In recent years there has been a greater focus on the role of coagulase-negative staphylococci (CNS) in bovine mastitis, despite authors having slightly different views of their impact on udder health (Piepers et al. Reference Piepers, Opsomer, Barkema, de Kruif and De Vliegher2010; Supré et al. Reference Supré, Haesebrouck, Zadoks, Vaneechoutte, Piepers and De Vliegher2011). This may be due to the fact that the frequency of isolation of the so called ‘major mastitis pathogens’ is decreasing (Makovec & Ruegg, Reference Makovec and Ruegg2003; Pitkälä et al. Reference Pitkälä, Haveri, Pyörälä, Myllys and Honkanen-Buzalski2004) with the ‘minor pathogens’ getting greater attention from the scientific community. In the past, individual species of CNS were grouped and it was assumed that all individual species presented the same characteristics in terms of pathogenicity. Several authors have studied the impact of different CNS species on mastitis, with evidence of some being more frequently isolated from cases of clinical mastitis (Waage et al. Reference Waage, Mørk, Røros, Aasland, Hunshamar and Ødegaard1999; Taponen et al. Reference Taponen, Simojoki, Haveri, Larsen and Pyörälä2006) and others found to lead to more persistent intramammary infection (IMI) (Taponen et al. Reference Taponen, Koort, Björkroth, Saloniemi and Pyörälä2007; Supré et al. Reference Supré, Haesebrouck, Zadoks, Vaneechoutte, Piepers and De Vliegher2011). It is well established that CNS are present in the environment (Piessens et al. Reference Piessens, Van Coillie, Verbist, Supré, Braem, Van Nuffel, De Vuyst, Heyndrickx and De Vliegher2011). However, the most frequent form of infection might differ for different CNS species. Taponen et al. (Reference Taponen, Björkroth and Pyörälä2008) found Staphylococcus chromogenes pulsotypes that were shared between isolates from mastitic milk samples and extramammary sites, whereas Staph. simulans was very seldom isolated from extramammary sites despite frequently being isolated from mastitic milk samples, indicating a stronger possibility of cow to cow transmission. This suggests scientific attention is needed on how often transmission of individual CNS species occurs in the milking parlour.
The objectives of the present study were to compare IMI due to different CNS species in terms of quarter somatic cell count and duration of infection, and to gain insight into diversity of staphylococcal species.
Materials and methods
Herds
This longitudinal observational study was performed in four Portuguese commercial dairy farms selected on their compliance and geographical location. All animals were Holstein-Friesian, zero-grazed, with year-round calving and housed in cubicles with sand bedding. Farm A milked 3 times daily and was a closed farm, whereas the other farms milked twice daily and would occasionally buy in primiparous animals. Post-milking teat dipping and antimicrobial dry cow therapy were routinely performed on every animal across farms. Mean data for SCC, milk production and number of animals in each farm is displayed in Table 1.
Table 1. Arithmetic mean for the number of lactating animals and milk yield, geometric mean for individual cow somatic cell count (SCC) for the duration of the study; number of cows and quarters sampled (initial sampling), number of cows and quarters with coagulase-negative staphylococcal intramammary infection (CNS IMI) and proportion of quarters affected (initial sampling)

Selection and sampling of animals
Quarter milk samples were taken according to NMC protocols (NMC, 1999) every 4 weeks on each farm for 12 visits, and a total of 48 weeks. In the initial visit, 12 cows were randomly selected from those with individual cow SCC showing an increase from below to above 200 000 cells/ml in the previous 2 milk recordings, to tentatively detect new infections at cow level. On subsequent visits, cows from which CNS had been isolated in at least 1 of the 2 previous visits, were resampled at quarter level; if these animals were not enough to total 12 cows, others were selected according to the criterion used in the first visit, to obtain samples from 12 cows per visit. None of the samples originated from clinical mastitis cases. Records of treatments performed during the study were analysed to exclude animals that received antimicrobial treatment during the study from the analysis or for right censoring of data.
