNPs potentially altering the phosphorylation patterns of proteins. Using this process, 165 SNPs were identified in the nine candidate genes in the IGF-axis.
Genotyping was performed at Centre for Applied Genomics (Toronto, Ontario, Canada). SNPs were uploaded to Illumina's Assay Design Tool (http://www.illumina.com/) for probe design resulting in a custom panel. A total of 5 μl of 50 ng/μl in 10 mM Tris-HCL pH 8.0, 1 mM EDTA of genomic DNA underwent an allele-specific oligonucleotide hybridization followed by extension and ligation. A universal PCR step for all 1536 loci followed with primers labelled with either Cy3 (primer 1) or Cy2 (primer 2). The amplified products were then hybridized to a sentrix array matrix and scanned using the Illumina BeadArray Reader (BAR) (Illumina, San Diego, CA, USA). The resulting data were analyzed with Beadstudio v.3.0 using the default parameters. Only SNPs with GenCall scores >0.25 were called and samples were discarded if call rates were below 85%.
SNPs with a minor allele frequency of <10% were excluded from analyses. SNPs that did not pass the Hardy–Weinberg equilibrium test were also excluded. Thus, a total of 145 (of 165 genotyped) SNPs were used in analyses.
In the replication cohort, cord blood for DNA isolation was available in 59% of all live-born participating children. Sex-mismatch rate between genome-based sex and midwife-record-based sex was low (<0.5%), indicating that possible contamination of maternal DNA was extremely low. Missing cord blood samples were mainly due to logistical constraints at the delivery. Individual genotype data were extracted from the genome-wide Illumina 610 Quad Array.
Statistical analysis
We have utilized previously published models to analyze antenatal growth.Reference Mook-Kanamori, Marsh and Warrington25 Covariates considered in these multivariate models included: parental height, weight, body mass index (BMI), age, socioeconomic status; maternal smoking status, parity, GA at birth; placental weight and function; and gender of the child. Parity and breast-feeding duration were factored to reflect the effect plateau in the higher categories. BMI was modeled longitudinally using linear mixed effect models from childhood to adolescence adjusting for the age that BMI was measured and the mothers’ smoking status during pregnancy.
All continuous covariates were mean-centered to remove potential correlations between model coefficients. The SNPs tagging the IGF-axis were coded according to the number of minor alleles (0, 1 or 2) using SimHapReference Carter, McCaskie and Palmer26 and analyzed under an additive model. Replication was performed where possible in the replication cohort (Generation R).
Analyses of antenatal measures
In the discovery cohort of 1162 subjects, antenatal analyses focussed on 588 subjects with repeated ultrasound measures at 18, 24, 28, 34 and 38 weeks gestation. This enabled both cross-sectional and longitudinal analyses to be performed on the same subset of the discovery cohort. Fetal AC and HC were analyzed using linear mixed effects models,Reference Laird and Ware27 including intercept and GA as random effects. SNPs significantly associated with these measures of antenatal growth (either intercept or change with time; trajectory) within the discovery cohort (data 18–38 weeks gestation) were analyzed longitudinally in the replication cohort (data 12–30 weeks gestation, n = 2642) and cross-sectionally using multivariate linear regression for the following three timeframes: 16–24 weeks, 26–32 weeks inclusive and >32 weeks for discovery cohort and >12 weeks, 16–24 weeks and 26–32 weeks in the replication cohort study to explore the time of onset of significant associations.
Analyses of postnatal measures
Height and weight were utilized as the primary predictors and determinants of postnatal growth within the discovery cohort. BMI was additionally analyzed. A maximum of 1162 subjects with height and weight data collected at ages 1, 2, 3, 5, 8, 10, 14 and 17 years were analyzed longitudinally using linear mixed effects modeling. These models adjusted for repeated measures over time. Our BMI model was analogous to those chosen for height and weight analyses and therefore did not adjust for the nonlinear relationship of BMI over time.
