Yogurt is a favorite fermented milk product as an important part of people's diet in many countries. Production of yogurt is achieved using starter cultures in industry, namely Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus. Yogurt quality is highly influenced by the strain differences of these cultures. Therefore, yogurt manufacturers and starter producers are still seeking, identifying, screening, and selecting strains as yogurt cultures from mixed populations. Those strains are mostly isolated from their natural habitats such as raw milk, naturally fermented yogurts (without using commercial starter cultures) (Holzapfel, Reference Holzapfel2002). However, identification and discrimination of these strains, particularly species of Lactobacillus genus has always been a problem because of the heterogeneity of the genus. The divergency among lactobacilli is so great that guanine and cytosine content of the species varies between 32–54%, which is about twice the amount expected in a well-defined genus (Nour, Reference Nour1998). For identification of Lb. delbrueckii subsp. bulgaricus and Strep. thermophilus from a mixed population, as in the case of naturally fermented yoghurt, the situation may even be more difficult, since several species and subspecies of lactic acid bacteria co-exist.
Conventional methods used for identification of yoghurt starter bacteria include growth at different temperatures, and pH values, carbohydrate fermentation tests, API kits, production of lactate isoforms, SDS-PAGE, etc. (Coeuret et al. Reference Coeuret, Dubernet, Bernardeau, Gueguen and Vernoux2003; Andrighetto et al. Reference Andrighetto, De Dea, Lombardi, Neviani, Rossetti and Giraffa1998). Such procedures are labour intensive, time consuming, and may result in misidentification because of the limitations of the methods, i.e. carbohydrate fermentation tests rely on colour changes, which may be difficult to interpret (Andrighetto et al. Reference Andrighetto, De Dea, Lombardi, Neviani, Rossetti and Giraffa1998).
The number of studies, based on molecular biology, to identify many bacteria including Lb. bulgaricus (Tilsala-Timisjarvi & Alatossava, Reference Tilsala-Timisjarvi and Alatossava1997; Giraffa, De Vecchi & Rosetti, Reference Giraffa, De Vecchi and Rossetti1998; Miteva et al. Reference Miteva, Boudakov, Ivanova-Stoyancheva, Marinova, Mitev and Mengaud2001) and Strep. thermophilus (Tilsala-Timisjarvi & Alatossava, Reference Tilsala-Timisjarvi and Alatossava1997; Lick et al. Reference Lick, Keller, Bockelmann and Heller1996), at species, subspecies level has increased in the last decade. However, our survey of the literature and preliminary studies have revealed the lack of a complete rapid method for accurate selective identification of both Strep. thermophilus and Lb. delbrueckii subsp. bulgaricus at species and subspecies level from a mixed population and results of trials are either unsatisfactory or ambiguous. As an example, identification of both Strep. thermophilus and Lb. delbruecki at species level was accomplished by PCR based method using species specific primer sets targeted intergenic spacer region of 16S-23S rRNA gene by Tilsala-Timisjarvi & Alatossava (Reference Tilsala-Timisjarvi and Alatossava1997). Yet, the differentiation of Lb. delbrueckii strains at subspecies level was not described in their study. On the other hand, identification of two Lb. delbrueckii subspecies; subsp. lactis and subsp. bulgaricus was reported by Delley & Germond (Reference Delley and Germond2002), while selective identification of these isolates from a mixed population was not mentioned in their study.
Our study was carried out to present a new method for selective identification of Strep. thermophilus and Lb. delbrueckii subsp. bulgaricus from mixed cultures. It has been reported by Bolotin et al. (Reference Bolotin, Quinquis, Renault, Sorokin, Ehrlich, Kulakauskas, Lapidus, Goltsman, Mazur, Pusch, Fonstein, Overbeek, Kyprides, Purnelle, Prozzi, Ngui, Masuy, Hancy, Burteau, Boutry, Delcour, Goffeau and Hols2004), that Strep. thermophilus and Lb. delbrueckii subsp. bulgaricus have 95% identical methionine biosynthesis genes. Thus, the proposed method is based on partial amplification of methionine biosynthesis gene region to differentiate yoghurt starter cultures, Strep. thermophilus and Lb. delbrueckii subsp. bulgaricus from mixed population.
Materials and Methods
Bacterial strains and growth conditions
The bacterial strains used in this study are listed in Table 1. These include, strains isolated from commercial starter cultures and from rural yoghurts, produced traditionally, by back-slopping (Holtzapfel, Reference Holzapfel2002), in rural areas of Turkey and reference strains. The strains were provided as freeze dried cultures, activated 3 times in MRS and M17 broth (Merck KGaA, Darmstadt, Germany) at 37°C and 42°C for use, and maintained on Microbanks (Pro-Lab Diagnostics) and 20% glycerol stocks at −80°C.