Bacteriology and somatic cell count determination
Samples were refrigerated, transported to the laboratory and processed on the sampling day. From each milk sample, 0·01 ml of milk were plated onto sheep blood agar (Columbia™, bioMérieux, France), incubated at 37 °C and observed after 24 and 48 h of incubation. Samples yielding more than 2 morphologically different bacterial colonies were considered contaminated. A single colony representative of colonies with similar morphology was selected for isolation and identification if there was evidence of growth of ⩾500 cfu/ml. Gram-positive catalase-positive cocci were initially identified to species level by use of a biochemical identification system (ID 32 Staph™, bioMérieux, France). Isolates with an identification probability <90% were excluded from the study. Milk samples in which CNS were found concurrently with a major mastitis pathogen were also excluded from the analysis. Individual quarter milk samples were submitted to flow cytometry (CombiFoss™, Foss, Denmark) for somatic cell count (SCC) determination.
Strain typing
Molecular fingerprinting of staphylococci to strain level was performed by pulsed-field gel electrophoresis (PFGE) as described previously (Chung et al. Reference Chung, de Lencastre, Matthews, Tomasz, Adamsson, Aires de Sousa, Camou, Cocuzza, Corso, Couto, Dominguez, Gniadkowski, Goering, Gomes, Kikuchi, Marchese, Mato, Melter, Oliveira, Palacio, Sá-Leão, Santos Sanches, Song, Tassios and Villari2000). Staphylococcal isolates submitted to PFGE (n=467) included all the CNS isolated from each quarter when these were found in 4 or fewer sampling visits in succession, or isolates isolated in alternating visits when these were found in 5 or more sampling visits in succession. Bacterial disks were prepared with SeaPlaque® GTG® agarose (Lonza, Rockland ME, USA). Cell lysis was performed with a solution of ribonuclease A (Sigma-Aldrich, St. Louis MO, USA), lysostaphin (Ambi Products LLC, Lawrence KS, USA) and lysozyme (Sigma-Aldrich, St. Louis MO, USA) for 5 h at 37 °C. Deproteinisation was performed by incubation in a solution with proteinase K (Roche Diagnostics GmbH, Mannheim, Germany) at 50 °C for 17 h. DNA was digested with SmaI (Invitrogen, Carlsbad CA, USA) overnight at 25 °C. A CHEF-DRIII apparatus (Bio-Rad Laboratories, Hercules CA, USA) was used for the PFGE, run at 11·3 °C with a voltage of 6 V/cm, an initial pulse time of 5 s and a final pulse time of 35 s, with a total run time of 23 h. A Lambda Ladder PFG marker (New England Biolabs, Ipswich MA, USA) was used as a molecular weight marker. The gel was stained with ethidium bromide and photographed under UV transillumination. PFGE patterns were analysed with BioNumerics v. 4.61 (Applied Maths NV, Sint-Martens-Latem, Belgium). Levels of similarity between profiles were estimated with the Pearson coefficient and an unweighted pair group method using arithmetic averages was used for clustering, to produce band-based dendrograms, with a band position tolerance of and optimisation of 0·5–2·0%. A cut-off value of 80% similarity was used to differentiate clusters of PFGE types.
