Staphylococcus spp. are the biggest cause of contagious mastitis in bovine species, and antibiotic therapy is considered the main form of treatment. However, the indiscriminate use of antimicrobials has encouraged the emergence of multi-drug-resistant isolates (Lin et al., Reference Lin, Nishino, Roberts, Tolmasky, Aminov and Zhang2015). Mechanisms of Staphylococcus spp. resistance include enzyme production, modification of the drug target and multi-drug efflux systems (Costa et al., Reference Costa, Viveiros, Amaral and Couto2013; Kumar et al., Reference Kumar, Mukherjee and Varela2013). Multi-drug efflux systems comprise of cytoplasmic proteins that are involved in the extrusion of toxic agents from inside the bacteria to the external environment. They are encoded in chromosomes or plasmids and perform active transport of the antimicrobial agent out of the cell (Truong-Bolduc et al., Reference Truong-Bolduc, Dunman, Strahilevitz, Projan and Hooper2005; Micas, Reference Micas2008). Moreover, they prevent the drug from reaching inhibitory concentrations inside the bacterial cell. In Staphylococcus spp., more than 20 efflux systems classified into five families of membrane proteins have already been identified: the major facilitator superfamily (MFS), the small multidrug resistance family, the multidrug and toxin extrusion family, the ATP-binding cassette (ABC) superfamily, and the resistance-nodulation-division superfamily (Truong-Bolduc et al., Reference Truong-Bolduc, Strahilevitz and Hooper2006; Kumar et al., Reference Kumar, Mukherjee and Varela2013; Jang, Reference Jang2016).
For the pathogen, the effectiveness of its multi-drug efflux systems is related to three mechanisms: elimination of endogenous metabolites harmful to the bacterium, secretion of virulence determinants, and responses to cellular stress (Poole, Reference Poole2008). As such, the drugs can be defined as ‘accidental substrates’ of these transporters. However, the ability of multi-drug efflux systems to expel antimicrobials, both synthetic and natural, presents problems for treatment of infections, especially those caused by the highly pathogenic Staphylococcus spp. (Truong-Bolduc et al., Reference Truong-Bolduc, Dunman, Strahilevitz, Projan and Hooper2005).
The emergence of antimicrobial resistance has stimulated research into possible alternatives for the affected drugs, such as polymers and herbal medicines (Peixoto et al., Reference Peixoto, Silva, Almeida, Branco and Costa2016; Sanchez Ramirez et al., Reference Sanchez Ramirez, Varesano, Carletto, Vineis, Perelshtein, Natan, Perkas, Banin and Gedanken2019). Bacteriophages, lactic acid bacteria, nanoparticles of chemical compounds and extracts or essential oils from plants are some of the alternative therapies reported against Staphylococcus spp. isolated from the agricultural environment (Quintero et al., Reference Quintero, Serna Cock and Campos2011; Costa Junior et al., Reference Costa Junior, André, Campos, Palácio, Macário and Cavalcanti2018; Berguenmaier de Olanda et al., Reference Berguenmaier de Olanda, Peripolli Bevilaqua, Damé Schuch, Souza Prestes and Job2019; Leite et al., Reference Leite, Pereira, Borges, Alves, Ramos, Martins and Arcuri2019; Sperandio et al., Reference Sperandio, Veleirinho, Honorato, Campestrini and Kuhnen2019).
The objective of this study was to detect multi-drug efflux system genes in Staphylococcus aureus and non-aureus spp. present in bovine mastitis, milkers, and the milking environment and evaluate the antimicrobial activity of polypyrrole nanoparticles (PPy-NPs) and aqueous extract of Moringa oleifera seed oil against these Staphylococcus spp.
Materials and methods
Ethical approval
This study was approved by the Animal Use Ethics Committee of the Universidade Federal Rural de Pernambuco, Recife, Brazil (license number 106/2017), the Ethics Committee on Animal Use of the Universidade Federal de Pernambuco (process number 23076.033782/2015-70) and the Research Ethics Committee of the Universidade de Pernambuco, Recife, Brazil (CAAE number: 28833619.7.0000.5207).