The statistical software package R version 2.8.028 was used to conduct all analyses. All SNPs selected have a minor allele frequency of >10% in the discovery cohort. All analyses were performed in males and females separately, as well as the combined dataset. P-values <0.05 indicate statistical significance. To adjust for multiple testing, a modified threshold for statistical significance of P ⩽ 0.00041 was utilized as described by the simpleM method.Reference Gao, Starmer and Martin29
Results
Population characteristics
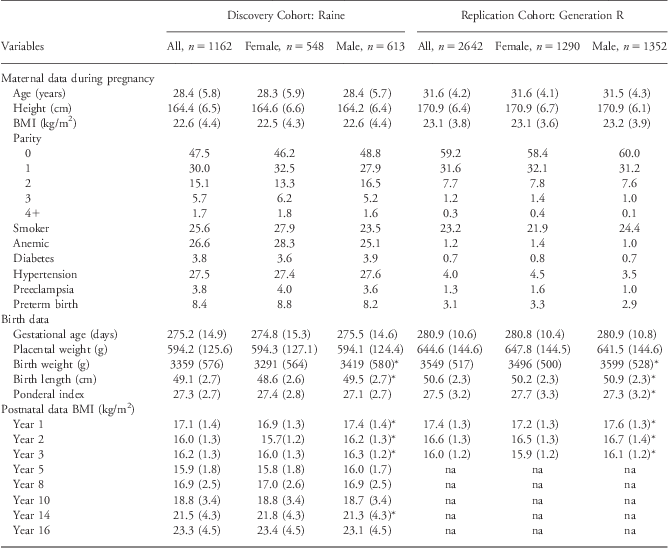
The demographic data for the discovery and replication cohort are presented in Table 1. The maternal demographic and obstetric data were similar between the two cohorts. Offspring of the replication cohort had, on average, a longer gestation (∼5 days) and birth length (∼1 cm), higher birth order and had greater placental weights (∼50 g) and birth weights (∼200 g) than members of the discovery cohort. Further, mothers in the replication cohort were observed to be slightly older (∼3 years), and with more tertiary education and had lower smoking rates than mothers in the discovery cohort. There were also fewer women with pregnancy complications, including anemia, diabetes and hypertension, during pregnancy in the replication cohort than the discovery cohort. Maternal characteristics were similar for both genders within a cohort; however, birth weight and length were significantly greater in male compared with female offspring in both the discovery and replication cohorts. In the discovery cohort, the subset that was randomly selected to undergo repeated ultrasound measures (n = 588) was representative of all discovery cohort members with genetic data (n = 1162; Supplementary Table 1a).
Table 1 Cohort demographic data

BMI, body mass index.
Maternal, pregnancy, birth and postnatal data presented as mean and standard deviation for continuous variables and percentages for categorical variables in the discovery cohort (Raine) and replication cohort (Generation R).
*Indicates that a statistically significant difference (P < 0.05) was observed between males and females within each individual cohort.
Genetic associations
Antenatal
A summary of the associations between SNPs in genes in the IGF-axis and antenatal growth is presented in Fig. 2. Utilizing a P-value threshold of 0.05, there were 58 significant associations identified, more than expected for the total number of SNPs analyzed. Almost half of the number of SNPs analyzed in each IGF-axis gene (34 of the 73 SNPs genotyped for IGF-1R and 13 of the 34 SNPs genotyped for IGF-2R) demonstrated significant associations with antenatal growth in the discovery cohort. Twenty-six of these SNPs were also significantly associated with postnatal growth (detailed later in this section). Similar patterns were observed with the replication data with 11 SNPs from IGF-2R also exhibiting significant associations with antenatal growth in the replication cohort.

Fig. 2 Significant associations between antenatal and postnatal growth for the insulin-like growth factor (IGF)-axis: IGF single-nucleotide polymorphisms (SNPs) significantly associated with antenatal (abdominal circumference and/or head circumference and/or fetal length) growth and postnatal height and/or weight are summarized here. Data from the Raine Cohort are presented in (a). There are more SNPs than expected with significant associations with antenatal and postnatal growth, especially in IGF-1R and IGF-2R. Data from Generation R are presented in (b) showing a similar pattern of association with the discovery cohort.