Table 1. The bacterial strains used in the study
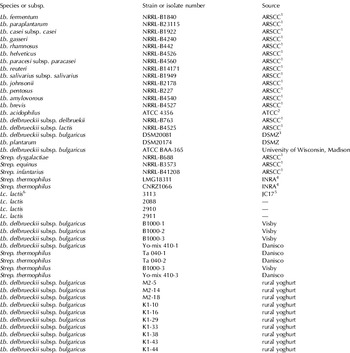
1 Agricultural Research Service Culture Collection, USA
2 American Type Culture Collections
3 German Culture Collection Center
4 National Institute of Agronomical Research, France
5 Piard et al. Reference Piard, Kuipers, Rollema, Desmazeaud and de Vos1993
6 Lc.–Lactococcus
Phenotypic identification of isolates
Isolates were identified according to Randazzo et al. Reference Randazzo, Torriani, Akkermans, de Vos and Vaughan2002. Briefly; colony morphology on agar, microscopic examination, gram staining, catalase production, reduction of nitrate, gas production, hydrolysis of arginine, hydrolysis of esculin, growth at different pH values (pH 2, pH 10) and different temperatures (10 and 45°C) were tested. Carbohydrate fermentation profiles were performed using microtiter plates according to Bulut et al. (Reference Bulut, Gunes, Okuklu, Harsa, Kilic, Coban and Yenidunya2004). The carbohydrates tested were arabinose, cellobiose, fructose, galactose, glucose, lactose, maltose, mannitol, melibiose, ribose, saccharose, salicin, sorbitol, trehalose and xylose. Fermentation patterns were also screened using appropriate API galleries (Biomerieux® S.A., Marcy-l Etoile, France). The results of API system were analysed with API identification software.
DNA isolation
Preparation of genomic DNA was performed according to modified method of Luchansky et al. (Reference Luchansky, Tennant and Klaenhammer1991) as follows: 2 ml of overnight grown culture was centrifuged at 25 000 g (Andreas Hettich GmbH, Tuttlingen, Germany) for 2 min, and washed twice in TE buffer (10 mm-TrisHCl (AppliChem GmbH, Darmstadt, Germany), 1 mm-EDTA (pH 8·0; Merck KGaA, Darmstadt, Germany). The pellet was dissolved in 300 μl TE buffer and 5 μl lysozyme (50 mg/ml; AppliChem GmbH) was added to lyse the cells. The solution is incubated at 37°C for 45 min. Thereafter, 20 μl EDTA (0·25 m; Merck KGaA), 25 μl SDS (10%; Merck KGaA, Darmstadt, Germany) and 4 μl Proteinase K (20 mg/ml; Fermentas, Vilnius, Lithuania) was added to the solution and incubated at 60°C for 60 min. Once digestion was complete, samples were extracted with phenol-chloroform three times, and ethanol precipitated with the addition of 0·1 V 3 m-sodium acetate (pH 5·5; Merck KGaA). DNA was precipitated; samples were washed with 70% ethanol (Merck KGaA), air-dried for 20 min and dissolved in distilled H2O. Rnase (Fermentas) was used to digest RNA, and samples were stored at −20°C until use. The quality of DNA isolations were tested both with A260 /A280 ratios and on agarose gels with comparison to decreasing concentrations of bacteriophage λ (Fermentas).
Primers and PCR conditions
For the methionine biosynthesis gene, primers cysmet2F (forward): 5′-GGAACCTGAAGGCTCAAT-3′, cysmet2R (reverse): 5′-GTCAACCACGGTAAAGGTC-3′ were designed using on-line tools, primer3 and oligocalc program. Nucleotide-nucleotide BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) was made to confirm primers' selectivity. Genbank accession numbers of methionine biosynthesis genes were; YP_141251 and YP_141252. For the amplification of 16S rRNA gene, 9699–9700 primer pair was used (Delley & Germond, Reference Delley and Germond2002). PCR amplifications were made on MJMini thermal cycler (BioRad, Hercules, USA). Reaction mixture and amplification conditions for the analysis of methionine biosynthesis gene were as follows: an initial denaturation at 94°C for 2 min, 45 cycles of denaturation at 94°C for 30 sec for 10 min. The reaction mixture was 30 μl, and consisted of 1·5 mm-MgCl2, annealing at 54°C for 40 sec, extension at 72°C for 45 sec, and final annealing of 72°C, 200 μm of each dNTP (Fermentas), 7 μm of each primer, 0·5 U Taq DNA polymerase (BIORON GmbH, Ludwigshafen, Germany), and 500 ng DNA.