Staphylococcus spp. isolates
Five dairy farms (A, B, C, D and E) located in different regions of the state of Pernambuco, Northeastern Brazil, were included in the present study. Milk samples collected from 676 udder quarters were subjected to the California Mastitis Tests (CMT) and those that were equal to or greater than one cross (+) (319 samples) or found positive in the routine strip cup test (16 samples) were selected for microbiological testing. Also, samples from the dairy environment (mechanical milking equipment and milking buckets) and milkers (hands and nasal cavities) were obtained using swabs and subjected to microbiological analysis. A total of 15 samples of milk and hand swabs from milkers, 14 samples from teat taps and nine samples of milking buckets were obtained. The distribution by farm of the collected samples is described in the online Supplementary Table S1.
Isolation and preliminary identification of Staphylococcus spp.
Isolation of Staphylococcus spp. was performed according to the methodology of Carter (Reference Carter1998) as described in the online Supplementary File.
Susceptibility to antimicrobials
Staphylococcus spp. underwent in vitro antimicrobial sensitivity tests using the disk diffusion technique on Mueller-Hinton agar (CLSI, 2018). The following antibiotic-impregnated disks were used oxacillin (1 μg), cefoxitin (30 μg) and penicillin G (10 U, vancomycin (30 μg), sulfamethoxazole (25 μg) + trimethoprim (25 μg), tetracycline (30 μg), gentamycin (10 μg) and neomycin (30 μg).
Molecular analysis
Isolates of S. aureus and non-aureus that showed resistance to at least two drugs were selected for molecular analysis. Genomic DNA was extracted by thermal extraction; method as described by Fan et al. (Reference Fan, Kleven and Jackwood1995).
Identification of S. aureus and of efflux system genes in S. aureus and non-aureus
To identify S. aureus, all isolates underwent amplification of the nuc gene region according to the methodology described by Brakstad et al. (Reference Brakstad, Aasbakk and Maeland1992). For detection of norA, norB, norC, msrA, mgrA, tet-38, and lmrS genes, all Staphylococcus spp. underwent PCR following the methodologies described by Martineau et al. (Reference Martineau, Lansac, Me, Roy, Ouellette and Bergeron2000), Truong-Bolduc et al. (Reference Truong-Bolduc, Zhang and Hooper2003), Truong-Bolduc et al. (Reference Truong-Bolduc, Dunman, Strahilevitz, Projan and Hooper2005), Truong-Bolduc et al. (Reference Truong-Bolduc, Strahilevitz and Hooper2006) Details are provided in the online Supplementary Table S2.
Determination of minimum inhibitory concentration
The minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of PPy-NPs in water and Moringa oleifera aqueous extract were determined using the broth microdilution methodology following recommendations of the Clinical Laboratory Standards Institute (CLSI, 2018). For PPy-NPs in water the method used was that of da Silva et al. (Reference da Silva, Queiroz, Macedo, Fernandes, Freire, da Costa and de Oliveira2016) and the concentrations of PPy-NPs tested were 2, 1, 0.5, 0.250, 0.125, and 0.062 mg/ml. The protein content of aqueous Moringa oleifera extract was determined according to Lowry et al. (Reference Lowry, Rosebrough, Farr and Randall1951). The presence of lectins was monitored according to Paiva and Coelho (Reference Paiva and Coelho1992) and Bing et al. (Reference Bing, Weyand and Stavinsky1967) (see online Supplementary File). Aqueous Moringa oleifera seed extract was used at initial concentrations of 6.1, 3.05, 1.525, 0.7625, 0.3812, and 0.1906 mg/ml.
Results
In this study, 162 Staphylococcus spp. were isolated from samples collected from dairy farms in the state of Pernambuco, Brazil. The results of isolation, molecular identification of Staphylococcus aureus and non-aureus samples from milk, milkers, and milking utensils are shown in the online Supplementary Table S3.