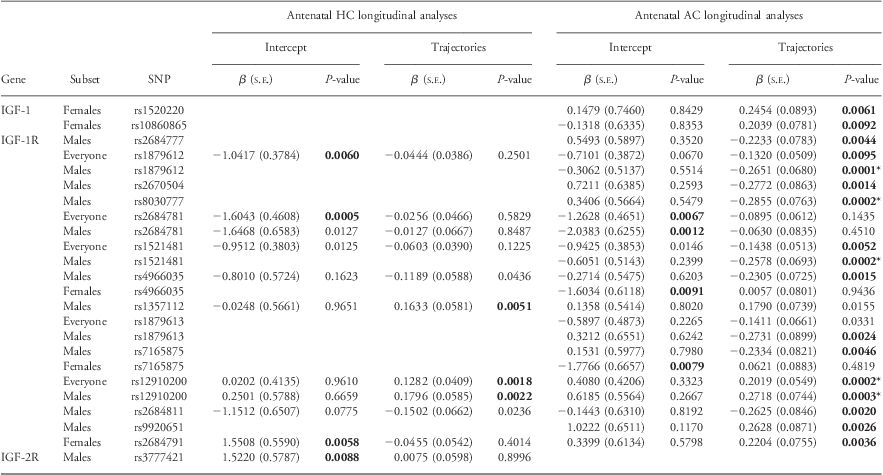
Table 2 summarizes the longitudinal analyses of fetal HC and AC in the discovery cohort (five biometry measures between 18 and 38 weeks gestation). Of the 19SNPs, which had significant associations with longitudinal analyses (P ⩽ 0.01), 15 of these SNPs had P-values ⩽0.005. After adjusting for multiple testing (P ⩽ 0.00041; Table 2 and Supplementary Table 4), four SNPs in IGF-1R and one SNP in IGF-2R remained statistically significant. When IGF–SNPs were evaluated in the replication cohort (three biometry measures between 12 and 30 weeks gestation) only four SNPs were significantly associated (P ⩽ 0.05) with HC/AC or FL intercept or trajectory (Supplementary Tables 2a and 4).
Table 2 Significant associations between longitudinal measures of antenatal growth and the IGF-axis in the Raine Cohort

IGF, insulin-like growth factor; HC, head circumference; AC, abdominal circumference; SNP, single-nucleotide polymorphisms.
IGF SNPs significantly associated (P-values ⩽ 0.005) with antenatal HC or antenatal AC from linear mixed-effects analyses in either gender combined (‘Everyone’) or gender stratified (‘Males’/‘Females’) are listed here.
β represents the regression coefficients for both the intercept and change with time (trajectory) within the discovery cohort (data 18–38 weeks gestation).
P-values ⩽0.010 are highlighted in bold text.
*Denotes P-values that remain statistically significant after adjusting for multiple testing (P ⩽ 0.00041).
Omitted from the table are values for AC or HC where intercept and trajectory have been found to be non-significant.
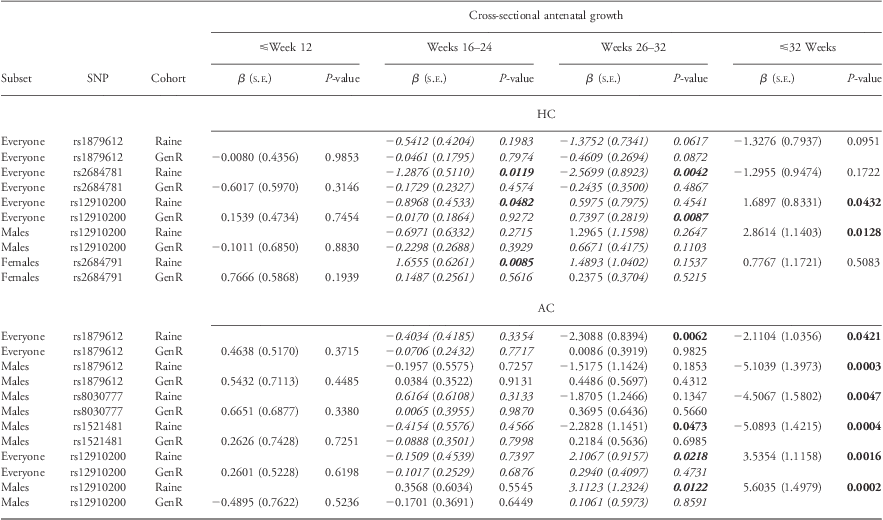
To explore the timing of onset of the effects on fetal growth, we focussed on six IGF-1R SNPs (rs1879612, rs8030777, rs1521481, rs2684781, rs2684791 and rs12910200) that were significant to P ⩽ 0.005 in the discovery cohort. A summary of the cross-sectional analyses of these SNPs is presented in Table 3. Results from cross-sectional analyses suggested that for the majority of these SNPs, the effects on fetal growth became statistically significant in the mid to late third trimester; hence, it is to be expected that smaller effect sizes were seen in replication cohort where their last ultrasound assessment was at 30 weeks gestation. It is interesting to note that the direction of effect was analogous between the discovery and replication cohorts in those cross-sectional timeframes where numerous measures were available (16–24 weeks).