Genotypic species identification
Genotypic identification was sought using an internal transcribed spacer PCR (ITS-PCR), as described previously (Couto et al. Reference Couto, Pereira, Miragaia, Santos-Sanches and Lencastre2001) with modifications. Briefly, the disks containing DNA used to perform the PFGE were melted at 65 °C for 10 min in 1×TE buffer. A 1·25-μl aliquot of bacterial DNA was combined with 10 μl of reaction buffer, 6 μl of 25 mm-MgCl2, 1 μl of 10 mm-dNTP mix, 0·25 μl of DNA polymerase, 29 μl of nuclease-free water (all reagents from Promega, Madison WI, USA) and 1·25 μl of the G1 (5′-GAAGTCGTAACAAGG) and of the L1 (5′-CAAGGCATCCACCGT) primers (Stabvida, Caparica, Portugal). PCR amplification products, obtained using a MyCycler™ thermal cycler (Bio-Rad Laboratories, Hercules CA, USA), were resolved in a 3% Sea Plaque agarose (Lonza, Rockland ME, USA) for 6 h at 80 volts. A 100 bp DNA step ladder was used as a molecular weight marker (Promega, Madison WI, USA). All the isolates with characteristic PFGE profiles were submitted to the ITS-PCR, with at least 2 representatives of each profile being selected whenever available. The ITS-PCR profiles of 94 isolates were compared with 19 type strains (Table 1) and with a Staph. pseudintermedius isolate previously identified and characterised genotypically (Couto et al. Reference Couto, Pomba, Moodley and Guardabassi2011). The 19 type strains were selected based on phenotypic identification results. The profile analysis was performed visually with the aid of the BioNumerics software v. 4.61 (Applied Maths NV, Sint-Martens-Latem, Belgium). Whenever ITS-PCR results did not allow for a clear differentiation between species or did not match type strain profiles, isolates (n=27) were submitted to PCR amplification of both sodA and rpoB housekeeping genes, as described previously by Poyart et al. (Reference Poyart, Quesne, Boumala and Trieu-Cuot2001) and Drancourt & Raoult (Reference Drancourt and Raoult2002), respectively. PCR products were sequenced at Macrogen (Seoul, Korea), and sequence data proofread using Bioedit v. 7.0.5 (Hall, Reference Hall1999), and compared with publicly available sequence data using nucleotide-nucleotide BLAST (See: http://www.ncbi.nlm.nih.gov/blast/).
Data analysis
Unless otherwise stated, only results based on genotypic identification were used in the data analysis. An IMI with a CNS was defined by isolation of CNS from at least 1 quarter milk sample. Spontaneous cure was defined as not detecting a specific strain in the same quarter in 2 subsequent visits in succession. Duration of CNS IMI was defined by the period of time a CNS with the same PFGE profile, was found in successive or alternate months and analysed by way of a Kaplan-Meier survival curve and a log-rank survival test. A general linear model was used with log10 transformation of individual quarter SCC as the outcome variable. Explanatory variables included CNS species, sampling number, quarter, cow and herd level:
where the subscripts i, j, k and n denote the ith CNS species, the jth quarter, the kth cow and the nth farm respectively.
Data were analysed using SPSS version 15.0 (survival analysis) and in Minitab 16.0 (general linear model). A significant difference was defined as a probability value of P<0·05.
Results
To detect infections by CNS, a total of 2302 quarter milk samples from 264 cows were collected. These represented 1021 functional quarters, of which 147 (14·4%) had a CNS IMI, according to phenotypic identification. The proportion of quarters with CNS IMI among sampled quarters for the first time, ranged from 6·4% in farm B to 347% in farm A (Table 1). Out of the 147 IMI, 36 were not followed for at least 3 successive samplings. From the remaining 111 CNS IMI (representing 51 cows), 63 were right-censored in order to be further studied. Both exclusion of IMIs and right censoring occurred because: (a) antimicrobial treatments were performed during the study; (b) it was not possible to follow the animals through time (some died, were culled or were missed by the operator during sampling visits); (c) some animals included in the final visits did not stay long enough in the study to allow for cure or maintenance of infection to be effectively evaluated.
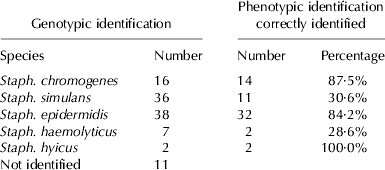
Genotypic identification of the 111 CNS revealed the presence of 38 Staph. epidermidis, 36 Staph. simulans, 16 Staph. chromogenes, 7 Staph. haemolyticus and 2 Staph. hyicus. It was not possible to identify genotypically 12 isolates. The ITS-PCR method did not allow for confident identification of several isolates, namely it was difficult to distinguish between Staph. epidermidis and Staph. chromogenes, and some isolates that had been identified phenotypically as Staph. intermedius had profiles that did not match any of the type strains used. Some of these were clarified through sequencing of housekeeping genes and revealed to be Staph. simulans (n=2) and Staph. haemolyticus (n=1). Comparison of genotypic and phenotypic identification is summarised in Table 2.