For 16S rRNA gene restriction analysis and sequencing, the reaction mixture was 50 μl, and consisted of 1·5 mm-MgCl2, 200 μm of each dNTP (Fermentas), 1 μm of each primer, 0·5 U Taq DNA polymerase (BIORON), and 500 ng DNA. Amplification conditions were, an initial denaturation at 95°C for 2 min, 35 cycles of denaturation at 95°C for 1 min, annealing at 56°C for 1 min, extension at 72°C for 1 min, and final annealing of 72°C for 10 min.
The PCR products were run on 1·5% agarose gels, by gel electrophoresis for 1 h at 80 V in 1× TAE (40 mm-Tris-acetate and 1 mm-EDTA, pH 8·0) and post-stained with ethidium bromide (1 μg/ml), and visualized under UV light in a gel documentation system (BioRad, Hercules, USA). DNA ladder of 100 bp plus (Fermentas) was used as a DNA molecular weight marker.
Restriction analysis of 16S rRNA gene product
Restriction of 16S rRNA gene product was carried on using EcoRI as the enzyme (Delley & Germond, Reference Delley and Germond2002). The reaction mixture contained 1× of the corresponding buffer, 8·5 μl of PCR product, and 0·5 μl EcoRI (Fermentas). The restriction products were run on 1·5% agarose gels, post-stained with ethidium bromide, and visualized under UV light.
Genotypic identification by 16S rRNA gene sequence analysis
For sequencing of the 16S rRNA gene, PCR product was amplified using forward primer 5′-AGAGTTTGATCCTGGCTCAG-3′ (Mora et al. Reference Mora, Fortina, Nicastro, Parini and Manachini1998) and reverse primer U926 (CCGTCAATTCCTTTRAGTTT) (Baker et al. Reference Baker, Smith and Cowan2003). PCR amplicons were recovered from PCR mixtures using DNA Extraction Kit (Fermentas) and further subjected to sequencing analysis (Iontek, Istanbul, Turkey). The sequences of each isolate were compared with those reported in the basic BLAST database (Altschul et al. Reference Altschul, Madden, Schaffer, Zhang, Zhang, Miller and Lipman1997; http://www.ncbi.nlm.nih.gov/BLAST/). Strains showing homology of at least 97% were considered to belong to the same species (Stackebrandt & Goebel, Reference Stackebrandt and Goebel1994).
Results and Discussion
The aim of this study was to identify yogurt starter bacteria (Lb. delbrueckii subsp. bulgaricus and Strep. thermophilus) rapidly and selectively from mixed populations containing other natural inhabitants of dairy products, a new PCR-based method has been tested. To achieve this goal, 16 Lb. bulgaricus and 6 Strep. thermophilus strains were used. The strains were initially identified by conventional methods. Confirmation of identification results for those strains was performed by genotypic identification using sequencing of V1-V3 region of 16S rRNA gene. Comparisons with 16S rRNA sequences held in Genbank verified that all of the studied strains, including commercial starter isolates and isolates from rural yoghurts, belonged to Strep. thermophilus and Lb. delbrueckii subsp. bulgaricus.
In order to achieve selective identification of Lb. delbrueckii subsp. bulgaricus and Strep. thermophilus strains by methionine biosynthesis gene, more than 10 primer pairs were designed and primer pair, cysmet2F and cysmet2R, was chosen to identify both yoghurt starters. Partial amplification of the methionine biosynthesis gene using these primers revealed a 700 bp product in all Lb. delbrueckii subsp. bulgaricus (n=16) and Strep. thermophilus isolates (n=6) as expected and also in two closely related bacteria, Lb. delbrueckii subsp. lactis NRRL-B4535 and Lb. helveticus NRRL-B442 (Fig. 1). All other species (n=20) including other lactobacilli, lactococci and streptococci species (Table 1) used in this study gave no PCR reaction (Fig. 2).

Fig. 1. Partial amplification of the methionine biosynthesis gene in commercial, rural yogurt isolates, some reference strains of Lb. delbrueckii subsp. bulgaricus and closely related lactobacilli and Strep. thermophilus. Lanes 1–2, 5–18: Lb. delbrueckii subsp. bulgaricus (1: DSM20081, 2: ATCC BAA-365, 5: Visby B1000-1, 6: Visby B1000-2, 7: Visby B1000-3, 8: Danisco Yo-mix 410-1, 9: M2-5, 10: M2-14, 11: M2-18, 12: K1-10, 13: K1-16, 14: K1-29, 15: K1-33, 16: K1-38, 17: K1-43, 18: K1-44), 3: Lb. delbrueckii subsp. lactis NRRL-B4525, 4: Lb. delbrueckii subsp. delbrueckii NRRL-B763, 19: Lb. helveticus NRRL B-4526, lanes 20–24: Strep. thermophilus (20: LMG18311, 21: CNRZ1066, 22: Visby B1000-3, 23: Danisco Yo-mix 410-3, 24: Danisco Ta 040-1), NT: Negative control (no DNA control), M: 100 bp DNA ladder.