Table 3 Cross-sectional antenatal HC and AC analyses of IGF-1R SNPs in Raine and Generation R

HC, head circumference; AC, abdominal circumference; IGF, insulin-like growth factor; SNP, single-nucleotide polymorphisms.
Cross-sectional analyses demonstrate that effects in fetal HC occur late in third trimester, with cross-sectional data at (26–32 weeks) demonstrating similar β coefficients in the discovery and replication cohorts for antenatal HC. Generation R do not have serial ultrasound scans at 34 and 38 weeks gestation; hence, we have limited ability to replicate our mid and late third trimester associations.
Bold text indicates effects observed to be statistically significant (P ⩽ 0.05), whereas effects going in the same direction in both cohorts are highlighted in italics.
Bold italics text indicates effects observed to be statistically significant (P ⩽ 0.05) and going in the same direction in both cohorts.
As a form of secondary analyses, FL was analyzed (Supplementary Table 4) and compared with postnatal height to assess the influence of IGF SNPs on developmental linear growth. The majority of SNPs effecting antenatal FL across gender-combined datasets belonged to IGF-2R, whereas male FL tended to be influenced by SNPs in IGF-1. Two SNPs, rs9347380 and RS9456497 (all in IGF-2R) replicated in GenR.
Postnatal
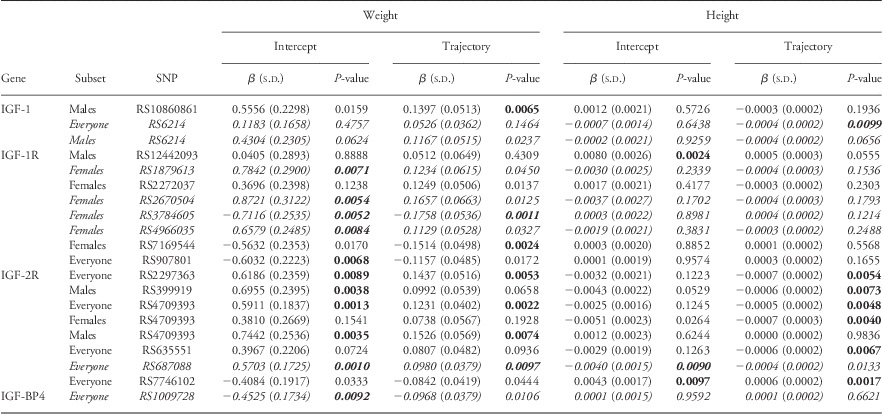
The longitudinal analyses of associations between SNPs in the IGF-axis and postnatal height and weight (using eight measures recorded over 17 years of life) are summarized in Table 4. Additional longitudinal postnatal analyses using BMI as an outcome are detailed in Supplementary Table 3. These measures were not available in the replication cohort because of the cohort's age. Similar to antenatal growth, the vast majority of the significant associations were in SNPs in the IGF receptors.
Table 4 Associations between postnatal weight and length with SNPs of the IGF-axis (Raine Cohort)

SNP, single-nucleotide polymorphisms; IGF, insulin-like growth factor; HC, head circumference; AC, abdominal circumference.
Longitudinal analyses of postnatal weight and height were performed using data from 1 to 17 years of age. Effects observed to be statistically significant (P ⩽ 0.01) are detailed in bold here.
SNPs significantly associated with antenatal growth (antenatal HC or antenatal AC; Table 2) are highlighted in italics.
