Kefir is a fermented milk produced by using a complex mixture of microorganisms known as kefir grains. The complex and variable microflora of grains includes yeasts, lactic acid bacteria (LAB), acetic bacteria and fungal mycelia (Witthuhn et al. Reference Witthuhn, Schoeman and Britz2005), held together with a matrix consisting mainly of proteins and polysaccharides. Lactobacilli are normally present as the largest portion (65–80%) of the microbial population. Typically, Lactobacillus acidophilus, Lb. brevis, Lb. casei, Lb. fermentum, Lb. kefir, Lb. parakefir and Lb. plantarum are LAB most frequently isolated from Kefir and Kefir grains (Bosch et al. Reference Bosch, Golowczyc, Abraham, Garrote, De Antoni and Yantorno2006; Farnworth & Mainville Reference Farnworth, Mainville and Farnworth2008; Golowczyc et al. Reference Golowczyc, Gugliada, Hollmann, Delfederico, Garrote, Abrahan, Semorile and De Antoni2008). The microbial composition may differ if the grains have different origins but, currently, phages have never been isolated. Other natural and complex dairy starters, such as the natural whey cultures and milk cultures used for cheese productions, are integrated by a very complex mixture of LAB and phages (Zago et al. Reference Zago, Comaschi, Neviani and Carminati2005, Reference Zago, Rossetti, Bonvini, Remagni, Perrone, Fornasari, Carminati and Giraffa2008). In these cases, lisogeny and spontaneous induction are considered as the origin of these phages, usually present at high numbers. It is accepted that the strong phage resistance of natural starters is due to the presence and the selective pressure of phages, which concur to select phage-resistant bacterial sub-populations. Kefir cultures are microbial systems similar to artisanal cheese cultures and, so, a complete description of their microflora should include phages, if their presence can be demonstrated.
The aim of this work was to investigate the presence of Lb. plantarum phages in Kefir grains and characterize them.
Materials and Methods
Selection of host strains
A total of 58 Lb. plantarum strains of different origins were used: 29 strains isolated from Argentinean artisanal cheeses, 19 strains isolated from Italian artisanal cheeses, 5 strains isolated from Kefir grains, and 4 collection strains. The wild strains used were identified by species-specific PCR according to Torriani et al. (Reference Torriani, Felis and Dellaglio2001). To investigate the genetic diversity, all the strains were tested by RAPD-PCR analysis according to Giraffa et al. (Reference Giraffa, Rossetti and Neviani2000). Thirty-six strains with different RAPD profiles and coming from different dairy sources (Table 1) were selected and used as host strains for bacteriophages.
Table 1. Host range of Lb. plantarum phages used in this study

a : Wild strains, Corrientes (Argentina); b: Wild strains, Santa Fe (Argentina); c: Wild strains, Lodi (Italia); d: Wild strains, La Plata (Argentina); e: Collection strains
(n° ): subculture number when the lysis was observed
Bacterial stocks were maintained at −80°C in MRS broth (Biokar, Beauvois, France) with the addition of 15% (v/v) of glycerol as a cryoprotective agent and routinely cultured overnight at 37°C in MRS broth.
Kefir samples
Two kefir grains, named AGK 1 (from Europe) and AGK 2 (from Argentina), were studied. Filtrates of kefir were obtained as follows: the grains were thawed from −80°C and cultured in reconstituted skim milk (RSM) (10% w/v) by incubating at 37°C for 24 h. These cultures were centrifuged (10 000 rpm – 10 min) and supernatants were filtered (Millipore, 0·45 μm pore size). The filtrates were named FAGK1 and FAGK2. They were immediately used to test the presence of phages.