In the antimicrobial susceptibility test, 35/162 (21.6%) of Staphylococcus spp. isolates comprising 23/35 (65.7%) S. aureus and 12/35 (34.3%) S. non-aureus, were resistant to at least two antimicrobials. In the molecular analysis of these isolates, norA, norC, tet-38, and msrA genes were all observed. Table 1 details the efflux system genes in Staphylococcus aureus and non-aureus according to origin.
Table 1. Frequency of the efflux system genes in S. aureus and S. non-aureus according to origin
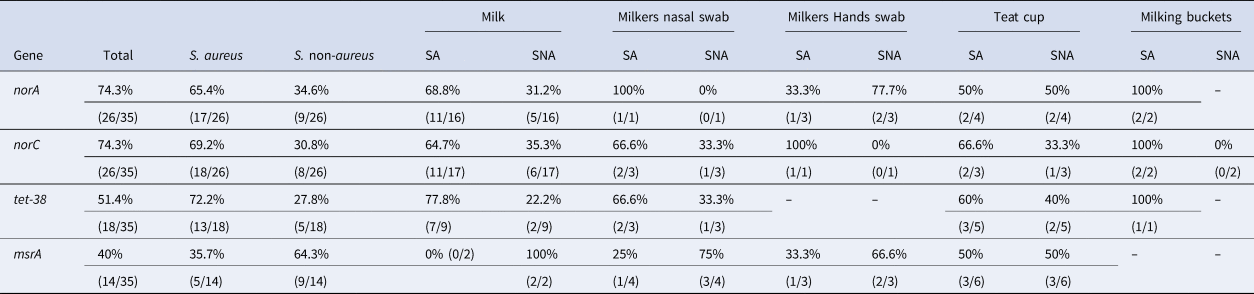
SA, Staphylococcus aureus; SNA, Staphylococcus non-aureus; –, not identified.
The Moringa oleifera extract showed a specific hemagglutination assay value of 32, indicating the presence of lectins. Previous assays revealed the presence of flavonoids, tannins, saponins, phenylpropanoids, alkaloids, and reducing sugars in the extract (unpublished data). The results of the antimicrobial activity of aqueous Moringa oleifera seed extracts and of polypyrrole nanoparticles against Staphylococcus spp. carrying multi-drug efflux system genes are presented in Table 2.
Table 2. Distribution by MIC of the aqueous extract of Moringa oleifera and PPy-NPs against Staphylococcus spp. carriers of norA, norC, tet-38 and msrA isolated from milk cows, Milkers and dairy farm environment

MIC, minimum inhibitory concentration; M, milk; HS, hands swab; SN, nasal swab; TC, Teat cup; MB, Milking buckets.
Discussion
In this study norA, norC, tet-38, and msrA were detected in our samples. The first three belong to MFS, the family with the largest number of studies in the context of staphylococci (Jang, Reference Jang2016); MsrA belongs to the ABC family (Martineau et al., Reference Martineau, Lansac, Me, Roy, Ouellette and Bergeron2000). In Staphylococcus aureus the reported identification frequencies of the efflux system genes ranges widely from 2 to 98.8% (Hassanzadeh et al., Reference Hassanzadeh, Ganjloo, Pourmand, Mashhadi and Ghazvini2020).
These results indicate the circulation of isolates with molecular mechanisms of resistance to quinolones, tetracyclines, erythromycin, and macrolides in the studied farms. The detected genes confer resistance to these antimicrobials and so, if they are expressed, multidrug resistance is observed (Truong-Bolduc et al., Reference Truong-Bolduc, Zhang and Hooper2003; Deng et al., Reference Deng, Sun, Ji, Liang, Missiakas, Lan and He2012; Duran et al., Reference Duran, Ozer, Duran, Onlen and Demir2012; Kumar et al., Reference Kumar, Mukherjee and Varela2013; Phillips-Jones and Harding, Reference Phillips-Jones and Harding2018). In our case the most frequently observed genes were norA and norC (Table 1), both of which are responsible for quinolone resistance, followed by tet-38, which confers resistance to tetracyclines. These findings might reflect the frequent use of these antimicrobials to treat mastitis at the studied farms. Their use has likely contributed to the selection pressure for the emergence of resistant isolates (Hawkey, Reference Hawkey2008; Holko et al., Reference Holko, Tančin, Vršková and Tvarožková2019).