Bold italics text indicates SNP effects associated with antenatal growth (Table 2) and postnatal growth (P ⩽ 0.01).
We observe that the majority of IGF-1R SNPs associated with postnatal growth are significantly associated with weight (Table 4) and subsequently BMI (Supplementary Table 3). Theses associations with weight appeared to be more prevalent in females, where six out of eight IGF-1R SNPs produced significant associations in female subsets with only one for males and gender-combined datasets, respectively. To illustrate the magnitude of effect of these SNPs on postnatal growth we use IGF-1R SNP rs3784605, which was significantly associated with reducing weight in females, decreasing weight 0.72 kg for the intercept and a further 0.18 kg for the trajectory effects. Several SNPs in IGF-2R were significantly associated with both weight gain and linear growth across children of the discovery cohort. Six out of ten SNPs significantly associated with postnatal height analyses (Table 4, P < 0.01) belonged to IGF-2R, where five SNPs were produced from analyses in the gender-combined dataset. To illustrate the effects of these SNPs on longitudinal height, rs7746102 in IGF-2R was significantly associated with increasing both the intercept and trajectory of height in males and females combined. At 17 years of age, this equated to an increase in height of 0.0145 m2 for the heterozygote (TC) and 0.029 m2 for the minor homozygote (CC).
Comparing antenatal and postnatal growth
The number of SNPs with significant associations with both antenatal growth and postnatal growth was similar for each of the ligands, receptors and binding proteins (results not published; details available upon request). Figure 3 illustrates 26 SNPs that were significantly associated with both antenatal and postnatal growth in the discovery study; 17SNPs (13 in IGF-1R) were associated with discordant growth patterns consistent with the Developmental Origins of Health and Disease (DOHaD) hypothesis. IGF-1R SNPs rs4966035, rs7165875, rs1521481, rs1879613 and IGF-2R SNP rs687088 were all associated with reduced antenatal AC, FL and increased weight gain. Nine SNPs were associated with concordant growth patterns, binding proteins rs9658238 (IGFBP1) consistently associated with increased growth and rs1009728 of IGFBP4 consistently associated with decreased growth.

Fig. 3 Insulin-like growth factor (IGF) single-nucleotide polymorphisms (SNPs) significantly associated with both antenatal and postnatal growth. Of the 58 SNPs associated with antenatal growth, 12 SNPs have demonstrated a U-shaped association relating antenatal growth restriction and postnatal growth. This is consistent with one of the underlying hypotheses of the Developmental Origins of Health and Disease paradigm that describes how fetal programming may operate across differing stages of nutrition with a U-shaped curve relating prenatal growth and nutrition to adult obesity and metabolic disease. SNPs listed here were significantly associated (P-values ⩽0.05) with both antenatal measures of growth (head circumference, HC; abdominal circumference, AC; or femur length, FL) and postnatal growth (height, weight or body mass index).
Discussion
The aim of the current study was to investigate the association between SNPs in genes within the IGF-axis and antenatal and postnatal growth from birth to adolescence in the discovery and replication cohorts adjusting for common environmental influences on IGF levels. We have shown that 40% (58/145) of the SNPs in genes within the IGF-axis were associated with measures of fetal growth. The majority of these SNPs were in the genes for the two cell-surface receptors, IGF-1R and IGF-2R [23% (34) and 9% (13), respectively]. Of the eight SNPs within the two ligands, IGF-1 had more SNPs associated with antenatal growth, accounting for a small proportion of the significant associations (4%; six SNPs, respectively) with the remaining two SNPs found in IGF-2. Three SNPs from the five IGFBP assessed (IGFBP 1–5) constituted a further 2%. These data suggest a potentially important role of genetic variants in IGF-1R (and to a lesser extent IGF-2R) in the regulation of fetal growth. This finding is consistent with the observation that both IGF-1R and insulin receptors are necessary for optimal pre- and postnatal growth and development, with the interaction of IGFs and their receptors playing a critical role in promoting and regulating embryonic growth from mid-gestation onwardsReference Rother and Accili4, Reference Robertson17 with IGF-1 and IGF-2 acting via IGF-1R to promote growth.Reference Rother and Accili4, Reference Massague and Czech30
In addition to playing an important role in antenatal growth, our data suggest that SNPs in genes within the IGF-axis may also play a role in postnatal growth with associations identified with 38% (55/145) of the SNPs investigated and longitudinal height and/or weight from 1 to 17 years. Similar to antenatal growth, the majority of these associations were in the genes for the two cell-surface receptors, with 21% in IGF-1R (31 SNPs) and 9% in IGF-2R (13 SNPs).