Table 2. Identification of coagulase-negative staphylococci (CNS) by the ID32 Staph® compared with the phenotypic identification (ITS-PCR and sequencing of rpoB or sodA)†

† The Staphylococcus sp. used for comparison with ITS-PCR isolate profiles and respective DSM numbers included Staph. lugdunensi (4804), Staph. sciuri subsp. carnaticus (15 613), Staph. sciuri subsp. rodentium (16 827), Staph. epidermidis (20 044), Staph. saprophyticus subsp. Saprophyticus (20 229), Staph. aureus subsp. aureus (20 231), Staph. haemolyticus (20 263), Staph. xylosus (20 266), Staph. warneri (20 316), Staph. simulans (20 322), Staph. capitis subsp. capitis (20 326), Staph. hominis subsp. hominis (20 328), Staph. intermedius (20 373), Staph. chromogenes (20 454), Staph. hyicus (20 459), Staph. auricularis (20 609), Staph. gallinarum (20 610), Staph. caprae (20 698), Staph. pseudintemedius
When comparing only the 4 most frequently isolated species, duration of infection (Table 3) was not significantly different (P=0·179) when the species were compared altogether. When performing a 2-way comparison in duration of infection between the species with the shortest and longest duration, the difference approached statistical significance (P=0·071). Quarter SCC was also not significantly different between CNS species. The relationship of log10 SCC with sampling occasion and with cow affected were dependent on CNS species considered (P=0·014 and P=0·015, respectively).
Table 3. Number of isolates (Isol), cows affected and pulsotypes (Puls) per farm, for different coagulase-negative staphylococcal (CNS) species. Different pulsotypes were defined based on less than 80% similarity. Number of infections followed per farm, geometric mean somatic cell count (SCC) and presence in successive months of CNS identified genotypically

† Geometric mean
‡ Kaplan-Meier survival analysis
Dendrograms showed that clusters within Staph. epidermidis and Staph. simulans isolates were generally grouped per farm (Fig. 1), whereas for Staph. chromogenes and Staph. haemolyticus this separation was not noticeable. The number of pulsotypes per species and per farm is displayed in Table 3. Cows with 2 or more quarters affected by CNS IMI (n=29) had the same species in multiple quarters (n=10), different species between quarters (n=9) or both (n=10). In cases where the same species was isolated in more than 1 quarter per cow (n=20), shared pulsotypes between quarters of the same cow were found in 17 animals.

Fig. 1. Dendrograms with PFGE profiles for Staph. epidermidis (above) and Staph. chromogenes (below), showing the 80% similarity line.
Discussion
CNS were the most frequently isolated bacteria from milk samples in several studies worldwide (Makovec & Ruegg, Reference Makovec and Ruegg2003; Pitkälä et al. Reference Pitkälä, Haveri, Pyörälä, Myllys and Honkanen-Buzalski2004). Considering only the first time a cow was sampled, 147 out of 1021 sampled quarters (14·4%) had a CNS IMI according to the ID 32 Staph™ identification system. Some CNS species were only found in 2 herds, notably Staph. epidermidis and Staph. simulans, the most frequently isolated, were found in farms A and D, whereas Staph. chromogenes and Staph. haemolyticus, which had the third and fourth greatest number of CNS isolates respectively, were isolated from milk samples on all 4 farms. Most studies on prevalence and persistence of CNS infection have focused on single farms or on a small number of farms, not allowing for clustering effect evaluation of CNS species per farm. Recently, Piessens et al. (Reference Piessens, Van Coillie, Verbist, Supré, Braem, Van Nuffel, De Vuyst, Heyndrickx and De Vliegher2011) found herd-to-herd differences in the distribution of individual CNS species isolated from milk and from the environment of 6 dairy herds. Despite the low number of farms in the current study, the predominance of 2 species in 2 of the farms also suggests a clustering effect for individual CNS species. As stated above, Staph. epidermidis and Staph. simulans were the most frequently isolated CNS species, followed by Staph. chromogenes and Staph. haemolyticus. Other studies resorting to genotypic identification methods have found Staph. simulans (Taponen et al. Reference Taponen, Simojoki, Haveri, Larsen and Pyörälä2006) or Staph. chromogenes (Piessens et al. Reference Piessens, Van Coillie, Verbist, Supré, Braem, Van Nuffel, De Vuyst, Heyndrickx and De Vliegher2011) to be the most frequently isolated CNS.