Isolation and manipulation of bacteriophages
The methods applied to detect bacteriophages were agar spot-test, turbidity test (Svensson & Christiansson, Reference Svensson and Christiansson1991) and microplate test (Zago et al. Reference Zago, Suárez, Reinheimer, Carminati and Giraffa2007). MRS broth or agar (Biokar, Beauvois, France), which had been supplemented with 10 mm-CaCl2 (MRS-Ca when specified), were routinely used to propagate and count phages, respectively. Phage enumerations (PFU/ml) were performed by the double-layer plate titration method (Svensson & Christiansson, Reference Svensson and Christiansson1991) using MRS-Ca with addition of 100 mm-glycine (Lillehaug, Reference Lillehaug1997). Phage stocks were prepared as described by Neviani et al. (Reference Neviani, Carminati and Giraffa1992) in MRS-Ca broth, stored at 4°C, and frozen at −80°C in the presence of 15% (v/v) of glycerol. Phages ATCC 8014-B1 and ATCC 8014-B2 (both replicated on Lb. plantarum ATCC 8014) were used as reference phages.
Electron microscopy
Micrographs of phages were obtained according to Bolondi et al. (Reference Bolondi, Gaggino and Monesiglio1995). Phage suspensions were concentrated by centrifugation (1 h, 70 000 g, 5°C) and then stained using uranyl acetate (2% w/v, pH 4·5) or phosphotungstic acid (2% w/v). Electron micrographs were taken with a JEOL 100-C electron microscope (Jeol USA, Inc. Peabody, MA) operating at 80 kV. Phage morphologies and dimensions (capsid diameter, tail length and width) were recorded.
Host range
A total of 36 strains (Table 1) was used to determine the phage host range by means of the turbidity test (Svensson & Christiansson, Reference Svensson and Christiansson1991). Strain cultures and bacteriophages were inoculated in MRS-Ca broth at a multiplicity of infection (m.o.i.) of approx 1. As a control, a tube without the bacteriophage was used. Three subcultures were performed to confirm the phage resistance (no lysis) or sensitivity (lysis) of strains.
One-step growth curves
A culture of Lb. plantarum ATCC 8014 in exponential growth (OD560nm=0·5) was harvested and suspended in one-fifth of the initial volume of MRS-Ca broth. Phages were added at m.o.i. of approx 2. After adsorption (30 min at 37°C), cells were harvested by centrifugation (10 000 g for 5 min), suspended in 10 ml MRS-Ca broth and decimal dilutions of this suspension were carried out and incubated at 37°C. At regular intervals, 100 μl of each dilution were collected for bacteriophage counts (Chow et al. Reference Chow, Batt and Sinskey1988). Latent periods, burst times and burst sizes were calculated from one-step growth curves. Values are the mean of three different assays.
Phage DNA manipulation and analysis
Phages were propagated in a volume of 100 ml MRS-Ca broth, treated for 30 min with DNase I (Sigma-Aldrich Corporation, St. Louis, MO, USA) (1 μg/ml) and RNase (USB Corporation, Cleveland, Ohio, USA) (1 μg/ml), centrifuged (10 min at 5 000 g) and filtered (Millipore membranes, 0·45 μm pore size). Phage particles were concentrated overnight at 4°C with PEG 8000 (10% w/v) and 0·5 m-NaCl (Yamamoto et al. Reference Yamamoto, Alberts, Benzinger, Lawhorne and Treiber1970), centrifuged (10 min at 10 000 g) and resuspended in TE buffer (10 mm-Tris-HCl and 1m-EDTA, pH 8). Phage DNAs were purified by three phenol-chloroform-isoamyl alcohol extractions and precipitated by addition of absolute ethanol. DNA pellets were suspended in double-distilled and nuclease-free water. Phage DNAs were detected by electrophoresis on agarose (0·8% w/v) gels (Quiberoni et al. Reference Quiberoni, Guglielmotti, Binetti and Reinheimer2004). Their visualization by ethidium bromide coloration was performed according to standard protocols (Sambrook et al. Reference Sambrook, Fritsch and Maniatis1989).
Phage DNAs were cleaved with PvuI, MluI, SalI and HindIII by using the instructions provided by the manufacturers, and restriction fragments were then treated for 10 min at 70°C (Quiberoni et al. Reference Quiberoni, Guglielmotti, Binetti and Reinheimer2004). Heated and unheated digested DNA fragments were separated on 0·8% (w/v) agarose gel and visualized by ethidium bromide staining according to standard protocols (Sambrook et al. Reference Sambrook, Fritsch and Maniatis1989) 1 Kbp plus DNA Ladder (Invitrogen, Milan, Italy) was used as DNA molecular weight marker. The approximate size of each phage genome was estimated by averaging the sums of the size of the fragments obtained after single endonuclease digestion.