Regarding isolate origin, milk samples gave the highest percentages of detected genes. This indicates a role for isolates with an efflux system in the etiology of bovine mastitis in the study region (Northeast Brazil). A similar situation has previously been reported in Egypt (Elsayed et al., Reference Elsayed, Roshdey, Salah, Tarabees, Younis and Eldeep2019). The exception was msrA, which was more prevalent in the liner samples and milkers (Table 1), although given the small number of samples, these results must be interpreted with considerable caution. The msrA gene supports resistance to erythromycin and macrolides, which are usually not antimicrobials of choice for mastitis in Brazil (unlike Argentina, where these drugs are commonly used: Srednik et al., Reference Srednik, Usongo, Lépine, Janvier, Archambault and Gentilini2018). This probably explains why it was detected more frequently in isolates from liners (utensil manipulated by humans), nasal swabs, and milkers' hands at the studied farms (Truong-Bolduc et al., Reference Truong-Bolduc, Dunman, Strahilevitz, Projan and Hooper2005; Jang, Reference Jang2016; Pereira and Scussel, Reference Pereira and Scussel2017).
Our detection of multi-drug efflux system genes in Staphylococcus spp. demonstrates that there is variability in the genetic components of resistance in the studied region. It is, therefore, pertinent to research alternatives to the affected drugs, such as plant compounds with antibacterial activity such as Moringa oleifera, and chemical compounds such as PPy.
At all concentrations except 6.1 mg/ml, aqueous Moringa oleifera extract showed activity for at least one isolate. Greatest inhibition was found at concentrations of 1.525 and 0.7625 mg/mL. This inhibitory effect was due to aqueous Moringa oleifera extract's bactericidal activity at these concentrations; this was verified by MBC testing. However, only msrA-carrying isolates were 100% inhibited, which indicates better bactericidal activity against Staphylococcus carriers of this gene.
Moringa oleifera (family Moringaceae) is a tree grown in tropical and subtropical regions (Pontual et al., Reference Pontual, Napoleão, Dias de Assis, de Souza Bezerra, Xavier, Navarro, Coelho and Paiva2012a). Its seeds are used to treat and reduce the turbidity of water intended for human consumption. In addition, its flowers are consumed and have hypoglycemic, tonic, and diuretic therapeutic actions (Moura et al., Reference Moura, Pontual, Gomes, Napoleão, Xavier, Paiva and Coelho2011; Santos et al., Reference Santos, Paiva and Coelho2012). It also has larvicidal activity against Aedes aegypti and is antiparasitic against Trypanosoma cruzi (Pontual et al., Reference Pontual, Napoleão, Dias de Assis, de Souza Bezerra, Xavier, Navarro, Coelho and Paiva2012a, Reference Pontual, Pires-Neto, Fraige, Higino, Carvalho, Alves, Lima, Zingali, Coelho, Bolzani, Figueiredo, Napoleão and Paiva2018). Further, it has technological potential in the food industry, due to its caseinolytic activity and milk-coagulating effects (Pontual, et al., Reference Pontual, Carvalho, Bezerra, Coelho, Napoleão and Paiva2012b). No previous studies have evaluated the antimicrobial activity of aqueous Moringa oleifera extract against Staphylococcus spp. isolated from the agricultural environment. This makes it difficult to establish comparative parameters for the results we obtained but also indicates this study's novelty. However, there are reports of Moringa oleifera activity against Staphylococcus aureus and Staphylococcus epidermidis (Viera et al., Reference Viera, Mourão, Ângelo, Costa and Vieira2010; Moura et al., Reference Moura, Pontual, Gomes, Napoleão, Xavier, Paiva and Coelho2011; Marrufo et al., Reference Marrufo, Nazzaro, Mancini, Fratianni, Coppola, De Martino, Agostinho and De Feo2013; Zaffer et al., Reference Zaffer, Ahmad, Sharma, Mahajan, Gupta and Agnihotri2014; Fayemi et al., Reference Fayemi, Ekennia, Katata-Seru, Ebokaiwe, Ijomone, Onwudiwe and Ebenso2018; Fouad et al., Reference Fouad, Abu Elnaga and Kandil2019), which corroborate this study.