Mean duration of infection for all CNS was 188 d, corresponding to 6·7 successive monthly visits. There were no significant differences in mean duration of infection between CNS species when these were compared altogether. When the species with the shortest mean duration (Staph. haemolyticus with 123 d) was compared with the species with the longest duration (Staph. simulans with 213 d) there was still no statistically significant difference. Despite having the shortest duration of infection Staph. haemolyticus still led to persistent infections, which is in agreement with what was observed by Piessens et al. (Reference Piessens, Van Coillie, Verbist, Supré, Braem, Van Nuffel, De Vuyst, Heyndrickx and De Vliegher2011) and Mørk et al. (2012). Persistence of CNS IMI has been reported by several authors, with 46% (Taponen et al. Reference Taponen, Koort, Björkroth, Saloniemi and Pyörälä2007), 66% (Seymour et al. Reference Seymour, Jones and McGilliard1989), 75·6% (Rainard et al. Reference Rainard, Ducelliez and Poutrel1990), 84·5% (Chaffer et al. Reference Chaffer, Leitner, Winkler, Glickman, Krukucks, Ezra and Saran1999) and 85% (Timms & Schultz, Reference Timms and Schultz1987) of infections persisting until dry-off or until the cows left the herd. Todhunter et al. (Reference Todhunter, Cantwell, Smith, Hoblet and Hogan1993) mentioned an average length of CNS IMI of 222 d and Rainard et al. (Reference Rainard, Ducelliez and Poutrel1990) of 236 d, despite neither of these authors resorting to molecular biology techniques for confirmation of IMI with the same strain. Supré et al. (Reference Supré, Haesebrouck, Zadoks, Vaneechoutte, Piepers and De Vliegher2011) recorded an average duration of infection of 148 d for Staph. chromogenes and of 176 d for Staph. simulans. It is impossible to establish more accurately the exact duration of the IMI followed in our study because there is no exact knowledge of when the infections were acquired or cleared.
Geometric mean SCC for CNS IMI followed through time was 132 000 cells/ml, with no significant differences observed between CNS species. Staph. chromogenes was associated with the highest mean SCC, with a geometric mean of 202 000 cells/ml and Staph. epidermidis with the lowest mean, with a SCC of 95 000 cells/ml. The geometric mean obtained in this study is similar to the 138 000 cells/ml reported by Djabri et al. (Reference Djabri, Bareille, Beaudeau and Seegers2002) in a meta-analysis for quarter milk samples infected with staphylococci other than Staph. aureus. Similarly to what was observed in the current study, both Nickerson et al. (Reference Nickerson, Owens and Boddie1995) and Supré et al. (Reference Supré, Haesebrouck, Zadoks, Vaneechoutte, Piepers and De Vliegher2011) observed that Staph. chromogenes led to the highest quarter SCC and Hogan et al. (Reference Hogan, White and Pankey1987) did not find significant differences in quarter SCC between CNS species. In the current study, the impact of sampling number and of cow on log10 SCC were both species-dependent. The reduced number of observations and the fact that not every species had the same average duration of IMI, render the sampling occasion relationship of questionable significance. The significant relationship for cow would be expected since individual animal have been shown to respond differently to infections with CNS (Simojoki et al. Reference Simojoki, Orro, Taponen and Pyörälä2009) or with other udder pathogens (Burvenich et al. Reference Burvenich, Van Merris, Mehrzad, Diez-Fraile and Duchateau2003).