Bacteriophage structural proteins
Phages were replicated in 100 ml MRS-Ca broth, collected by a two-step centrifugation at 15 000 g for 1 h at 4°C and dialyzed against distilled water at 4°C for 24 h using a cellulose membrane (cut off 3500 Da) (Orange Scientific, Belgium). The dialyzed suspensions were concentrated by SpeedVac SV100 (Savant Instrument, NY, USA) and boiled for 5 mins to disrupt phage particles after resuspension in distilled water. After treatment, the samples were examined by SDS-PAGE using the Phast System (GE Healthcare, Italy) and stained with Comassie blue (Gatti et al. Reference Gatti, Fornasari and Neviani1997). Low molecular weight calibration kit for SDS electrophoresis (GE Healthcare, Italy) was used to estimate the molecular weight of the major phage proteins.
Results
Isolation of bacteriophages and host range
Preliminary screening of kefir filtrates by agar spot test showed a positive signal on the strains Lb. plantarum O36 and ATCC 8014. The capacity of these presumptive active bacteriophages (named as FAGK1 and FAGK2) to propagate on strain ATCC 8014 was verified using the turbidity test. This strain was chosen as indicator for both the phages. Host ranges were very similar for the two phages (Table 1). The only difference was the ability of FAGK2 to propagate on strain O75. Moreover, host ranges were different from those determined for the two collection phages ATCC 8014-B1 and ATCC 8014-B2 (Table 1).
Electron microscopy
Both phages, FAGK1 and FAGK2, showed isometric capsids (62·5±1·0 nm and 63·6±1·0 nm diameter, respectively) and long non-contractile tails (167±2·0 nm and 181·8±1·8 nm length, and 6·0±0·9 nm and 6·8±0·9 nm width), with transversal striations (Fig. 1). Basal plaques could be observed, smaller for phage FAGK1. Both phages belonged to the Siphoviridae family (morphotype B1) (Ackermann, Reference Ackermann2001).

Fig. 1. Electron micrographs of Lactobacillus plantarum phages isolated from Kefir grains. Panel A: phage FAGK1; panel B: phage FAGK2. Bars represent 50 nm.
One-step growth curves
Multiplication parameters of the lytic cycle of FAGK1 and FAGK2 were determined from one-step growth curves (Fig. 2). The two phages showed latent periods of approx 30 min and burst periods of approx 80 min. Burst size values of 12·0±0·3 and 10·8±0·4 PFU per infected cell were obtained for FAGK1 and FAGK2, respectively.
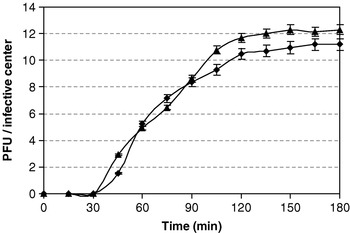
Fig. 2. One-step growth curves of Lactobacillus plantarum phages FAGK1 (▴) and FAGK2 (◆). Values are the mean of three determinations.
Genetic analysis
Genome sizes of both FAGK1 and FAGK2 were calculated by summing of the fragments originating from the action of the MluI (Fig. 3) and SalI (profile not shown) restriction enzymes. The molecular weight obtained for both phages was 34·8±3·2 Kbp.
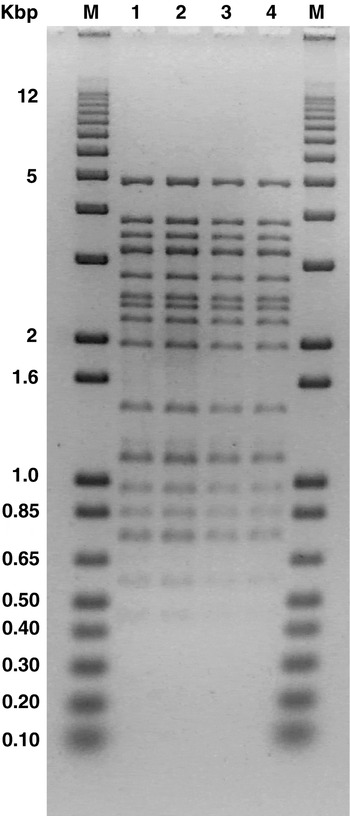
Fig. 3. Agarose gel electrophoresis of MluI generated DNA fragments of phages FAGK1 (lines 1 and 2) and FAGK2 (lines 3 and 4). Lines 2 and 4: fragments treated at 70°C for 10 min. M: 1 Kbp plus DNA Ladder (Invitrogen, Milan, Italy).