Results from this study illustrated that SNPs from IGF-2R were associated with reduced antenatal FL but increased postnatal weight gain/BMI.
Further, the associations we identified between SNPs in IGFBP-4 were consistent with published literature, which state that it has an inhibitory effect on growth by preventing the binding of IGF-1 with IGF-1R.Reference Jones and Clemmons31, Reference Rechler32 We have illustrated that rs1009728 in IGF-BP4 was significantly associated with reducing both antenatal growth and postnatal weight (and BMI) in males and females.
The underlying principle of DOHaD is founded upon decades of research in both human and animal studies illustrating the relationship between antenatal growth and postnatal obesity, where early life growth restriction predisposes individuals to diseases such as obesity, diabetes, metabolic syndrome, stroke and cardiovascular disease during adulthood.Reference Barker33–Reference Barker, Osmond, Golding, Kuh and Wadsworth38 It is widely accepted that genetic and epigenetic pathways mediate much of the relationship between the environment and the DOHaD paradigm where both mechanisms likely contribute to the variation seen in an individual's response to the environment as complex interactions between genes and the early environment modulate developmental programming of adult disease.Reference Newnham, Pennell, Lye, Rampono and Challis39 Genetic differences not only regulate gene–environment interactions underlying disease onset but also regulate disease susceptibility following environmental alterations.Reference Newnham, Pennell, Lye, Rampono and Challis39
There is growing evidence that the relationship between antenatal/postnatal growth and disease is mediated by genetic variants in the IGF-axis.Reference Monzavi and Cohen2, Reference Rother and Accili4, Reference Gohlke, Huber and Hecher10, Reference Smith, Gunnell and Holly15, Reference Abuzzahab, Schneider and Goddard40–Reference Pirazzoli, Cacciari and De Iasio44 We have demonstrated that a large number of SNPs from the IGF pathway were significantly associated with anthropomorphic measures taken from ultrasounds and postnatal growth. Further, we have provided data suggesting that 26 SNPs in the IGF-axis were associated with both antenatal and postnatal growth patterns consistent with the DOHaD hypothesis. Seventeen SNPs were associated with discordant growth patterns consistent with the lower end of the U-shaped curve described between antenatal growth and postnatal obesity.Reference Newnham, Pennell, Lye, Rampono and Challis39, Reference Freathy, Mook-Kanamori and Sovio45, Reference Hattersley and Tooke46
Genetic variants in the IGF receptors accounted for the majority of the associations with growth observed in our study. The next step in understanding the mechanism responsible for the association between variants in the IGF pathway and early human growth and development is to investigate the interactions between insulin-like factors, their receptors and their associated IGFBP, which fundamentally control the factor–receptor interaction.Reference Monzavi and Cohen2–Reference Wolf, Lahm, Wu, Wanke and Hoeflich7, Reference Westwood13, Reference Robertson17, Reference Firth and Baxter19, Reference Salih, Tripathi and Holding20, Reference Massague and Czech30 Further investigations of the entire IGF pathway and the interactions within genetic components of the pathway and the environment would provide greater detail as to the underlying function and influence on growth and metabolic diseases. Investigating SNPs individually (i.e. one-by-one) may miss important information regarding essential biological pathways.Reference Weng, Macciardi and Subramanian47, Reference Peng, Luo and Siu48 This is highlighted by publications detailing genetic analyses utilizing both candidate gene approaches and Genome-Wide Association Study (GWAS) where single SNPs account for only a small amount of the genetic variation in obesity and diabetes.Reference Mook-Kanamori, Marsh and Warrington25, Reference Freathy, Mook-Kanamori and Sovio45 Therefore, new analytic methods are required in order to better analyze and understand this key metabolic pathway to include how the IGF SNPs work in conjunction. This is particularly important as levels of phosphorylation and methylation of these SNPs also contributes heavily to the overall functioning of the pathway affecting anthropomorphic outcomes by activating, mediating or inhibiting the interaction between IGFs and receptors.