Fig. 2. PCR results for partial amplification of the methionine biosynthesis gene in other reference strains of lactoccocci, lactobacilli and streptococci. Lanes: 1–4: Lc. lactis (1: 2910, 2: 2911, 3: 2088, 4: 3113), 5: Negative control (no DNA control), 6: Lb. delbrueckii subsp. bulgaricus DSM20081, 7: Lb. reuteri NRRLB-14171, 8: Lb. pentosus NRRLB-227, 9: Lb. brevis NRRLB-4527, 10: Lb. gasseri NRRLB-4240, 11: Lb. amylovorous NRRLB-4540, 12: Lb. casei subsp. casei NRRLB-1922, 13: Lb. johnsonii NRRLB-2178, 14: Lb. salivarius subsp. salivarius NRRLB-1949, 15: Lb. paraplantarum NRRLB-23115, 16: Lb. fermentum NRRLB-1840, 17: Lb. rhamnosus NRRLB-442, 18: Lb. paracasei subsp. paracasei NRRLB-4560, 19: Strep. dysgalactiae NRRLB 688, 20: Strep. equinus NRRLB-3573, 21: Strep. infantarius NRRLB-41208, 22: Lb. delbrueckii subsp. delbruekii NRRLB-763, M: 100 bp DNA ladder.
Additionally, Lb. acidophilus ATCC 4356 did not give an amplicon by those primers (data not shown). As predicted from BLAST analysis, the primer set synthesized for partial amplification of methionine biosynthesis gene was able to identify Strep. thermophilus and Lb. delbrueckii subsp. bulgaricus (Fig. 1). This protocol has provided additional discrimination of Lb. delbrueckii subsp. delbrueckii from yoghurt starters and from Lb. delbrueckii subsp. lactis (Fig. 1). The unexpected production of 700 bp fragment in Lb. delbrueckii subsp. lactis and Lb. helveticus, however, coincides with the horizontal transfer of genes sharing the same niche (Bolotin et al. Reference Bolotin, Quinquis, Renault, Sorokin, Ehrlich, Kulakauskas, Lapidus, Goltsman, Mazur, Pusch, Fonstein, Overbeek, Kyprides, Purnelle, Prozzi, Ngui, Masuy, Hancy, Burteau, Boutry, Delcour, Goffeau and Hols2004), since Lb. delbrueckii subsp. lactis, Lb. helveticus, Lb. delbrueckii subsp. bulgaricus, and Strep. thermophilus do exist in similar environments in dairy products (Hannon et al. Reference Hannon, Deutsch, Madec, Gassi, Chapot-Chartier and Lortal2006). Lb. helveticus genome sequence has, lately (03-December-2007), become available in Genbank with accession number CP000517. The analysis of Lb. helveticus genome confirmed our result that high homology in methionine biosynthesis gene exists for Lb. helveticus as well. However, the same methionine biosynthesis gene product we obtained for Lb. delbrueckii subsp. lactis remained unconfirmed since the genome sequence of this organism is still unknown.