Restriction patterns obtained when DNA from FAGK1 and FAGK2 were treated with PvuI, MluI, SalI and HindIII restriction enzymes did not show any difference among them (Fig. 3, digestion with MluI is shown as example). Apparently, neither of the two phages seems to contain cohesive extremities, because the restriction profiles did not show differences with and without heating for 10 min at 70°C with all enzymes assayed (Fig. 3).
Bacteriophage structural proteins
Patterns of structural proteins obtained for kefir and reference and bacteriophages were identical (Fig. 4). Two major (32·9 and 35·7 kDa) bands were observed for each phage. Two additional, less intense bands (43·0 and 66·2 kDa) were also detected.
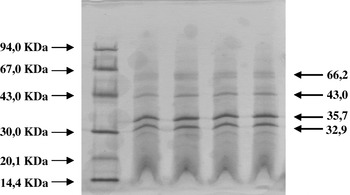
Fig. 4. Major structural protein profiles of phages FAGK1 (lines 1 and 2) and FAGK2 (lines 3 and 4). M: Molecular Weight Marker (GE Healthcare, Italy).
Discussion
Some temperate and virulent phages that can infect Lb. plantarum strains were previously isolated and characterized, but there are no reports of phages isolated from kefir grains. In our study, we report the isolation and characterization of two bacteriophages of Lb. plantarum isolated from kefir grains. This finding has scientific relevance because it underlines that phages belong to the very complex microbial composition present in kefir fermentations. Probably, in kefir grains there are lysogenic strains of Lb. plantarum that release temperate phages in the medium which could act as virulent phages towards other strains. According to the morphology, these new phages isolated form kefir belong to the Syphoviridae family (morphotype B1) (Ackermann, Reference Ackermann2001). The same morphology was obtained for the majority of the other reported Lb. plantarum phages such as Φ-JL-1 (Lu et al. Reference Lu, Breidt, Fleming, Altermann and Klaenhammer2003), Φ-B2 (Nes et al. Reference Nes, Brendehaug and Von Husby1988), Φ-SC 921 (Yoon et al. Reference Yoon, Kim, Breidt and Fleming2001), Φ-LP1 and Φ-LP2 (Caso et al. Reference Caso, De los Reyes-Gavilán, Herrero, Montilla, Rodríguez and Suárez1995), and Φ-gle (Kakikawa et al. Reference Kakikawa, Oki, Tadokoro, Nakamura, Taketo and Kodaira1996). Also, Chibani-Chennoufi et al. (Reference Chibani-Chennoufi, Dillmann, Marvin-Guy, Rami-Shojaei and Brüssow2004) reported the morphology of twenty phages, sixteen of them belonging to the Syphoviridae family and the others to the Myoviridae family. Another well characterized Myoviridae phage is Φ-fri (Trevors et al. Reference Trevors, Holley and Kempton1983). All the Syphoviridae family members show similar dimensions, with the exception of phage ATCC-B2, whose dimensions are almost double the values determined for other phages (Nes et al. Reference Nes, Brendehaug and Von Husby1988).
Host spectra of both bacteriophages were relatively narrow (17 and 19% of the tested strains were infected by the phages FAGK1 and FAGK2, respectively) and very similar to that obtained for the phage ATCC-B1. On the contrary, the bacteriophage ATCC-B2 exhibited the greatest virulence since it was able to infect 36% of strains used (13 out of 36). The lytic capacity reported for other Lb. plantarum phages is variable and ranges from 11% (e.g. phage LP2) to 55% (e.g. phage SC92) of the strains tested (Caso et al. Reference Caso, De los Reyes-Gavilán, Herrero, Montilla, Rodríguez and Suárez1995; Yoon et al. Reference Yoon, Kim, Breidt and Fleming2001).