These results with aqueous Moringa oleifera extract are promising, however, further studies are needed to identify the active components and develop effective therapies. Although there is no clear mechanism, there is evidence that the plant's antimicrobial activity is related to a wide spectrum of bactericidal substances produced by the plant (Kostova and Dinchev, Reference Kostova and Dinchev2005; Viera et al., Reference Viera, Mourão, Ângelo, Costa and Vieira2010). However, the seeds of M. oleifera also contain a water-soluble lectin called (WSMoL), which was previously reported to have antibacterial effects (Moura et al., Reference Moura, Napoleão, Coriolano, Paiva, Figueiredo and Coelho2015, Reference Moura, Trentin, Napoleão, Primon-Barros, Xavier, Carneiro, Paiva, Macedo and Coelho2017; Coriolano et al., Reference Coriolano, Brito, Ferreira, Moura, Melo, Soares, Lorena, Figueiredo, Paiva, Napoleão and Coelho2019).
For PPy-NPs, it was possible to determine MIC at two concentrations, 0.125 and 0.062 mg/ml (Table 2), the first of which delivered the greatest inhibition. The bactericidal activity of PPy-NPs at these concentrations (0.125 and 0.062 mg/ml) was responsible for the inhibitory effect found in the tests to determine MBC. The results differ from those obtained with the aqueous Moringa oleifera extract, where there was variability in inhibition across the concentrations. However, PPy-NPs did not inhibit all isolates with the four detected genes, indicating a narrower range of activity as compared with aqueous Moringa oleifera extract.
PPy is a polymer obtained through the oxidative chemical or electrochemical polymerization of monomer solutions (Sajesh et al., Reference Sajesh, Jayakumar, Nair and Chennazhi2013). It is used in engineering and biomedical sciences (de Oliveira and de Oliveira, Reference de Oliveira and de Oliveira2014; Xue et al., Reference Xue, Zhou, Shi, Feng, Xin and Song2014). The antimicrobial activity of PPy against Staphylococcus aureus, as well as other gram-positive and negative bacteria has already been reported (Sayyah et al., Reference Sayyah, Mohamed and Shaban2014; da Silva et al., Reference da Silva, Alcaraz-Espinoza, da Costa and de Oliveira2017). No other reports have investigated Staphylococcus spp. isolated from the agricultural environment. Unlike aqueous Moringa oleifera extract, in PPy the mechanism responsible for antimicrobial activity has been elucidated. The polymer causes collapse of the cytoplasmic membrane resulting in bacterial death (Varesano et al., Reference Varesano, Vineis, Aluigi, Rombaldoni, Tonetti and Mazzuchetti2013; Sanchez Ramirez et al., Reference Sanchez Ramirez, Varesano, Carletto, Vineis, Perelshtein, Natan, Perkas, Banin and Gedanken2019). This study demonstrates that PPy-NPs could be used as an alternative to traditional antimicrobials. For this to occur, it is necessary to carry out further studies aimed at drug development.
In conclusion, we have shown that there are genes from multi-drug efflux systems in Staphylococcus spp. isolated from the agricultural environment in Northeast Brazil. Aqueous Moringa oleifera extract and PPy-NPs both showed bactericidal activity against these isolates carrying multi-drug efflux system genes.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0022029920000874.
Acknowledgements
To the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for granting research grants to the first author. To Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for the financial support (Process number 409107/2018-2).