Comparison of genotypic and phenotypic identification showed 61·6% correct species identification with phenotypic methods, which is higher than the 41·3% found by Sampimon et al. (Reference Sampimon, Zadoks, De Vliegher, Supré, Haesebrouck, Barkema, Sol and Lam2009) using the same phenotypic identification system and sequencing of the rpoB gene. This difference may be partly explained by the fact that only isolates with a phenotypic identification probability ⩾90% were included in the present study.
In the current study, PFGE was used to evaluate duration of infection and strain diversity, which has also been used by other authors (Taponen et al. Reference Taponen, Björkroth and Pyörälä2008; Mørk et al. Reference Mørk, Jørgensen, Sunde, Kvitle, Sviland, Waage and Tollersrud2012). Choice of a cut-off value to differentiate between different PFGE pulsotypes is not unanimous, with some authors resorting to the lowest reproducibility value to define the cut-off (Silva et al. Reference Silva, Gaivão, Leitão, Jost, Carneiro, Vilela, Lopes da Costa and Mateus2008) and others defining similarity coefficients after reviewing epidemiological data associated with each cluster of isolates (McDougal et al. Reference McDougal, Steward, Killgore, Chaitram, McAllister and Tenover2003). The criteria for PFGE profile interpretation that seem to gather more agreement were defined by Tenover et al. (Reference Tenover, Arbeit, Goering, Mickelsen, Murray, Persing and Swaminathan1995). In the current study, the 80% cut-off value was used according to a publication by the same research group, analysing the same genus and using the same restriction enzyme (McDougal et al. Reference McDougal, Steward, Killgore, Chaitram, McAllister and Tenover2003). However, a lower cut-off value could be viewed as more appropriate because not all the criteria defined by Tenover et al. (Reference Tenover, Arbeit, Goering, Mickelsen, Murray, Persing and Swaminathan1995) were fulfilled in the current study; namely the analysed strains were obtained from different geographical areas and over a time span close to a year in which natural variation would likely add to the observed DNA fingerprint differences. Different criteria could be considered to evaluate within-herd and between-herd diversity (Zadoks et al. Reference Zadoks, van Leewen, Kreft, Fox, Barkema, Schukken and van Belkum2002). Dendrograms showed clustering of Staph. epidermidis and Staph. simulans per farm, whereas Staph. chromogenes and Staph. haemolyticus showed less clonality within each farm. This suggests that cow to cow transmission in the milking parlour is more likely to occur with Staph. epidermidis and Staph. simulans. Taponen et al. (Reference Taponen, Björkroth and Pyörälä2008) found the same pulsotypes of Staph. chromogenes isolated from milk samples and extramammary sites suggesting an environmental source of infection. They also found that Staph. simulans was very commonly isolated from milk samples but seldom found in environmental samples, indicating that this species was likely to be a specific mastitis pathogen. On the contrary, Piessens et al. (Reference Piessens, Van Coillie, Verbist, Supré, Braem, Van Nuffel, De Vuyst, Heyndrickx and De Vliegher2011) found that Staph. chromogenes and Staph. epidermidis were rarely found in the environment, whereas Staph. simulans and Staph. haemolyticus were commonly isolated from environmental samples. The low number of isolates per farm in the present study did not allow for a more extensive inter-species comparison within each farm. The use of such methodologies across different farms raises the question whether the differences observed in diversity between species are a feature of the species, a feature of the farm (including host and farm management factors) or a combination of both.
Based on the genetic heterogeneity observed in each CNS species, the predominant mode of infection seems to vary between species. Irrespective of that, the impact of different species on udder health seems to be similar, as there were no statistically significant differences between individual CNS species in terms of duration of IMI and mean quarter SCC.
R. Bexiga was financed by grant number SFRH/BD/36759/2007 from the Foundation for Science and Technology. The financial support of the Interdisciplinary Centre of Research in Animal Health (CIISA) is gratefully acknowledged. The authors acknowledge the access given to farms by farmers and practitioners, the technical assistance of staff at the Scottish MRSA Reference Laboratory, Dr Suvi Taponen for kindly providing staphylococcal type strains and Natacha Couto for providing the S. pseudintermedius isolate.