One-step growth parameters of kefir phages were, in general, similar to those reported in the literature, except for the phage Φ-fri (Trevors et al. Reference Trevors, Holley and Kempton1983) that yielded 200 PFU/infected cell at 30°C, a value unusually high for phages of this species.
Regarding the genome size, they were identical for both kefir phages and this value (34·8±3·2 Kbp) is similar to those reported for other Syphoviridae phages. However, the genome sizes of Myoviridae phages Φ-fri and Φ-LP65 are considerably higher (133 and 131 Kbp, respectively; Trevors et al. Reference Trevors, Holley and Kempton1983; Chibani-Chennoufi et al. Reference Chibani-Chennoufi, Dillmann, Marvin-Guy, Rami-Shojaei and Brüssow2004). Myoviridae bacteriophages with large genomes have previously been reported in Gram positive bacteria (Jarvis et al. Reference Jarvis, Collins and Ackermann1993). The more complex structure of this family of phages could be the reason for this great difference in genome sizes.
Identical structural proteins were exhibited by both kefir bacteriophages. The sizes of the major proteins were within the values reported for the other Lb. plantarum phages, including those belonging to the Myoviridae family. A detailed analysis of structural proteins was performed for the JL-1 phage, which show three bands (34, 45 and 61 KDa) for head proteins and three bands (28, 50 and 76 KDa) for tail proteins (Lu et al. Reference Lu, Altremann, Breidt, Predki, Fleming and Klaenhammer2005). However, sizes of FAGK1 and FAGK2 phage proteins were very similar to those reported for Φ-SC91 (Yoon et al. Reference Yoon, Kim, Breidt and Fleming2001). These authors reported two major bands (32 and 34 KDa) and minor bands that ranged between 13 and 112 KDa. The two native Lb. plantarum phages described in our study seem strongly related since only slight differences were found in their dimensions and host spectra.
Lb. plantarum is a versatile species, which can persist in a wide variety of ecological niches and can be found in dairy (especially cheeses and fermented milks), meat, and vegetable products (de Vries et al. Reference de Vries, Vaughan, Kleerebezem and de Vos2006). Besides, it is a natural inhabitant of the human and animal gastrointestinal tracts. These characteristics open the possibility to use this microorganism as a potential probiotic starter. As a direct consequence, however, the use of this species as starter may raise concerns about bacteriophage attacks. Phage infections are the most important cause of slow acid production by lactic acid bacteria (LAB) during industrial fermentations (Neve Reference Neve, Cogan and Accolas1996; Moineau Reference Moineau, Konings, Kuipers and Huis in 't Veld1999; Suárez et al. Reference Suárez, Quiberoni, Binetti and Reinheimer2002). The economic losses and the public health consequences incurred when a phage infection occurs may be very significant (Josephsen & Neve Reference Josephsen, Neve, Salminen and von Wright1998; Forde & Fitzgerald Reference Forde, Fitzgerald, Konings, Kuipers and Huis in ′t Veld1999). Only a few virulent and temperate Lb. plantarum bacteriophages have been isolated and characterized so far (Trevors et al. Reference Trevors, Holley and Kempton1983; Nes et al. Reference Nes, Brendehaug and Von Husby1988; Caso et al. Reference Caso, De los Reyes-Gavilán, Herrero, Montilla, Rodríguez and Suárez1995; Kakikawa et al. Reference Kakikawa, Oki, Tadokoro, Nakamura, Taketo and Kodaira1996; Lu et al. Reference Lu, Breidt, Fleming, Altermann and Klaenhammer2003, Reference Lu, Altremann, Breidt, Predki, Fleming and Klaenhammer2005; Chibani-Chennoufi et al. Reference Chibani-Chennoufi, Dillmann, Marvin-Guy, Rami-Shojaei and Brüssow2004). So, to increase the knowledge on Lb. plantarum phages may be useful to predict the nature of phage – bacterium interactions and minimize their effects.