The discovery cohort (Raine) and the replication cohort (Generation R) have been shown to be representative of the populations in Western Australia and Holland, respectively. Both cohorts have multiple measures of growth that have enabled longitudinal analyses, increasing the power and uniqueness of this study. The large number of available covariates associated with fetal growth has allowed us to focus on the effects due to genetic variants as we were able to account for ∼72% of the variance observed using epidemiologic data. We do, however, note important limitations of our study, including our limited sample size and concomitant modest statistical power to detect small genetic effects and the lack of multiple ultrasound scans in the third trimester for the replication cohort. The younger age of the replication cohort has also meant that we are unable to replicate our postnatal findings in this cohort.
Future research will need to be conducted in larger and ethnically diverse studies where growth has been measured on multiple occasions across the life-course. Further investigation into the association between IGF SNPs and growth will need to emphasize not only the genetic influence but also the complexities of environmental exposures and gene–environment interactions. Such exposures include diet and nutrition of mother and child as well as the level of physical activity by the children and its subsequent influence on postnatal growth measures.
In the future, we hope that lifestyle modifications can be implemented following the identification of those children whose antenatal growth patterns and individual genetic variation predispose them to the development of obesity. Further research is required to evaluate targeted interventions (including diet, exercise and medication) for those at the greatest risk of obesity.
Acknowledgments
The authors are grateful to the Raine Study participants and their families and the Raine Study Research staff. The authors gratefully acknowledge the assistance of the Western Australian DNA Bank (both National Health and Medical Research Council of Australia National Enabling Facilities). The authors also acknowledge the support of the Healthway Western Australia, the National Health and Medical Research Council of Australia (Grants 572613, 403981, 963209, 211912, 003209, 353514) and the Canadian Institutes of Health Research (Grant MOP 82893). We gratefully acknowledge the assistance of the University of Western Australia (UWA), Raine Medical Research Foundation, Wind Over Water Foundation, The Telethon Institute for Child Health Research, UWA Faculty of Medicine, Dentistry and Health Sciences, Women and Infants Research Foundation and Curtin University. The Generation R Study is conducted by the Erasmus Medical Center in close collaboration with the School of Law and Faculty of Social Sciences of the Erasmus University Rotterdam, the Municipal Health Service Rotterdam area, Rotterdam, the Rotterdam Homecare Foundation, Rotterdam and the Stichting Trombosedienst & Artsenlaboratorium Rijnmond (STAR-MDC), Rotterdam. They gratefully acknowledge the contribution of children and parents, general practitioners, hospitals, midwives and pharmacies in Rotterdam. The generation and management of GWAS genotype data for the Generation R Study were done at the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, The Netherlands. They would like to thank Karol Estrada, Dr Tobias A. Knoch, Anis Abuseiris, Luc V. de Zeeuw and Rob de Graaf, for their help in creating GRIMP, BigGRID, MediGRID and Services@MediGRID/D-Grid (funded by the German Bundesministerium fuer Forschung und Technology; grants 01 AK 803 A-H, 01 IG 07015 G) for access to their grid computing resources. The authors thank Mila Jhamai, Manoushka Ganesh, Pascal Arp, Marijn Verkerk, Lizbeth Herrera and Marjolein Peters for their help in creating, managing and QC of the GWAS database. Also, they thank Karol Estrada and Carolina Medina-Gomez for their support in creation and analysis of imputed data. The Generation R Study is made possible by financial support from the Erasmus Medical Center, Rotterdam, the Erasmus University Rotterdam and the Netherlands Organization for Health Research and Development (ZonMw 21000074). Vincent Jaddoe received an additional grant from the Netherlands Organization for Health Research and Development (ZonMw 90700303). Priyakumari Parmar is supported by the Women and Infants Research Foundation of Western Australia PhD Scholarship grant. Additional support was provided by a grant from the Dutch Kidney Foundation (C08.2251 – H. Rob Taal). Vincent Jaddoe reports receipt of funding from the Netherlands Organization for Health Research and Development (ZonMw 90700303, 916.10159).