The aim of the study was to solely identify yoghurt starters. Thus, presence of amplicons of the methionine biosynthesis genes also in Lb. delbrueckii subsp. lactis and Lb. helveticus necessitated further analysis for differentiation of those organisms. The differentiation of Lb. delbrueckii subsp. bulgaricus, Lb. delbrueckii subsp. lactis and Lb. helveticus, was achieved by restriction analysis method of 16S rRNA gene (ARDRA: Amplified Ribosomal DNA Restriction Analysis) by EcoRI as reported by Delley & Germond (Reference Delley and Germond2002). The amplification sizes of the 16S rRNA gene was approximately 1500 bp in all isolates of Lb. delbrueckii subsp. bulgaricus (n=16) and Strep. thermophilus (n=6) from commercial starter cultures, rural yoghurts and reference strains as well as Lb. delbrueckii subsp. lactis NRRL-B4525, Lb. helveticus NRRL-B4526 (Fig. 3). Restriction endonucleases EcoRI digested all Lb. delbrueckii subsp. bulgaricus and Strep. thermophilus isolates in two fragments of 650, 850 bp and 700, 850 respectively (Fig. 4a & b). The Lb. delbrueckii subsp. bulgaricus and Lb. delbrueckii subsp. lactis strains used in our experiment confirmed findings of Delley & Germond (Reference Delley and Germond2002) (Fig. 4a). The enzyme digested the PCR product of Lb. delbrueckii subsp. lactis NRRL-B4525 in one fragment of about 1440 bp (Fig. 4a). However, differing from their finding of complete digestion, Lb. helveticus NRRL-B4526 was partially digested. The digestion products were faint bands restricted at the middle of the gene (Fig. 4a & 4b). The results have been confirmed also on another Lb. helveticus starter stain from Christian and Hansen (data not shown). The reason for this situation might result from the presence of several 16S rRNA gene copies in a Lb. helveticus genome in some of which complete digestion was not generated by EcoRI due to the lack EcoRI recognition site (Giraffa et al. Reference Giraffa, Gatti, Rossetti, Senini and Neviani2000). Nevertheless, the method of Delley & Germond (Reference Delley and Germond2002) still provided differentiation between Lb. delbrueckii subsp. bulgaricus and Lb. helveticus. It is noteworthy that Strep. thermophilus has given a different profile than that of Lb. delbrueckii subsp. bulgaricus when ARDRA with EcoRI was performed. Partial digestion of Lb. helveticus has differentiated Strep. thermophilus from Lb. helveticus (Fig. 4b). In case of complete digestion, microscopic examination of cells could be necessary to differentiate Strep. thermophilus having the same banding pattern with Lb. helveticus.

Fig. 3. Amplification of 16S rRNA gene from isolates that amplified methionine biosynthesis gene product. Lanes 1–2, 4–16, 21: Lb. delbrueckii subsp. bulgaricus (1: DSM20081, 2: ATCC BAA-365, 4: M2-14, 5: M2-18, 6: K1-10, 7: K1-16, 8: K1-29, 9: K1-33, 10: K1-38, 11: K1-43, 12: K1-44, 13: M2-5, 14: Visby B1000-1, 15: Visby B1000-2, 16: Visby B1000-3, 21: Danisco Yo-mix 410-1), 3: Lb. delbrueckii subsp. lactis NRRL-B4525, 17: Lb. helveticus NRRL B-4526, lanes 18–20, 22–24: Strep. thermophilus (18: CNRZ1066, 19: LMG18311, 20: Visby B1000-3, 22: Yo-mix 410-3, 23: Danisco Ta 040-1, 24: Danisco Ta 040-2), 25: Negative control (no DNA control). M: 100 bp DNA ladder.

Fig. 4. Restriction analysis of 16S rRNA gene with EcoRI. (A) Restriction analysis of 16S rRNA gene with EcoRI for differentiation of Lb. delbrueckii subsp. bulgaricus. Lanes 1–16: Lb. delbrueckii subsp. bulgaricus (1: DSM20081, 2: ATCC BAA-365, 3: K1-10, 4: K1-16, 5: K1-29, 6: K1-33, 7: K1-38, 8: K1-43, 9: K1-44, 10: M2-5, 11: M2-14, 12: M2-18, 13: Danisco Yo-mix 410-1, 14: Visby B1000-1, 15: Visby B1000-2, 16: Visby B1000-3), 17: Lb. delbrueckii subsp. lactis NRRL-B4525, 18: Lb. helveticus NRRL B-4526, M: 100 bp DNA ladder (B) Restriction analysis of 16S rRNA gene with EcoRI for differentiation of Strep. thermophilus. Lanes 1–6: Strep. thermophilus (1: LMG18311, 2: CNRZ1066, 3: Visby B1000-3, 4: Danisco Yo-mix 410-3, 5: Danisco Ta 040-1, 6: Danisco Ta 040-2), 7: Lb. helveticus NRRL B-4526, 8: Lb. delbrueckii subsp. bulgaricus DSM20081. M: 100 bp DNA ladder.
The study presented here offered rapid, selective identification of Lb. delbrueckii subsp. bulgaricus and Strep. thermophilus from a mixed population using a specific PCR method of methionine biosynthesis gene and ARDRA analysis of 16S rRNA gene using EcoRI (Delley & Germond, Reference Delley and Germond2002).
The authors thank to Agricultural Research Service Culture Collection (USA), National Institute of Agronomical Research (France), and Dr. James L. Steele and Dr. Peter Smeianov from University of Wisconsin, Madison for providing us valuable strains used in this study. This study was partially supported by Scientific Research Projects of METU (Grant no. BAP-2006-03-14-02) and DPT-YUUP (2005 K 120570).







